Abstract
Acyl carrier proteins (ACPs) have been reported to play a crucial role in responding to biotic and abiotic stresses, regulating growth and development. However, the biological function of the ACP gene family in the Brassica genus has been limited until now. In this study, we conducted a comprehensive analysis and identified a total of 120 ACP genes across six species in the Brassica genus. Among these, there were 27, 26, and 30 ACP genes in the allotetraploid B. napus, B. juncea, and B. carinata, respectively, and 14, 13, and 10 ACP genes in the diploid B. rapa, B. oleracea, and B. nigra, respectively. These ACP genes were further classified into six subclades, each containing conserved motifs and domains. Interestingly, the majority of ACP genes exhibited high conservation among the six species, suggesting that the genome evolution and polyploidization processes had relatively minor effects on the ACP gene family. The duplication modes of the six Brassica species were diverse, and the expansion of most ACPs in Brassica occurred primarily through dispersed duplication (DSD) events. Furthermore, most of the ACP genes were under purifying selection during the process of evolution. Subcellular localization experiments demonstrated that ACP genes in Brassica species are localized in chloroplasts and mitochondria. Cis-acting element analysis revealed that most of the ACP genes were associated with various abiotic stresses. Additionally, RNA-seq data revealed differential expression levels of BnaACP genes across various tissues in B. napus, with particularly high expression in seeds and buds. qRT-PCR analysis further indicated that BnaACP genes play a significant role in salt stress tolerance. These findings provide a comprehensive understanding of ACP genes in Brassica plants and will facilitate further functional analysis of these genes.
1. Introduction
Acyl carrier proteins (ACPs) are acidic proteins found in organisms, and they play important roles in regulating growth and development, as well as in responding to biotic and abiotic stresses [1,2]. ACPs have a tertiary structure comprising a short transverse α-helix and three parallel α-helices. This structure creates a hydrophobic cavity and allows for significant structural adaptability. Consequently, ACPs can accommodate acyl chains of varying lengths, bind to acyl intermediates, and transfer between the active sites of different enzymes [3,4].
Studies have shown that ACPs play a crucial role in fatty acid synthesis by carrying acyl chains and facilitating enzymatic reactions. ACP acts as a cofactor in acyl ACP desaturation reactions and performs plastid acyl transferase-like actions with fatty acids of varying acyl chain lengths [5]. Moreover, ACPs are essential proteins for the cycle of fatty acid chain elongation and reduction. They effectively bind to the acyl chain and transport it to the active sites of various enzymes [6]. In a study where the gene expression of ACP4 was inhibited using RNAi, there was a decrease in the leaf fat content of Arabidopsis (Arabidopsis thaliana) and a decrease in the proportion of 16:3 in the total fatty acid composition [7]. Fibre-specific ACP in cotton (Gossypium hirsutum) positively regulates cotton fibre elongation by improving the synthesis of membrane lipids [8]. In the pepper plant (Capsicum chinense), the ACP gene plays a crucial role in the production of branched fatty acid chains, which are necessary for the synthesis of capsaicin [9]. Additionally, a study conducted on Brassica napus revealed that multiple ACPs can fulfil the requirements for fatty acid synthesis [10]. Recent research has also highlighted the essential role of CsACP1 (ACP1 in Coriandrum sativum) in the synthesis of distinct monoenoic fatty acids found in seed oil [11].
Recently, several studies have revealed that ACP also plays crucial roles in enhancing plant tolerance to various stresses [12,13,14,15]. The ACP genes that exhibit tolerance to abiotic stresses have been extensively studied in Arabidopsis. Notably, AtACP1, AtACP2, and AtACP3 have shown high gene expression levels in response to drought, indicating that these genes are involved in drought tolerance. In the presence of salt stress, the atacp5 mutant displays increased sensitivity compared to that of the wild type, while overexpression of AtACP5 enhances salt stress tolerance in transgenic Arabidopsis. Moreover, recent studies have identified certain SbACP (ACP in Sorghum bicolor) genes that contribute to drought and salt stress tolerance [16].
The Brassica genus is highly valued for its economically important species, including those used for food, oil, vegetables, ornamentals, and fodder [17,18]. Within this genus, there are six closely related species that are particularly significant and can be represented by the U triangle [19]. The U triangle consists of three fundamental diploid species: B. rapa (AA, 2n = 2x = 20), B. nigra (BB, 2n = 2x = 16), and B. oleracea (CC, 2n = 2x = 18). Additionally, there are three amphidiploids: B. napus (AACC, 2n = 4x = 38), B. juncea (AABB, 2n = 4x = 36), and B. carinata (BBCC, 2n = 4x = 34). These amphidiploids were created through hybridization between any two of the three diploid species. Moreover, these amphidiploids can be resynthesized using interspecific crosses involving diploid species, which makes them an excellent and widely used model for allopolyploid analysis. All of the genomes of the species in the triangle of U model have been well sequenced [20,21,22,23,24,25] and serve as valuable resources for studies involving gene function and genome evolution in Brassica. At the same time, it is also convenient for us to study the biological function of the ACP genes. At present, the function of ACP genes in the fatty acid synthesis pathway of Brassica species has been less explained, but the potential function in response to abiotic stress has not been investigated. Although the ACP gene family has been extensively studied in numerous species, the evolutionary history and gene functions of ACP genes in the six cultivated Brassica species have not been investigated thoroughly. The accessibility of multigenome sequencing allows us to identify potential ACP orthologous and paralogous genes. Additionally, RNA-seq data facilitate the examination of functional correlations among ACP genes. In this study, we systematically identified and analyzed the ACP gene family across the six cultivated species in the triangle of U. Furthermore, we conducted a comprehensive analysis of ACP expression patterns in B. napus under salt stress, which provides a solid foundation for further investigation into the molecular mechanisms underlying ACP responses to salt stress.
2. Results
2.1. Identification and Characteristics of ACP Genes in the Brassica Genus
To identify putative ACP genes in six cultivated Brassica species, we used ACP protein sequences from Arabidopsis and the PP-binding domain of ACP proteins (PF00550) as queries to search for ACP proteins across the genomes of the Brassica species. In total, 120 ACP genes were identified from six Brassica species (Table S1), including 14, 10, and 13 genes in the diploid progenitors B. rapa, B. nigra, and B. oleracea and 26, 27, and 30 ACP genes in the allotetraploid B. juncea, B. napus, and B. carinata. These identified ACP genes in six Brassica species were uniformly named according to their homologous relationship with the ACP genes of Arabidopsis and their origin from different Brassica species. For example, “BcamtACP2.1” indicates that this gene from B. carinata is highly homologous to mtACP2 in Arabidopsis.
The physicochemical properties of ACP family proteins in Brassica were analyzed. This analysis included the molecular weight (MW), theoretical isoelectric point (pI), instability coefficient (II), and protein average hydrophobicity index (GRAVY) (Table S1). The MW of these ACP proteins ranged from 12.93 kDa (BniACP2) to 21.57 kDa (BcamtACP3.2), while the theoretical isoelectric points ranged from 4.42 (BnaACP4.3) to 7.09 (BjumtACP2.3). Except for BjumtACP2.3, most ACP proteins were acidic (pI < 7), indicating that the ACP proteins in Brassica species had low molecular weight acidic proteins. Approximately 25.83% of the ACP proteins in Brassica were classified as stable proteins (instability index < 40), with the protein instability coefficient ranging from 29.41 to 61.86. The average hydrophobicity index of these ACP proteins ranged from −0.38 to 0.124, with the majority (96.67%) exhibiting weak hydrophilicity.
2.2. Subcellular Localization Analysis of ACP Family Proteins in B. napus
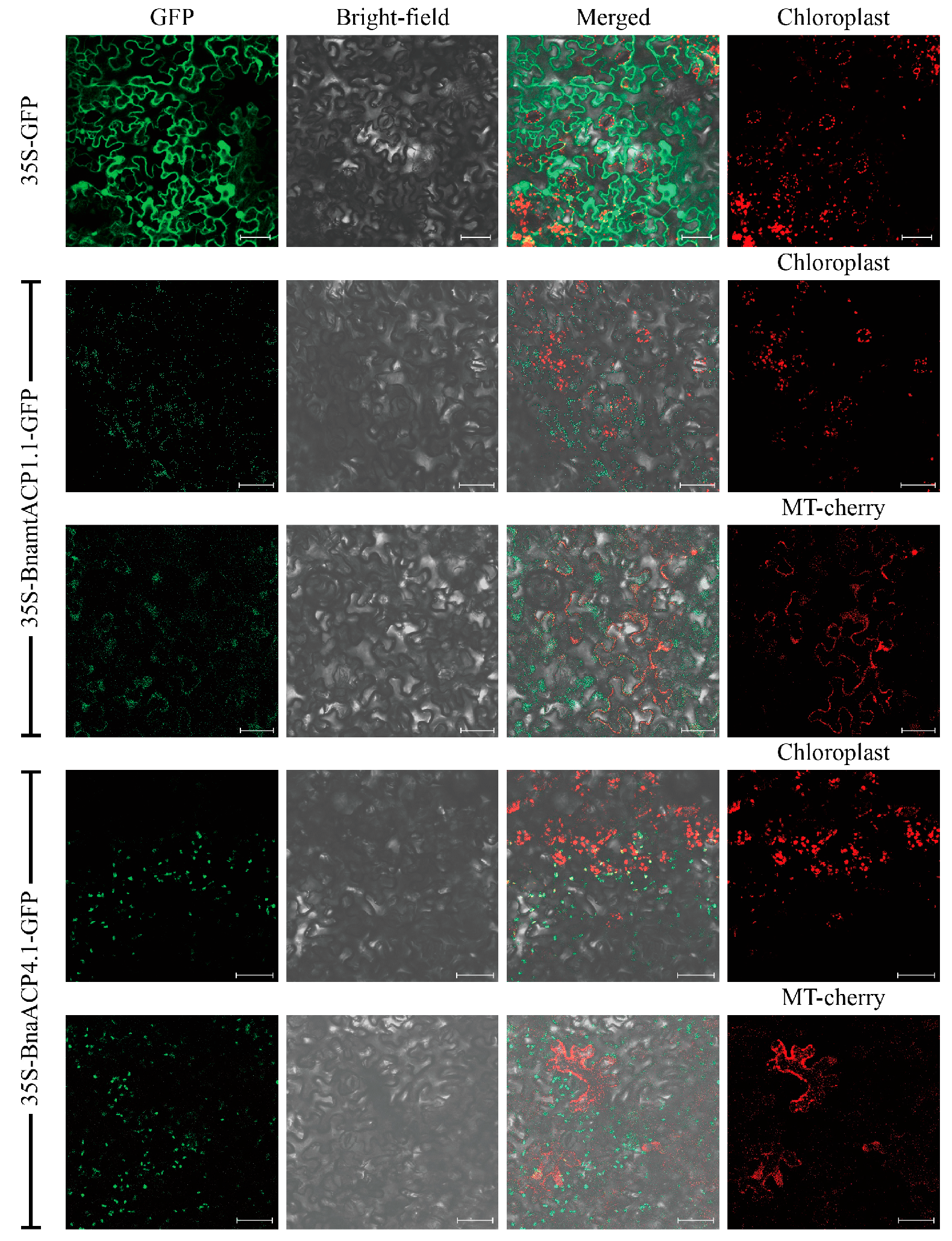
The subcellular localization of ACP proteins in Brassica plants was investigated, revealing that 22 ACP proteins were located in mitochondria, while 98 ACP proteins were localized in chloroplasts (Table S1). To confirm their subcellular localization, six BnaACPs tagged with green fluorescent protein (GFP) (BnaACP-GFP) were constructed. These constructs, along with a recombinant plasmid labelled with a mitochondria-specific marker, were used to infect the leaf epidermal cells of tobacco. Confocal microscopy was used to determine the expressed localizations of the BnaACPs. The results showed that BnaACP3.1, BnaACP4.1, and BnaACP5.1 GFP signals were observed only in combination with chloroplast autofluorescence signals. On the other hand, BnamtACP1.1, BnamtACP2.1, and BnamtACP3.1 GFP signals were observed only in combination with mitochondrial-specific marker signals. These findings indicate that the selected BnaACPs were located in both chloroplasts and mitochondria (Figure 1 and Figure S1).
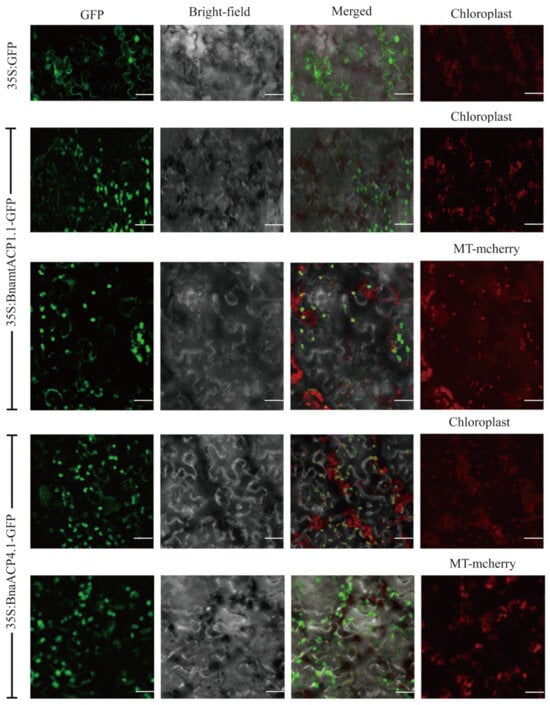
Figure 1.
Subcellular localization of two BnaACP proteins in tobacco. Chloroplast: chloroplast auto-fluorescence; MT-mcherry: mitochondria-specific marker-labelled recombinant plasmid. Scale bar = 50 µm.
2.3. Phylogenetic Analysis of ACP Proteins
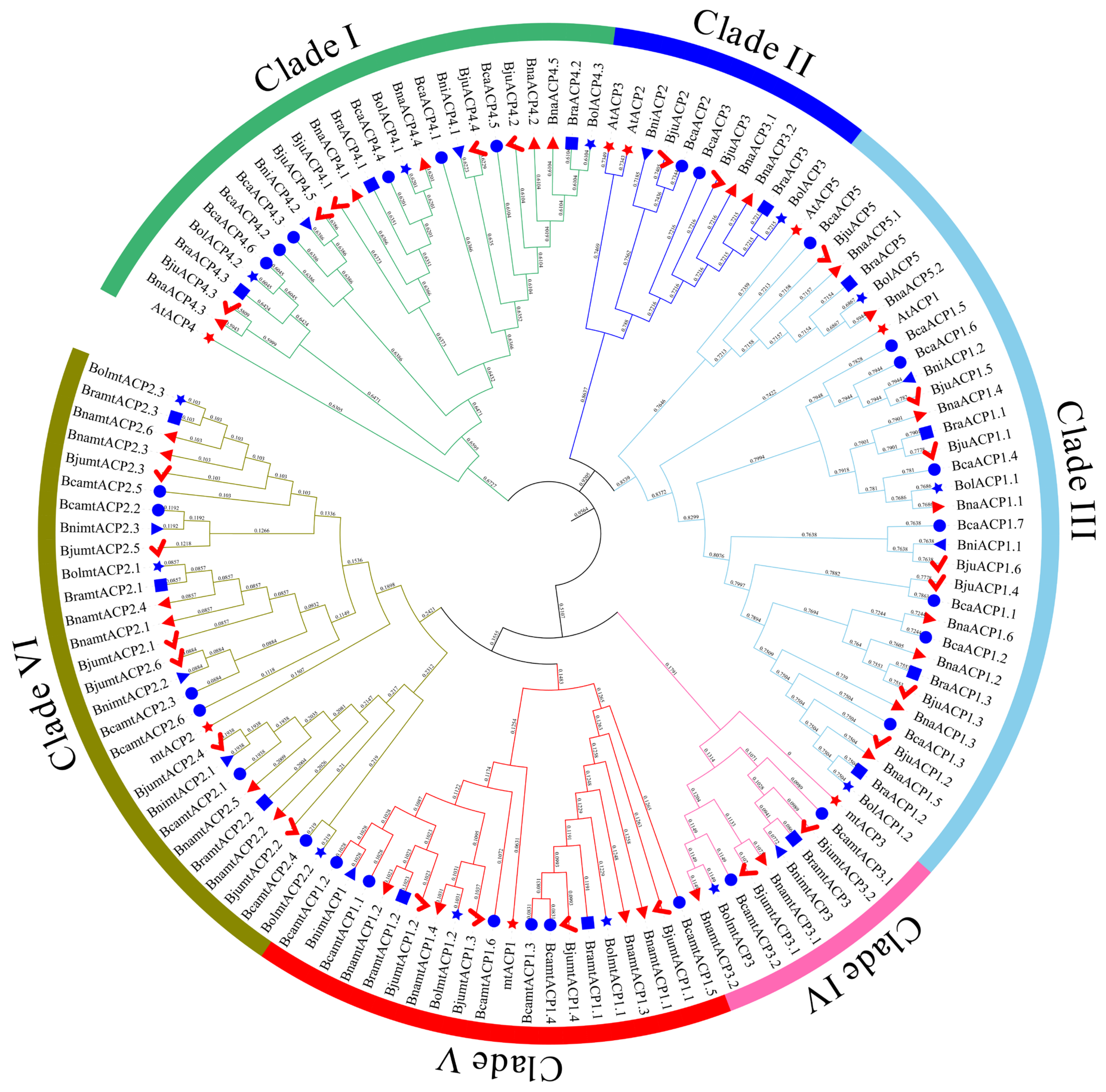
To investigate the evolutionary relationship of ACP proteins in Brassica, a phylogenetic tree was constructed using the maximum likelihood (ML) method. The ML tree included 120 ACP proteins from six Brassica species and eight ACP proteins from Arabidopsis. The analysis revealed that all the ACP proteins in Brassica and Arabidopsis could be categorized into six subclades (Clade I - Clade VI). Most orthologues were found to be clustered together in the same subclade (Figure 2). Clade I consisted solely of ACP4 orthologous proteins, with a total of 25 ACP proteins. Clade II contained the ACP3 (7 ACP proteins) and ACP2 (4 ACP proteins) orthologous proteins. Clade III had the highest number of ACP family protein members, including the 3 BolACP, 2 AtACP, 2 BniACP, 4 BraACP, 7 BjuACP, 8 BcaACP, and 8 BnaACP proteins. Furthermore, clades IV, V, and VI consisted of the 10 mtACP3, 20 mtACP1, and 28 mtACP2 orthologous proteins, respectively.
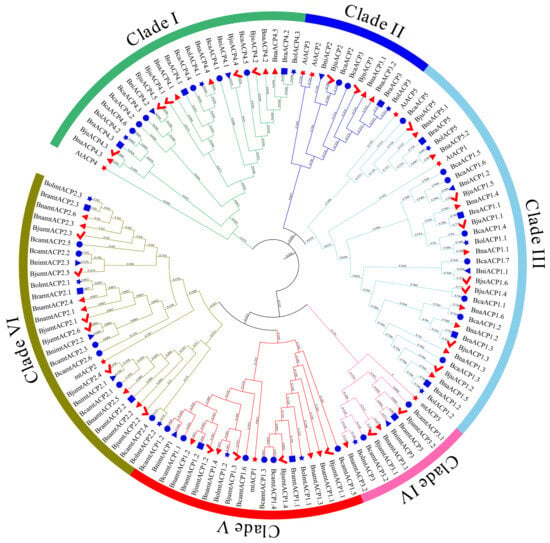
Figure 2.
Phylogenetic tree of ACP family proteins from Arabidopsis and six Brassica species. Red star, blue circle, red checkmark, red right pointing triangle, blue left pointing triangle, blue star, and blue rectangle represent ACP family proteins from Arabidopsis, B. carinata, B. juncea, B. napus, B. nigra, B. oleracea, and B. rapa, respectively.
2.4. Conserved Motif, Conserved Domain, and Gene Structure Analysis of ACP Family Proteins
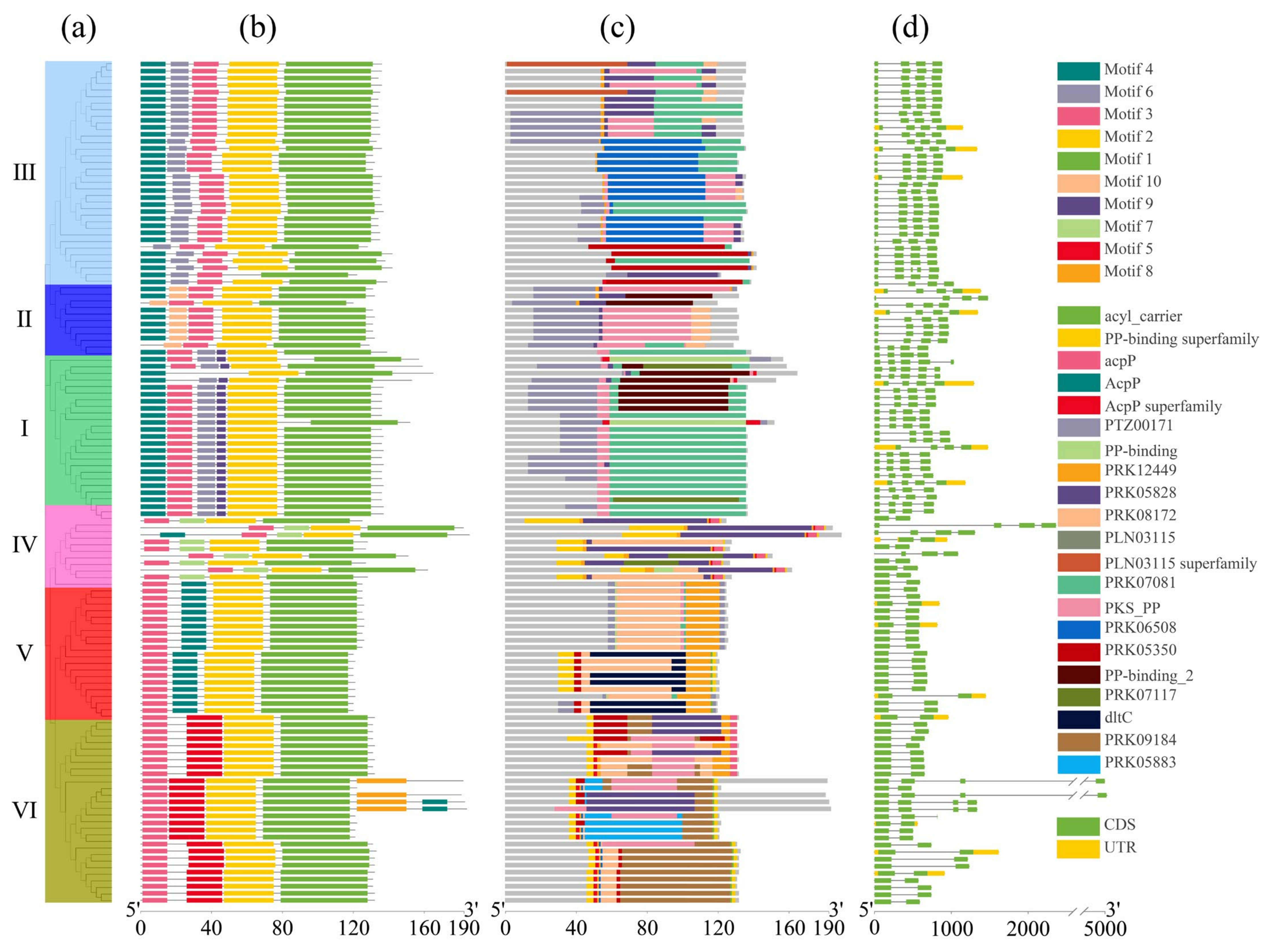
The conserved motifs within the ACP family proteins in Brassica were analyzed, and 10 conserved amino acid sequence regions (motifs) were identified across these ACP proteins (Figure 3a,b). The detailed amino acid sequence of these motifs is listed in Figure S2. Notably, we observed a higher genetic relationship corresponded to a greater similarity in motif structure. Family members within the same clade exhibited higher similarity, indicating that the structure of motifs in orthologous ACP proteins was more conserved than that in paralogous ACP proteins. Motif 4, motif 6, motif 3, motif 2, and motif 1 were present in all of these ACP proteins clustered in clade III, except in BcaACP5 (missing motif 4) and in BnaACP5.2 (missing motif 2). Motifs 10, 9, 7, and 5 were specific to the proteins clustered in clade II, clade I, clade IV, and clade VI, respectively. Moreover, motif 8 is an infrequent motif that was detected only in four ACP proteins (BolmtACP2.3, BnamtACP2.6, BnamtACP2.3, and BjumtACP2.3). The ACP proteins in clade IV showed an unstable arrangement and different motifs compared to the ACP proteins in the other clades. Conserved domain analysis (Figure 3c) revealed that the PTZ00171 domain was present in all the ACP proteins, indicating its specific structure for ACP family proteins. In addition, we also investigated the structural diversity of the identified ACP genes and analyzed their exon–intron structure, which demonstrated significant variation in the number of introns among the ACP families in Brassica compared to that among their orthologues in Arabidopsis (Figure 3d). Moreover, the similarity in gene structure among the ACP genes is positively correlated with their genetic relationship.
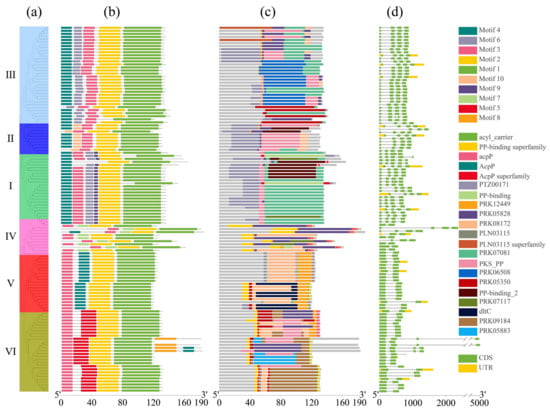
Figure 3.
Detailed structure of ACP family proteins in six cultivated species of Brassica. (a) Phylogenetic tree, (b) conserved motif, (c) conserved domain, (d) gene structure.
2.5. Chromosomal Localization Analysis of ACP Genes
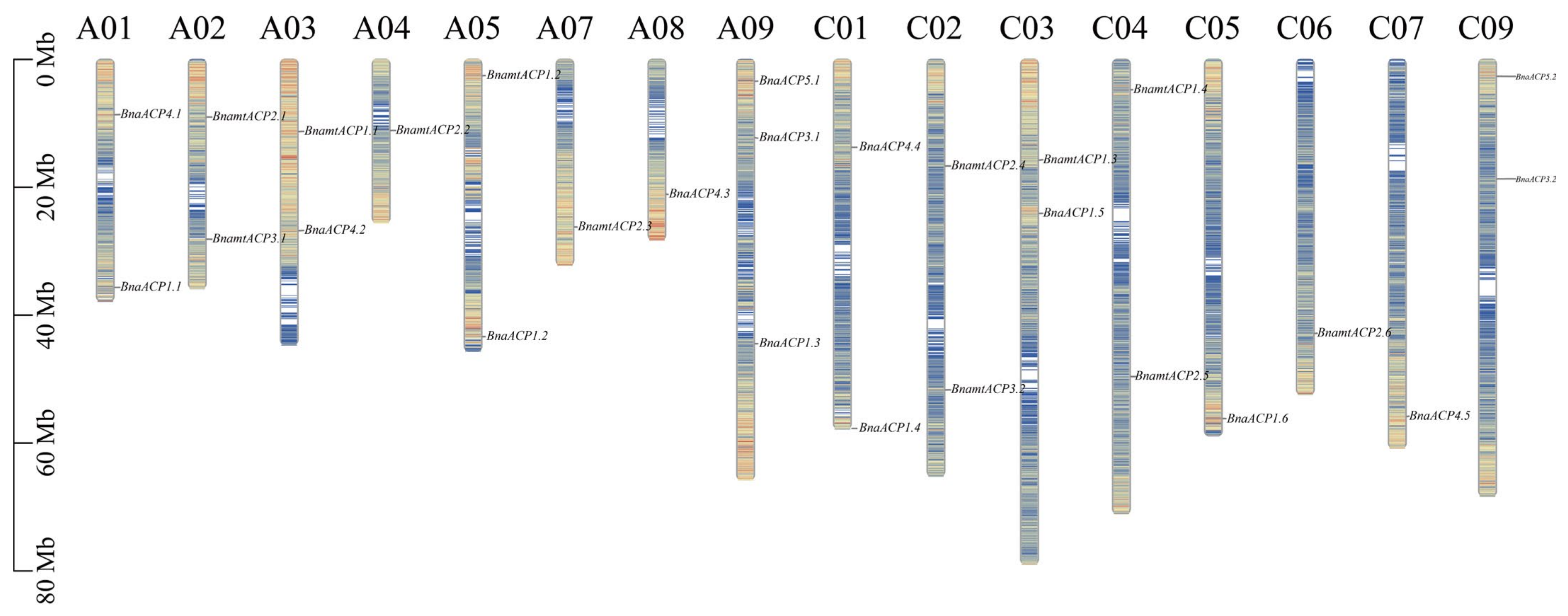
To visualize the location of ACP genes in chromosomes, the physical location of these genes was determined based on the Brassica genome annotation (gff) files. Out of 120 ACP genes, 119 were successfully mapped to assembled chromosomes (Figure 4 and Figure S3 and Table S1). However, BjuACP1.1 was found on unassembled scaffolds. We observed that the majority of the ACP genes and their locations in amphidiploid B. napus and B. juncea corresponded to those in the ancestral diploid genome, but the gene position and gene number were relatively unstable in B. carinata, in which several ACP genes were lost in the C subgenome and some ACP genes were duplicated in the B subgenome when compared to their orthologues in the diploid genomes. In addition, most ACP genes were located in regions with a high density of genes.
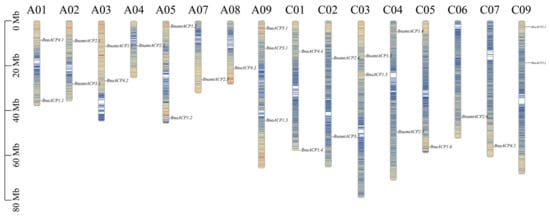
Figure 4.
Chromosome distribution of ACP genes in B. napus. Chromosomes without ACP gene distribution (A06, A10, and C08) are not shown. The different colours on the chromosomes represent the gene density.
2.6. Cis-Acting Elements of ACP Genes
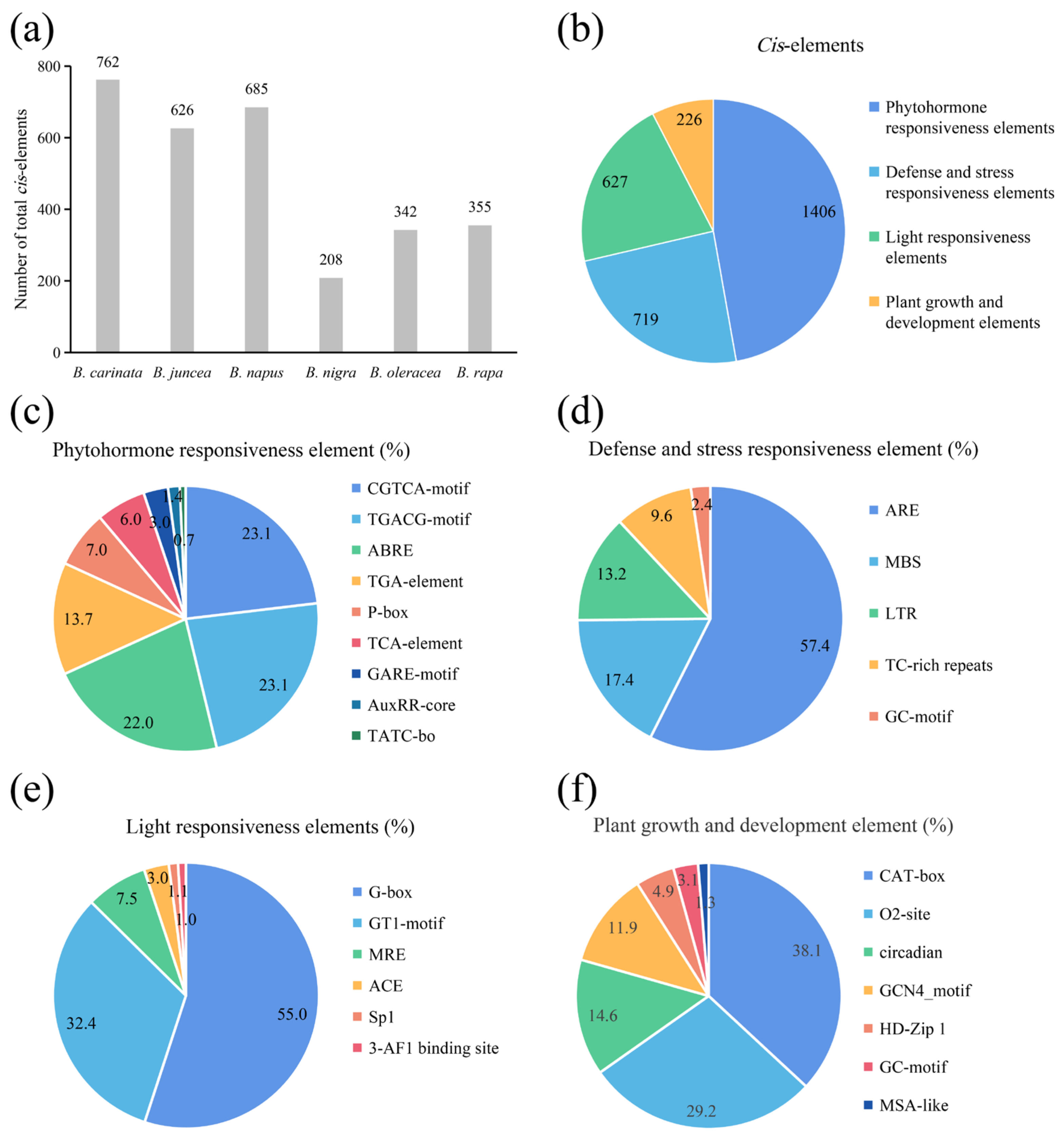
To detect the potential molecular functions of the ACP genes in Brassica, the 1.5 kb promoter sequence upstream of each ACP was extracted, and the distributions of cis-acting elements were analyzed. The results showed that a total of 2987 cis-acting elements ranging from 208 (B. nigra) to 762 (B. carinata) in the ACP genes were identified in these Brassica species (Figure 5a). These elements were primarily associated with hormone regulation (1406 elements of 2987 in total), stress resistance (719), light responsiveness (627), and growth and development (226) in sequence (Figure 5b). Within the hormone regulation group (Figure 5c), the CGTCA-motif (23.1%), TGACG-motif (23.1%), and ABRE (22.0%) were the most overrepresented elements. In the stress resistance group (Figure 5d), these elements were predominantly represented by AREs (57.4%), followed by MBSs (17.4%). In the light responsiveness group (Figure 5e), the G-box (55.0%) and GT1-motif (32.4%) were the dominant elements. In the plant growth and development group (Figure 5f), these elements were overrepresented by CAT-box (38.1%), O2-site (29.2%), and circadian (14.6%). The presence of multiple cis-acting elements indicates that the ACP genes in Brassica plants serve multiple functions, particularly the function of stress responsiveness.
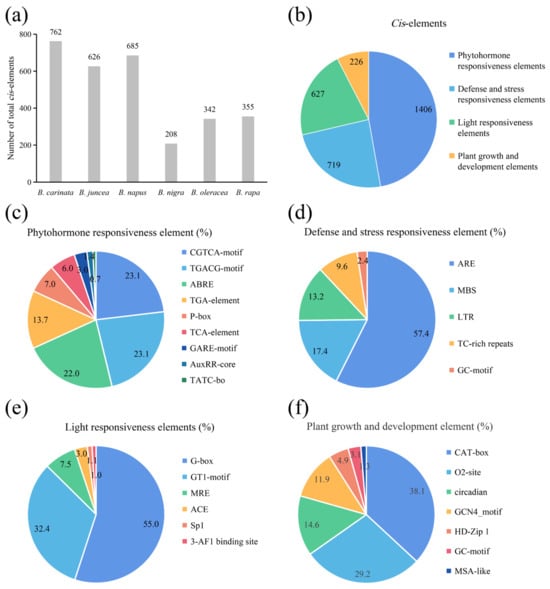
Figure 5.
The prediction results of the cis-acting elements of the ACP gene family in Brassica. (a) Promoter elements of all ACP genes in each species. (b) Classification of all promoter elements in six species. (c–f) Details of all promoter classifications.
2.7. Gene Duplication, Synteny, and Evolution Analysis of ACP Genes in Six Brassica Species
Angiosperms have been demonstrated to undergo multiple rounds of whole-genome duplication, resulting in the expansion of gene families. To investigate the replication events of ACP genes in Brassica species, we employed DupGen_finder [26] to analyze the occurrence of tandem duplication (TD), whole-genome duplication (WGD), proximal duplication (PD), transposed duplication (TRD), and dispersed duplication (DSD) genes in the ACP family of six Brassica species (Table S2). We did not find any TD, WGD, PD, or TRD events in the six Brassica species, but DSD events were observed in all of them. Among the DSD events in the ACP gene family across the six species, the highest proportion was 26.42% (28/106) in B. carinata, while the lowest proportion was 7.55% (8/106) in B. nigra compared to the other five species. This finding suggests that in the genomes of the six Brassica species, the ACP genes primarily contribute to the expansion of the gene family through DSD events.
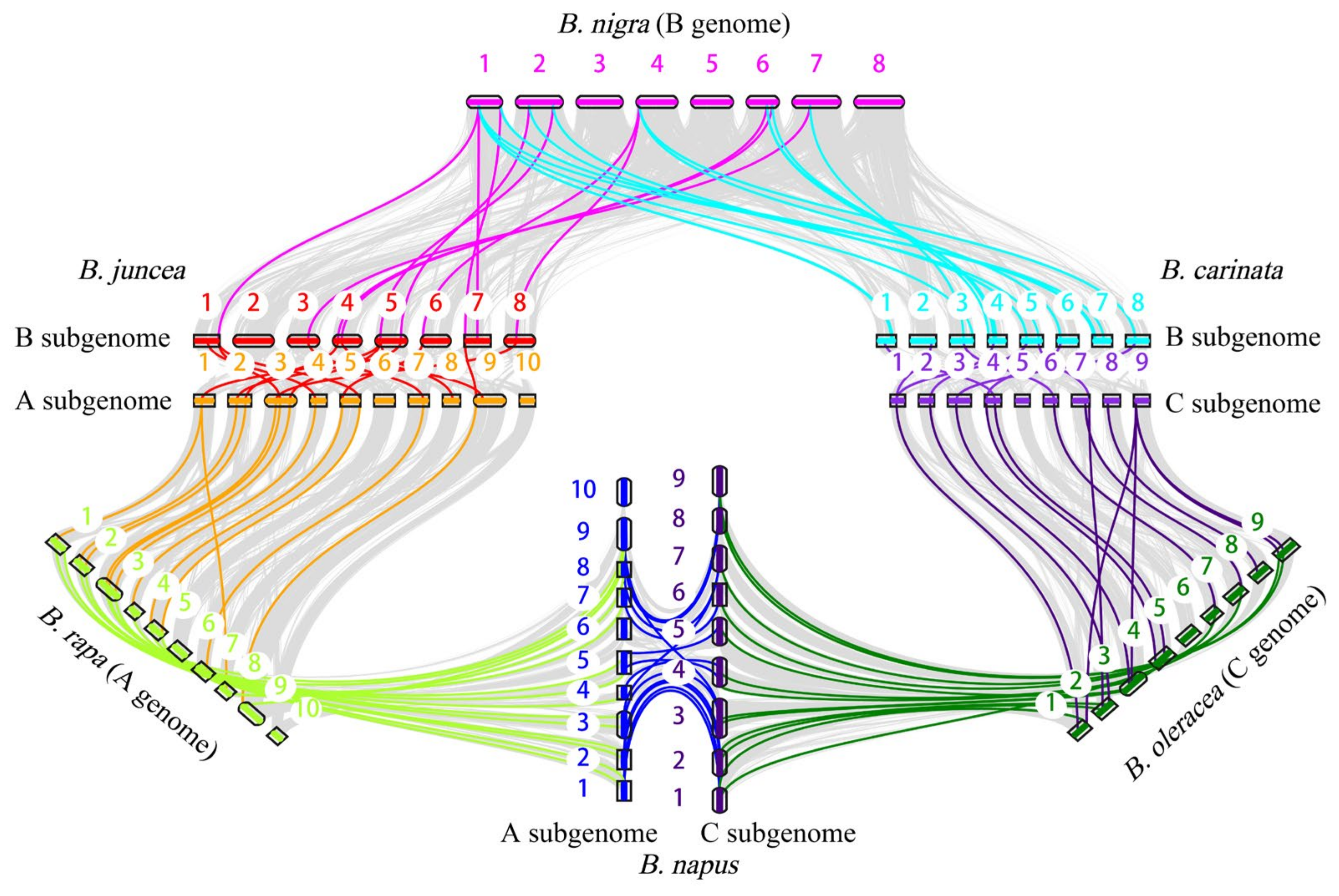
To identify the intraspecific and interspecific collinear gene pairs between three diploid species and their corresponding tetraploid species, we compared the ACP genes of the diploid species with those of the three allotetraploid species. We also identified collinear gene pairs within each species. The collinear gene pairs of ACP genes were then visualized (Figure 6). The intraspecific collinear gene pairs included 7 gene pairs in B. rapa, 4 in B. oleracea, 2 in B. nigra, 17 in B. napus, 18 in B. juncea, and 21 in B. carinata (Table S3). In addition, 13 pairs of syntenic orthologous genes were found in B. nigra and B. oleracea, but only 11 pairs were found in the two subgenomes of B. carinata, suggesting that some of the collinear ACP genes might have been lost or that homologous exchange (HEs) might have occurred during the process of polyploidization.
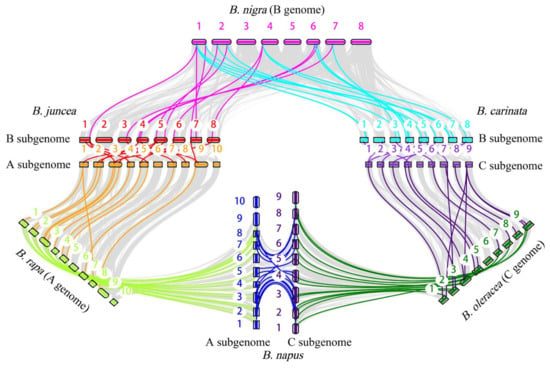
Figure 6.
Collinearity analysis of ACP genes in six Brassica species.
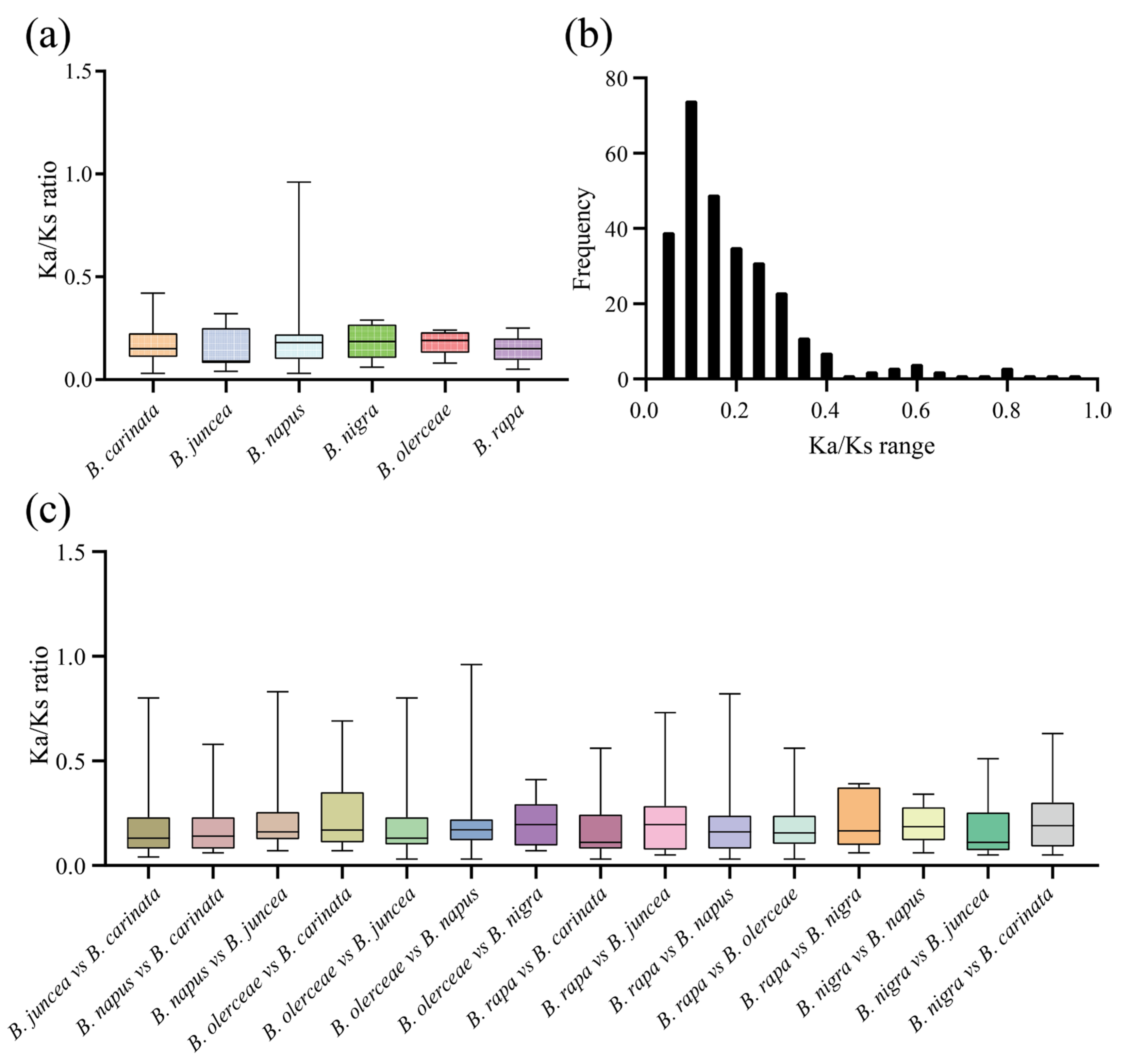
The nonsynonymous (Ka) and synonymous (Ks) nucleotide substitution patterns of protein-coding genes are important indicators of gene evolution. The Ka/Ks ratio is used in genetics to determine whether there are selective pressures on protein-coding genes and evaluate the gene divergence rates. A Ka/Ks ratio > 1 indicates positive selection, a Ka/Ks ratio = 1 indicates neutral selection, and a Ka/Ks < 1 indicates purifying selection [27]. In this study, we calculated the Ka/Ks values for duplicated gene pairs to investigate whether the ACP genes were associated with selective pressure after duplication events. First, we obtained the intraspecies Ka/Ks values in six Brassica ACP families (Figure 7a; Table S4). After removing genes with a Ka or Ks of 0, the average Ka/Ks ratio was 0.17. The Ka/Ks values of the ACP genes among the different species ranged from 0.03 to 0.96, with the majority falling within the range of 0.05–0.3 (Figure 7b). Next, we calculated the interspecies Ka/Ks values in the six Brassica ACP families (Figure 7c; Table S5). Similarly, genes with a Ka or Ks of 0 were removed. These results implied that these ACP genes were subject to strong purifying selection pressures, both intraspecies and interspecies, in the six Brassica species.
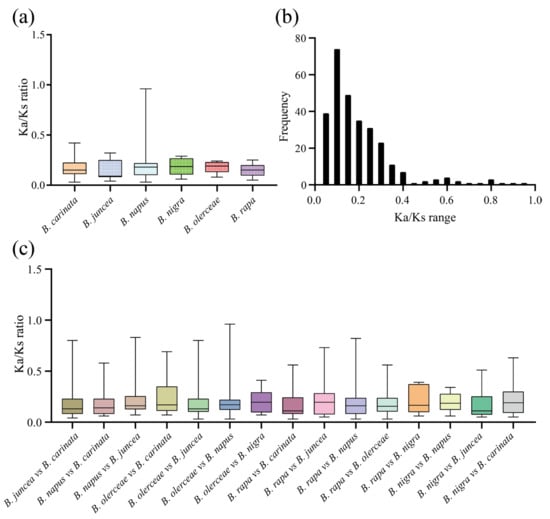
Figure 7.
The Ka/Ks values of ACP genes in six Brassica species. (a) Analysis of intraspecific Ka/Ks ratios. (b) Frequency distribution of the Ka/Ks values. (c) Analysis of interspecies Ka/Ks ratios.
2.8. Tissue-Specific Expression of ACP Genes
The B. napus multi-omics information resource (BnIR) database provides a comprehensive knowledge base of expression data for BnaACP (ACP genes in B. napus) genes from various tissues, including rosette, roots, stems, leaves, cotyledons, buds, sepals, petals, filaments, pollen, siliques, and seeds. This database allows for the detection of tissue expression patterns of ACP genes. The expression heatmap (Figure S4) revealed distinct temporal and spatial expression characteristics of BnaACP genes among different tissues. In general, two-thirds of mtACPs exhibited relatively low expression in all of the tested tissues. Notably, all of the BnaACP4 paralogues except BnaACP4.3 showed high expression levels in cotyledons and vegetative rosettes, suggesting that ACP4 genes actively function in vegetative growth. BnaACP4 genes are primarily expressed in vegetative tissues, and BnaACP4.3 is also enriched in vegetative tissues. However, BnaACP1.1 and BnaACP1.4 appear to express specifically in bud and seeds. Moreover, the majority of BnaACP1 and BnaACP3 paralogues exhibited relatively high expression in buds and seeds compared to other tissues, suggesting that these genes play a significant role in fatty acid synthesis.
2.9. Expression Patterns of ACP Genes in B. napus under Salts
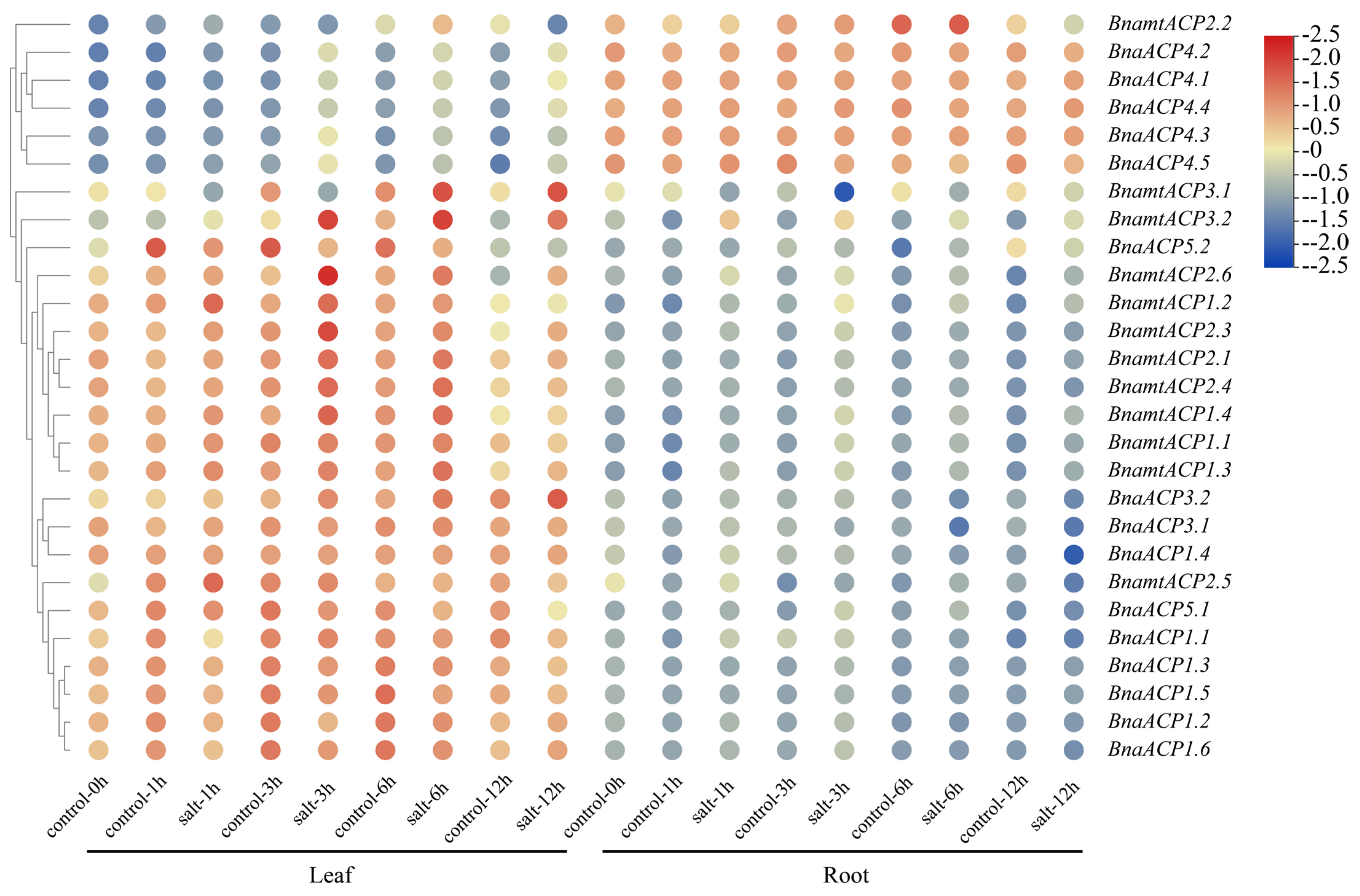
The analysis of the cis-elements of ACP genes in Brassica showed that BnaACP genes may respond to various stresses. To further detect the functions of ACP genes, we constructed a heatmap of BnaACPs expression (Figure 8) under salt stress based on the gene expression data from BnIR database. We found that approximately 80% of BnaACPs exhibited a relatively high expression in leaves but a low expression in roots. Moreover, almost all the BnaACPs genes in the leaves and roots showed different gene expression profiles, suggesting that the ACP genes were tissue-specific expression genes. Under salt stress, the expression level of most BnaACPs did not show significant changes in roots and leaves when compared to the controls. However, we observed that the expression levels of some BnaACPs were increased (such as BnaACP 5.1), while the expression levels of some BnaACPs were decreased (such as BnaACP 4.1) under salt stress, suggesting these genes are likely to be involved in responding to salt stress.
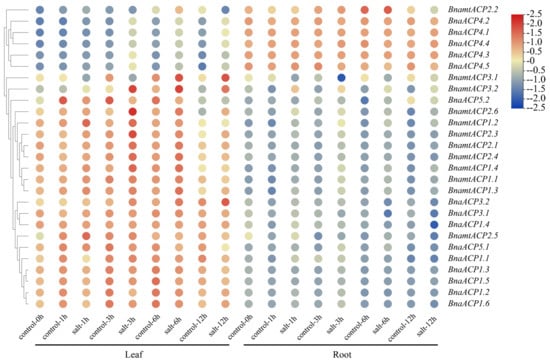
Figure 8.
Expression patterns of the BnaACP genes under salt stress. The expression level of BnaACP genes in leaves and roots after 1 h, 3 h, 6 h, and 12 h of treatment with salt stress.
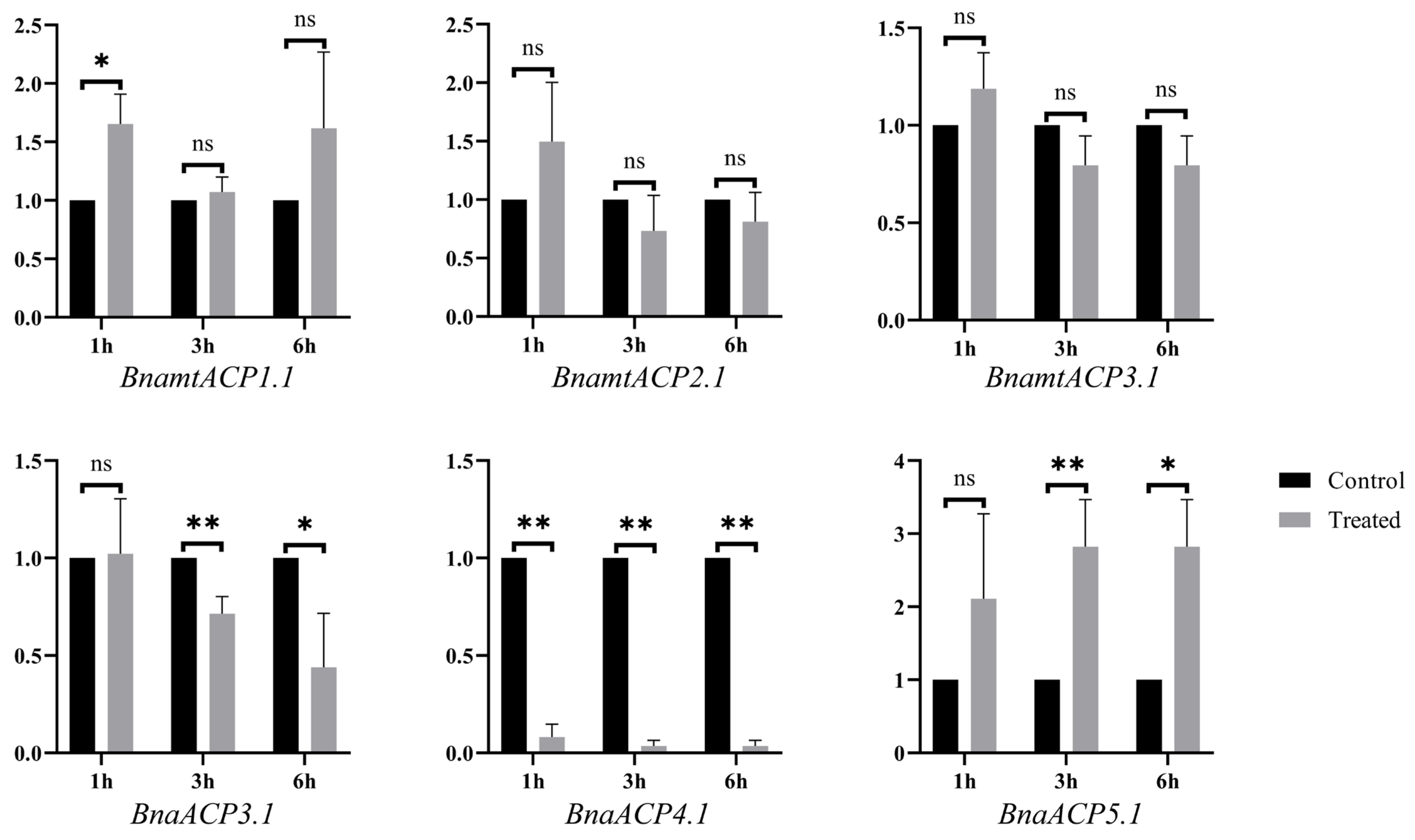
To further verify the functions of ACP genes, a qRT–PCR analysis of BnaACP genes of seedlings subjected to salt stress was conducted. According to the homology with the ACP genes in A. thaliana, we selected three BnaACP genes and three BnamtACP genes for qPCR verification (Table S6). The results (Figure 9) showed that except for BnamtACP1.1 and BnaACP4.1, these selected genes did not change significantly after salt treatment for 1 h. Under treatment for 3 h and 6 h, the expression levels of both BnaACP3.1 and BnaACP4.1 were significantly down-regulated, while the opposite was observed for the BnaACP5.1. It is worth noting that BnaACP4.1 showed a significantly decrease (p < 0.01) at all stress time points. Similar result for AtACP4 was observed under iron deficiency and nitrogen starvation stresses [14]. These results demonstrate that the expression patterns of BnaACP genes are dynamically altered in response to salt stress. Specifically, BnaACP4.1 may act as a negative regulator in B. napus salt stress response.
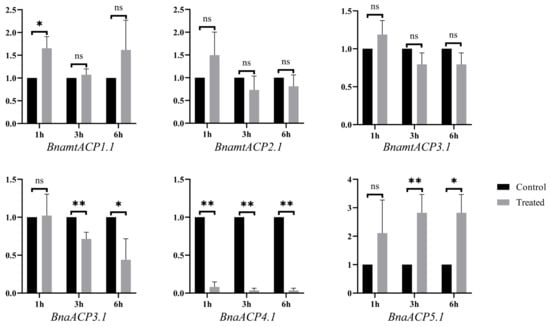
Figure 9.
The relative expression of BnaACP genes under salt stress treatment. * and ** indicate significant differences at the 5% and 1% level by t-test. ns indicates not significant by t-test.
3. Discussion
The six main cultivars of the Brassica genus in the triangle of U are important sources of vegetable oil and emerging sources of biodiesel and biofuel for industrial production [28,29]. Fatty acids, which are the main constituents of lipid molecules, are essential nutrients for the survival of living organisms and play a crucial role in the development and growth of plants. They also provide the energy needed by biological bodies and serve as key components of cell tissues. ACP family proteins are involved in the synthesis of plant fatty acids and facilitate the transfer of acyl intermediates during the synthesis process. They play a crucial role in the direct synthesis pathway of long-chain fatty acids and also have significant involvement in other cellular metabolic processes [30,31]. To date, numerous studies have focused on identifying and analyzing ACP family proteins in various plant species [16,32,33,34], but there is a lack of research on the function and potential role of ACP gene families, specifically in Brassica. It is worth noting that the genomes of six Brassica species have already been sequenced and assembled [20,21,22,23,24,25]. The use of the three natural allotetraploids and their diploid progenitors in the triangle of U has been instrumental in studying scientific problems related to polyploidization. Therefore, it is crucial to conduct a comprehensive investigation to identify and analyze the ACP gene families in Brassica.
In this study, a total of 120 ACP genes were identified from the triangle of U model. These genes were divided into six subclades based on phylogenetic relationships. The structural diversity of the genes reflects their functional diversity and plays a significant role in the evolution of gene families. Among these subclades, each subclade contained the same conserved motifs and domains but with some degree of variation between different subclades (Figure 2). These results showed that these ACP genes were highly conserved in key regions but with a degree of variation in sequence structure. This indicates that the ACP gene families might have undergone changes in gene structure and function during evolution. Notably, the number of ACP family members in subclade I and subclade III of the three diploid species was approximately 1.5 times higher than that in Arabidopsis, while the number of ACP genes from the three double diploid species was approximately 4 times higher than that in Arabidopsis. Additionally, these findings revealed that the gene families exhibited varying numbers of exons and introns and that certain Brassica ACP genes lacked introns. Furthermore, RNA-Seq analysis of B. napus in different tissues revealed diverse and tissue-specific expression patterns of BnaACP genes, highlighting the functional diversity of BnaACP.
The Brassica genus shares a common ancestor with Arabidopsis that underwent three whole-genome duplication (WGD) events (α, β, and γ) [21]. After diverging from the Arabidopsis lineage, Brassica plants experienced a lineage-specific whole-genome triplication (WGT) event approximately 15.9 million years ago (MYA). According to the U model triangle, a typical ancestral region in Arabidopsis would correspond to three regions in the Brassica genome [21,35,36,37]. The quantitative analysis revealed that the ACP gene family underwent significant expansion during the process of allopolyploidy. However, the total number of ACP genes in B. rapa, B. oleracea, and B. nigra is 14, 13, and 10, respectively, which is not three times the number found in Arabidopsis. This indicates that the ACP genes experienced a loss of copy number following the whole-genome triplication. For example, the ACP2 gene was lost during allopolyploidy in B. napus. Similar phenomena have been observed in the MADS-box [38] and WRKY [39] transcription factor families of Chinese cabbage, as well as the MYB28 [40] family of Brassica. Fragment repeats and tandem duplication (TD) events have been reported to contribute to the amplification of ZmPRXs [41] in maize and PbPRXs [42] in Chinese pear (Pyrus bretschneideri). Our observations indicate that ACP genes in Brassica primarily undergo gene family expansion through dispersed duplication (DSD) events, suggesting that DSD events have played a major role in the expansion of the ACP gene family in Brassica. Functionally, these genes may be lost or undergo new functionalization and subfunctionalization [43]. Moreover, these new gene functions may help plants better adapt to changing natural environments, thereby preventing extinction [44].
In bacteria, ACP exists as an independent soluble protein in the type II fatty acid synthase system, while in mammals, it exists as a domain within the type I fatty acid synthase [45]. ACP is a critical member of the carrier protein family and directly participates in the synthesis of long-chain fatty acids. Plant cells possess fatty acid synthase complexes, particularly in plastids and mitochondria. The type II fatty acid synthase complex in plastids gas been extensively studied. ACP plays a crucial role in the process of fatty acid synthesis in Brassica crops; for example, B. napus could encode multiple copies (this study found 27) of ACP to meet the demands of fatty acid synthesis that occurred during oilseed development [10]. In transgenic Brassica, the prime increase was found for linolenic (C18:3) in leaves. Interestingly, transgenic approaches could be used to improve the seed oil quality of Brassica, mainly by increasing the ratio of monounsaturated (C18:1)/saturated fatty acids and reducing the content of erucic acid (C22:1) [46]. Overexpression of AtACP5 also led to changes in fatty acid composition, including a decrease in C18:1 and an increase in palmitic acid (C16:0) [14]. Therefore, by altering the expression of the ACP gene, the composition and fatty acid content of Brassica seed oil can be changed, which is a convenient way to improve the quality of seed oil in Brassica crops.
In Arabidopsis, the function of ACP genes has also been extensively reported, and these ACP genes have been divided into two types (plastidial ACP and mitochondrial ACP) [33,47]. In this study, we also predicted the expression of ACP genes in chloroplasts and mitochondria, which has been validated in tobacco. Based on our analysis of cis-acting elements, the ACP gene of Brassica plants plays a crucial role in light response, hormone regulation, stress resistance, and growth and development [48]. However, the number of cis-acting elements may vary among different species due to gene loss during differentiation and other factors [49]. These findings further support the positive role of the ACP gene in stress resistance. Tissue expression data and qRT-PCR analysis revealed distinct expression levels of BnaACP genes throughout the entire growth period of B. napus in all tissues. Under salt stress treatment, the expression pattern of BnaACP genes varied at different time points. We noticed that the most pronounced response was observed for BnaACP4.1 and BnaACP5.1, which showed particular sensitivity to salt stress. After exposed to salt stress, the relative expression level of BnaACP4.1 was significantly decreased, while that of BnaACP5.1 showed an increase. Similar results were observed in orthologous AtACPs. In Arabidopsis, exposure to abiotic stress such as iron deficiency and nitrogen starvation led to down-regulation of AtACP4 [14]. Knocking out AtACP5 resulted in increased sensitivity to salt stress, while overexpressing AtACP5 in transgenic lines exhibited improved salt tolerance compared to the wild-type [14]. Additionally, this result aligns with the predicted involvement of cis-acting elements in response to stress and defence. The diversity in the expression patterns of BnaACP genes suggests the involvement of various mechanisms in plant development and defence responses.
4. Materials and Methods
4.1. Plant Materials and Data Sources
The seeds of the tetraploid B. napus accession ‘Zhongshuang 11’ were preserved by our laboratory. The seeds were sterilized and germinated in Petri dishes with wet filter paper until the cotyledon was fully expanded. They were then transplanted to 1/2 Hoagland nutrient solution for a duration of 10 days. The temperature and light cycle were maintained at 22 °C/19 °C (light and darkness) with a 16 h light and 8 h darkness cycle. Subsequently, the experimental group was subjected to salt stress treatment by adding 150 mM NaCl [50,51]. Three biological replicates were set up for this purpose. Fresh leaves were collected at 1, 3, and 6 h after treatment. These leaves were immediately frozen in liquid nitrogen and stored at −80 °C for subsequent experiments.
To obtain information on the Arabidopsis ACP (AtACP) gene family, we retrieved eight protein sequences and nucleic acid sequences of the AtACP gene family (AtACP1~AtACP5, mtACP1~mtACP3) from the TAIR database (http://www.arabidopsis.org, accessed on 14 August 2023). The hidden Markov model (HMM) of the ACP PP-binding domain (PF00550) was downloaded from the Pfam database (http://pfam.Xfam.org/, accessed on 14 August 2023). The genome data of B. carinata was available from CoGe (https://genomevolution.org/coge/, accessed on 10 August 2023) under id 63922 [25], and the other five Brassica genomes were obtained from the BRAD database (http://brassicadB.org/brad/, accessed on 10 August 2023) [52]. The transcriptome data were downloaded from the BnIR database https://yanglab.hzau.edu.cn/, accessed on 23 August 2023) [53].
4.2. Identification and Characterization of ACP Genes
The ACP genes in allotetraploids (B. juncea, B. napus, and B. carinata) and their diploid progenitors (B. rapa, B. nigra, and B. oleracea) were comprehensively identified. In the identification process, eight ACP protein sequences from Arabidopsis were used as queries to perform BLASTp searches (E-value < 1 × 10−5) with all proteins from these six species. Furthermore, the protein sequence of the six Brassica species was searched using the PP-binding domain (PF00550) as a query through the hmmersearch subroutine of HMMER v3.0 [54]. The candidate genes obtained from hmmersearch and BLASTp were merged, and any repeated candidate genes were removed. These candidate genes were then verified using the SMART database (http://smart.embl-heidelberg.de/, accessed on 25 August 2023), NCBI conserved domain database (CDD, https://www.ncbi.nlm.nih.gov/cdd, accessed on 25 August 2023), and Pfam database. The physicochemical properties of the ACP gene family members in the six Brassica species, including MW, pI, II, and GRAVY, were analyzed using the online software ProtParam v3.0 tool (http://web.ExPASy.org/protparam/, accessed on 25 August 2023). The subcellular localization of the ACP proteins in the six Brassica species was predicted using the Cell-PLoc 2.0 online software [55].
4.3. Phylogenetic Relationship Analysis
To construct the phylogenetic tree of the ACP proteins, we aligned 120 full-length protein sequences from six Brassica species and eight protein sequences from Arabidopsis using the MUSCLE program [56]. The alignment was performed with default parameter values. The MGEA v11.0.13 [57] software was used with the ML method and the Tamura-Nei nucleotide substitution model. The stability of the phylogenetic tree was confirmed using the bootstrap method with 1000 replicates. Finally, the web service iTOL (http://itol.embl.de/, accessed on 28 August 2023) [58] was utilized to annotate the phylogenetic tree.
4.4. Conserved Motif and Gene Structure Analysis
The full-length ACP protein sequences of the six Brassica species were analyzed using the online software MEME v5.5.4 [59] to identify conserved sequences and important functional sites. Ten motifs were set, while other options were kept at default parameters. To determine the gene structure of ACP, GSDS 2.0 software (Gene Structure Display Server 2.0, https://gsds.gao-lab.org/, accessed on 29 August 2023) [60] was used to detect exon/intron composition information, following the default parameters.
4.5. ACP Gene Promoter Sequences and Chromosomal Localization Analysis
The promoter sequences of the ACP genes in the six Brassica species were extracted using TBtools v1.098 [61]. The cis-regulatory elements (CREs) in the promoter regions of the ACP genes were predicted using the PlantCARE server (Plant cis-Acting Regulatory Element, http://bioinformatics.psb.ugent.be/webtools/plantcare/html/, accessed on 29 August 2023) [62]. The chromosome localization of the ACP gene family members in the six Brassica species was mapped using the Gene Location Visualize feature of TBtools software, based on the chromosome annotation information of ACP genome files.
4.6. Gene Duplication, Synteny, and Evolution Analysis of ACP Genes
Gene duplication is a common occurrence in plant species and plays a significant role in the expansion of gene families. In this study, we investigated the duplication events of ACP genes using DupGen_finder. Subsequently, we identified interspecific and intraspecific collinear gene pairs within the ACP gene family across the six Brassica species using JCVI v1.3.5 [63]. Finally, we calculated the Ka/Ks ratio of the intraspecies and interspecies ACP gene family genes in these six Brassica species using the Simple Ka/Ks Calculator module of TBtools.
4.7. Analysis of Tissue Expression Characteristics and qRT–PCR
To investigate the expression levels of BnaACP gene family members in various tissues of B. napus, we downloaded transcriptome data for roots, stems, leaves, cotyledons, buds, sepals, petals, filaments, pollen, siliques, and mature seeds of B. napus. The expression levels of the BnaACP gene family members were standardized and visualized using TBtools.
Total RNA was extracted from the leaves of B. napus seedlings using an RNA extraction kit (Aidlab Biotech, Beijing, China). First-strand cDNA was synthesized by reverse transcription using a HiFiScript gDNA Removal cDNA Synthesis Kit. For real-time quantitative PCR analysis, six BnaACP genes with the highest homology to Arabidopsis ACP genes were selected. Specific quantitative analysis primers were designed using NCBI, with Actin (used by [64]) serving as a control (Table S6). The FQD-48 X real-time fluorescence quantitative PCR system was employed to assess gene expression levels in this study, following the method of [65]. Three biological replicates and three technical repeats per gene were prepared. The 2−∆∆CT method [66] was used for the relative quantitative analysis of the gene, revealing the expression pattern of BnaACP genes in B. napus under salt stress treatment.
4.8. Subcellular Localization
In this experiment, the fusion protein expression method was used as follows: the GFP was fused with the C-terminal of the BnaACP protein, and the location of the BnaACP protein was determined by observing the fluorescence signal of the GFP. The specific primers (Table S6) were used to amplify the entire coding sequences (CDS) of the BnaACP. The vector was linearized using BamHI. The Hieff Clone® Plus One Step Cloning Kit was utilized to subclone the CDS of six selected BnaACPs into the target vector with the GFP. The target gene, which contained the GFP fusion protein, was then introduced into tobacco leaves using Agrobacterium (GV 3101) injection. Additionally, the mitochondria-specific marker (pSuper 1300-pFAY-mCherry, Protein Interaction) was transformed using Agrobacterium (GV 3101), and the target gene with the GFP fusion protein was co-injected into the tobacco. The tobacco plants were cultured in dark conditions at 28 °C for 24 h, followed by normal cultivation for 48 h. Finally, the fluorescence signal was observed using laser confocal microscopy.
5. Conclusions
In this study, a total of 120 ACP genes with PP-binding domains were identified in the six Brassica species, which were mainly divided into six subclades according to the phylogenetic relationships of these ACP genes, and each subclade contained the same conserved motifs and domains. A comprehensive and systematic study of these genes was conducted from the aspects of gene location, gene evolution, gene structure, cis-acting elements, protein physical properties, gene expression patterns, and subcellular localization. Additionally, qRT-PCR was used to preliminarily explore the expression level and potential role of the BnaACPs gene under salt stress. By employing gene family identification and comparative genomics, this study contributes to the bioinformatics analysis of the ACP gene family in the six Brassica species, providing a theoretical foundation for future investigations on ACP family genes.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/plants13070950/s1. Figure S1: Subcellular localization of four BnaACP proteins. GFP: green fluorescent protein. Scale bar = 20 μm. Figure S2: Conserved motifs of ACP genes in six Brassica species. Figure S3: Chromosome distribution of ACP genes in six cultivated species of Brassica. (a) B. rapa, (b) B. nigra, (c) B. juncea, (d) B. oleracea, (e) B. carinata. The different colours on the chromosomes represent the gene density. Figure S4: The gene expression levels of ACP genes in different tissues of B. napus. Table S1: Summary of ACP family proteins in six cultivated Brassica species. Table S2: Dispersed duplication events of ACP genes in six Brassica species. Table S3: Identify the interspecific and intraspecific collinear gene pairs. Table S4: The intraspecific Ka/Ks ratios analysis of duplicate gene pairs of ACP gene family in six Brassica species. Table S5: The interspecies Ka/Ks values of ACP families in the six Brassica species. Table S6: The primer sequences of ACP genes used for qRT-PCR and subcellular localization.
Author Contributions
Conceptualization, H.W. and J.O.; Formal analysis, F.Q.; Investigation, T.Z., L.G. and X.D.; Methodology, F.Q. and D.Z.; Resources, B.Z.; Software, F.Q.; Validation, D.Z.; Visualization, F.Q.; Writing—original draft, D.Z.; Writing—review and editing, F.Q., H.W., B.Z. and J.O. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Natural Science Foundation of China (NSFC, Grant No. 32060463 and 32260460), Guizhou Provincial Science and Technology Plan Project (Qian Kehe Support [2022] key 026), and the Scientific and Technological Key Program of Guizhou province (No. Qiankehezhicheng [2022] Key 031).
Data Availability Statement
All the data generated or analyzed during this study are included in this published article and its Supplementary Information Files. The genome data of B. carinata were available from CoGe (https://genomevolution.org/coge/, accessed on 10 August 2023), and the five other Brassica genomes were obtained from the BRAD database (http://brassicadB.org/brad/, accessed on 10 August 2023). The transcriptome data of B. napus were downloaded from the B. napus multiomics information resource database (BnIR, https://yanglab.hzau.edu.cn/, accessed on 23 August 2023). The A. thaliana sequences in this article were downloaded from the TAIR database (http://www.arabidopsis.org, accessed on 14 August 2023). The seeds of the tetraploid B. napus (cv. Zhongshuang 11) were preserved by our laboratory.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Hannapel, D.J.; Ohlrogge, J.B. Regulation of acyl carrier protein messenger RNA levels during seed and leaf development. Plant Physiol. 1988, 86, 1174–1178. [Google Scholar] [CrossRef]
- Li, M.J.; Wang, X.J.; Su, L.; Bi, Y.P.; Wan, S.B. Characterization of five putative acyl carrier protein (ACP) isoforms from developing seeds of Arachis hypogaea L. Plant Mol. Biol. Rep. 2010, 28, 365–372. [Google Scholar] [CrossRef]
- Misra, A.; Surolia, N.; Surolia, A. Catalysis and mechanism of malonyl transferase activity in type II fatty acid biosynthesis acyl carrier proteins. Mol. BioSyst. 2009, 5, 651–659. [Google Scholar] [CrossRef]
- Nguyen, C.; Haushalter, R.W.; Lee, D.J.; Markwick, P.R.; Bruegger, J.; Caldara-Festin, G. Trapping the dynamic acyl carrier protein in fatty acid biosynthesis. Nature 2014, 505, 427–431. [Google Scholar] [CrossRef]
- Branen, J.K.; Chiou, T.J.; Engeseth, N.J. Overexpression of acyl carrier protein-1 alters fatty acid composition of leaf tissue in Arabidopsis. Plant Physiol. 2001, 127, 222–229. [Google Scholar] [CrossRef]
- Liu, X.; Hicks, W.M.; Silver, P.A.; Way, J.C. Engineering acyl carrier protein to enhance production of shortened fatty acids. Biotechnol. Biofuels 2016, 9, 1–7. [Google Scholar] [CrossRef]
- Branen, J.K.; Shintani, D.K.; Engeseth, N.J. Expression of antisense acyl carrier protein-4 reduces lipid content in Arabidopsis leaf tissue. Plant Physiol. 2003, 132, 748–756. [Google Scholar] [CrossRef]
- Song, P.; Allen, R.D. Identification of a cotton fiber-specific acyl carrier protein cDNA by differential display. Biochim. Biophys. Acta (BBA)-Gene Struct. Expr. 1997, 1351, 305–312. [Google Scholar] [CrossRef]
- Aluru, M.R.; Mazourek, M.; Landry, L.G.; Curry, J.; Jahn, M.; O’Connell, M.A. Differential expression of fatty acid synthase genes, Acl, Fat and Kas, in Capsicum fruit. J. Exp. Bot. 2003, 54, 1655–1664. [Google Scholar] [CrossRef] [PubMed]
- Silva, J.; Loader, N.M.; Jarman, C.; Windust, J.H.; Hughes, S.G.; Safford, R. The isolation and sequence analysis of two seed-expressed acyl carrier protein genes from Brassica napus. Plant Mol. Biol. 1990, 14, 537–548. [Google Scholar] [CrossRef]
- Suh, M.C.; Schultz, D.J.; Ohlrogge, J.B. Isoforms of acyl carrier protein involved in seed-specific fatty acid synthesis. Plant J. 1999, 17, 679–688. [Google Scholar] [CrossRef] [PubMed]
- Finking, R.; Marahiel, M.A. Biosynthesis of nonribosomal peptides. Ann. Rev. Microbiol. 2004, 58, 453–488. [Google Scholar] [CrossRef]
- Zhang, D.; Song, H.; Cheng, H.; Hao, D.; Wang, H.; Kan, G. The acid phosphatase-encoding gene GmACP1 contributes to soybean tolerance to low-phosphorus stress. PLoS Genet. 2014, 10, e1004061. [Google Scholar] [CrossRef]
- Huang, J.; Xue, C.; Wang, H.; Wang, L.; Schmidt, W.; Shen, R.; Lan, P. Genes of ACYL CARRIER PROTEIN family show different expression profiles and overexpression of ACYL CARRIER PROTEIN 5 modulates fatty acid composition and enhances salt stress tolerance in Arabidopsis. Front. Plant Sci. 2017, 8, 987. [Google Scholar] [CrossRef] [PubMed]
- Nazari, M.; Moosavi, S.S.; Maleki, M.; Jamshidi Goharrizi, K. Chloroplastic acyl carrier protein synthase I and chloroplastic 20 kDa chaperonin proteins are involved in wheat (Triticum aestivum) in response to moisture stress. J. Plant Interact. 2020, 15, 180–187. [Google Scholar] [CrossRef]
- Ge, H.; Xu, J.; Hua, M.; An, W.; Wu, J.; Wang, B. Genome-wide identification and analysis of ACP gene family in Sorghum bicolor (L.) Moench. BMC Genom. 2022, 23, 538. [Google Scholar] [CrossRef]
- Al-Shehbaz, I.A.; Beilstein, M.A.; Kellogg, E.A. Systematics and phylogeny of the Brassicaceae (Cruciferae): An overview. Plant Syst. Evol. 2006, 259, 89–120. [Google Scholar] [CrossRef]
- Warwick, S.I.; Francis, A.; Al-Shehbaz, I.A. Brassicaceae: Species checklist and database on CD-Rom. Plant Syst. Evol. 2006, 259, 249–258. [Google Scholar] [CrossRef]
- Nagaharu, U.; Nagaharu, N. Genome analysis in Brassica with special reference to the experimental formation of B. napus and peculiar mode of fertilization. Jpn. J. Bot. 1935, 7, 389–452. [Google Scholar]
- Wang, X.; Wang, H.; Wang, J.; Sun, R.; Wu, J. The genome of the mesopolyploid crop species Brassica rapa. Nat. Genet. 2011, 43, 1035–1039. [Google Scholar] [CrossRef]
- Chalhoub, B.; Denoeud, F.; Liu, S.; Parkin, I.A.; Tang, H.; Wang, X. Early allopolyploid evolution in the post-Neolithic Brassica napus oilseed genome. Science 2014, 345, 950–953. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Liu, Y.; Yang, X.; Tong, C.; Edwards, D.; Parkin, I.A. The Brassica oleracea genome reveals the asymmetrical evolution of polyploid genomes. Nat. Commun. 2014, 5, 3930. [Google Scholar] [CrossRef]
- Yang, J.; Liu, D.; Wang, X.; Ji, C.; Cheng, F.; Liu, B. The genome sequence of allopolyploid Brassica juncea and analysis of differential homoeolog gene expression influencing selection. Nat. Genet. 2016, 48, 1225–1232. [Google Scholar] [CrossRef] [PubMed]
- Perumal, S.; Koh, C.S.; Jin, L.; Buchwaldt, M.; Higgins, E.E.; Zheng, C. A high-contiguity Brassica nigra genome localizes active centromeres and defines the ancestral Brassica genome. Nat. Plants 2020, 6, 929–941. [Google Scholar] [CrossRef] [PubMed]
- Yim, W.C.; Swain, M.L.; Ma, D.; An, H.; Bird, K.A.; Curdie, D.D. The final piece of the Triangle of U: Evolution of the tetraploid Brassica carinata genome. Plant Cell 2022, 34, 4143–4172. [Google Scholar] [CrossRef]
- Qiao, X.; Li, Q.; Yin, H.; Qi, K.; Li, L.; Wang, R. Gene duplication and evolution in recurring polyploidization–diploidization cycles in plants. Genome Biol. 2019, 20, 1–23. [Google Scholar] [CrossRef]
- Hurst, L.D. The Ka/Ks ratio: Diagnosing the form of sequence evolution. Trends Genet. 2002, 18, 486–487. [Google Scholar] [CrossRef]
- Lu, K.; Wei, L.; Li, X.; Wang, Y.; Wu, J.; Liu, M. Whole-genome resequencing reveals Brassica napus origin and genetic loci involved in its improvement. Nat. Commun. 2019, 10, 1154. [Google Scholar] [CrossRef]
- Bus, A.; Körber, N.; Snowdon, R.J.; Stich, B. Patterns of molecular variation in a species-wide germplasm set of Brassica Napus. Theor. Appl. Genet. 2001, 123, 1413–1423. [Google Scholar] [CrossRef] [PubMed]
- Chan, D.I.; Vogel, H.J. Current understanding of fatty acid biosynthesis and the acyl carrier protein. Biochem. J. 2010, 430, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Murugan, E.; Kong, R.; Sun, H.; Rao, F.; Liang, Z.X. Expression, purification and characterization of the acyl carrier protein phosphodiesterase from Pseudomonas Aeruginosa. Protein Expres. Purif. 2010, 71, 132–138. [Google Scholar] [CrossRef]
- Bonaventure, G.; Ohlrogge, J.B. Differential regulation of mRNA levels of acyl carrier protein isoforms in Arabidopsis. Plant Physiol. 2002, 128, 223–235. [Google Scholar] [CrossRef]
- Meyer, E.H.; Heazlewood, J.L.; Millar, A.H. Mitochondrial acyl carrier proteins in Arabidopsis thaliana are predominantly soluble matrix proteins and none can be confirmed as subunits of respiratory Complex I. Plant Mol. Biol. 2007, 64, 319–327. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Tóth, K.; Tanaka, K.; Nguyen, C.T.; Yan, Z.; Brechenmacher, L. A soybean acyl carrier protein, GmACP, is important for root nodule symbiosis. Mol. Plant-Microbe Interact. 2014, 27, 415–423. [Google Scholar] [CrossRef] [PubMed]
- Town, C.D.; Cheung, F.; Maiti, R.; Crabtree, J.; Haas, B.J.; Wortman, J.R. Comparative genomics of Brassica oleracea and Arabidopsis thaliana reveal gene loss, fragmentation, and dispersal after polyploidy. Plant Cell 2006, 18, 1348–1359. [Google Scholar] [CrossRef]
- Yang, T.J.; Kim, J.S.; Kwon, S.J.; Lim, K.B.; Choi, B.S.; Kim, J.A. Sequence-level analysis of the diploidization process in the triplicated FLOWERING LOCUS C region of Brassica rapa. Plant Cell 2006, 18, 1339–1347. [Google Scholar] [CrossRef]
- Blanc, G.; Hokamp, K.; Wolfe, K.H. A recent polyploidy superimposed on older large-scale duplications in the Arabidopsis genome. Genome Res. 2003, 13, 137–144. [Google Scholar] [CrossRef] [PubMed]
- Saha, G.; Park, J.I.; Jung, H.J.; Ahmed, N.U.; Kayum, M.A.; Chung, M.Y. Genome-wide identification and characterization of MADS-box family genes related to organ development and stress resistance in Brassica rapa. BMC Genom. 2015, 16, 178. [Google Scholar] [CrossRef]
- Tang, J.; Wang, F.; Hou, X.L.; Wang, Z.; Huang, Z.N. Genome-wide fractionation and identification of WRKY transcription factors in Chinese cabbage (Brassica rapa ssp. pekinensis) reveals collinearity and their expression patterns under abiotic and biotic stresses. Plant Mol. Biol. Rep. 2014, 32, 781–795. [Google Scholar] [CrossRef]
- Chen, D.; Chen, H.; Dai, G.; Zhang, H.; Liu, Y.; Shen, W. Genome-wide identification of R2R3-MYB gene family and association with anthocyanin biosynthesis in Brassica species. BMC Genom. 2022, 23, 441. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, Q.; Zhao, Y.; Han, G.; Zhu, S. Systematic analysis of maize class III peroxidase gene family reveals a conserved subfamily involved in abiotic stress response. Gene 2015, 566, 95–108. [Google Scholar] [CrossRef]
- Cao, Y.; Han, Y.; Meng, D.; Li, D.; Jin, Q.; Lin, Y.; Cai, Y. Structural, evolutionary, and functional analysis of the class III peroxidase gene family in Chinese pear (Pyrus bretschneideri). Front. Plant Sci. 2016, 7, 1874. [Google Scholar] [CrossRef]
- Force, A.; Lynch, M.; Pickett, F.B.; Amores, A.; Yan, Y.L.; Postlethwait, J. Preservation of duplicate genes by complementary, degenerative mutations. Genetics 1999, 151, 1531–1545. [Google Scholar] [CrossRef]
- Smith, S. Architectural options for a fatty acid synthase. Science 2006, 311, 1251–1252. [Google Scholar] [CrossRef] [PubMed]
- Byers, D.M.; Gong, H. Acyl carrier protein: Structure–function relationships in a conserved multifunctional protein family. Biochem. Cell Biol. 2007, 85, 649–662. [Google Scholar] [CrossRef] [PubMed]
- Jha, J.K.; Sinha, S.; Maiti, M.K.; Basu, A.; Mukhopadhyay, U.K.; Sen, S.K. Functional expression of an acyl carrier protein (ACP) from Azospirillum brasilense alters fatty acid profiles in Escherichia coli and Brassica juncea. Plant Physiol. Biochem. 2007, 45, 490–500. [Google Scholar] [CrossRef]
- Yang, X.; Liu, X.; Zhou, Y.; Zhang, F.; Huang, L.; Wang, J. New insights on the function of plant acyl carrier proteins from comparative and evolutionary analysis. Genomics 2021, 113, 1155–1165. [Google Scholar] [CrossRef] [PubMed]
- Zhao, K.; Chen, S.; Yao, W.; Cheng, Z.; Zhou, B.; Jiang, T. Genome-wide analysis and expression profile of the bZIP gene family in poplar. BMC Plant Biol. 2021, 21, 122. [Google Scholar] [CrossRef]
- Li, X.; Zhang, X.; Shi, T.; Chen, M.; Jia, C.; Wang, J. Identification of ARF family in blueberry and its potential involvement of fruit development and pH stress response. BMC Genom. 2022, 23, 329. [Google Scholar] [CrossRef]
- Lang, L.; Xu, A.; Ding, J.; Zhang, Y.; Zhao, N.; Tian, Z.; Liu, Y. Quantitative trait locus mapping of salt tolerance and identification of salt-tolerant genes in Brassica napus L. Front. Plant Sci. 2017, 8, 1000. [Google Scholar] [CrossRef]
- Raees, N.; Ullah, S.; Nafees, M. Interactive effect of tocopherol, salicylic acid and ascorbic acid on agronomic characters of two genotypes of Brassica napus L. under induced drought and salinity stresses. Gesunde Pflanz. 2022, 75, 1905–1923. [Google Scholar] [CrossRef]
- Chen, H.; Wang, T.; He, X.; Cai, X.; Lin, R.; Liang, J. BRAD V3.0: An upgraded Brassicaceae database. Nucleic Acids Res. 2022, 50, D1432–D1441. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Wang, S.; Wei, L.; Huang, Y.; Liu, D.; Jia, Y. BnIR: A multi-omics database with various tools for Brassica napus research and breeding. Mol. Plant 2023, 16, 775–789. [Google Scholar] [CrossRef] [PubMed]
- Mistry, J.; Finn, R.D.; Eddy, S.R.; Bateman, A.; Punta, M. Challenges in homology search: HMMER3 and convergent evolution of coiled-coil regions. Nucleic Acids Res. 2013, 41, e121. [Google Scholar] [CrossRef] [PubMed]
- Chou, K.C.; Shen, H.B. Cell-PLoc: A package of Web servers for predicting subcellular localization of proteins in various organisms. Nat. Protoc. 2008, 3, 153–162. [Google Scholar] [CrossRef]
- Edgar, R.C. Muscle5: High-accuracy alignment ensembles enable unbiased assessments of sequence homology and phylogeny. Nat. Commun. 2022, 13, 6968. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive tree of life (iTOL) v3: An online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 2016, 44, W242–W245. [Google Scholar] [CrossRef]
- Bailey, T.L.; Johnson, J.; Grant, C.E.; Noble, W.S. The MEME suite. Nucleic Acids Res. 2015, 43, W39–W49. [Google Scholar] [CrossRef]
- Hu, B.; Jin, J.; Guo, A.Y.; Zhang, H.; Luo, J.; Gao, G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 2015, 31, 1296–1297. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant. 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef] [PubMed]
- Goll, J.; Rusch, D.B.; Tanenbaum, D.M.; Thiagarajan, M.; Li, K.; Methé, B.A.; Yooseph, S. METAREP: JCVI metagenomics reports—An open source tool for high-performance comparative metagenomics. Bioinformatics 2010, 26, 2631–2632. [Google Scholar] [CrossRef] [PubMed]
- Yan, G.; Lv, X.; Gao, G.; Li, F.; Li, J.; Qiao, J. Identification and characterization of a glyoxalase I gene in a rapeseed cultivar with seed thermotolerance. Front. Plant Sci. 2016, 7, 150. [Google Scholar] [CrossRef]
- Gan, C.; Liu, Z.; Pang, B.; Zuo, D.; Hou, Y.; Zhou, L. Integrative physiological and transcriptome analyses provide insights into the Cadmium (Cd) tolerance of a Cd accumulator: Erigeron canadensis. BMC Genom. 2022, 23, 778. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).