Abstract
Floral bud growth influences seed yield and quality; however, the molecular mechanism underlying the development of floral buds in alfalfa (Medicago sativa) is still unclear. Here, we comprehensively analyzed the transcriptome and targeted metabolome across the early, mid, and late bud developmental stages (D1, D2, and D3) in alfalfa. The metabolomic results revealed that gibberellin (GA), auxin (IAA), cytokinin (CK), and jasmonic acid (JA) might play an essential role in the developmental stages of floral bud in alfalfa. Moreover, we identified some key genes associated with GA, IAA, CK, and JA biosynthesis, including CPS, KS, GA20ox, GA3ox, GA2ox, YUCCA6, amid, ALDH, IPT, CYP735A, LOX, AOC, OPR, MFP2, and JMT. Additionally, many candidate genes were detected in the GA, IAA, CK, and JA signaling pathways, including GID1, DELLA, TF, AUX1, AUX/IAA, ARF, GH3, SAUR, AHP, B-ARR, A-ARR, JAR1, JAZ, and MYC2. Furthermore, some TFs related to flower growth were screened in three groups, such as AP2/ERF-ERF, MYB, MADS-M-type, bHLH, NAC, WRKY, HSF, and LFY. The findings of this study revealed the potential mechanism of floral bud differentiation and development in alfalfa and established a theoretical foundation for improving the seed yield of alfalfa.
1. Introduction
The development of floral buds encompasses the initiation of plant reproductive growth, and they directly affect the yield and quality of seeds [1,2]. Researchers usually classify the stages of flower bud development by observing the morphological changes during flower bud growth [3,4]. Additionally, studying the dynamic changes in the physiological indexes and gene expression in each stage of flower bud growth can provide a theoretical basis for uncovering the potential mechanism of flower organ formation, improving the seed yield [5,6].
Alfalfa (Medicago sativa) is a perennial leguminous plant cultivated worldwide [7]. It is the most important forage in stockbreeding, and it has good quality, high yield, and strong resistance to stress [8,9]. However, low seed yields have limited the long-term development of the alfalfa industry. Alfalfa inflorescences are typical racemes with small flowers growing on the peduncle. Previous studies have revealed that the floret count determines the number of seeds, and the number of seeds per inflorescence is positively correlated with the seed yield [10,11]. Moreover, the floral bud growth affects the quantity and quality of florets and seeds in plants [12]. However, a few investigations have been performed on the inherent mechanisms of floral bud development in alfalfa.
The external environment and internal factors commonly influence the development of floral buds [13,14,15]. Endogenous phytohormones play an important role in regulating flower bud growth [16,17]. Various endogenous hormones, such as auxins (IAA), cytokinin (CK), abscisic acid (ABA), jasmonate (Ja), gibberellin (GA), and ethylene (ETH), coordinate with each other, thereby regulating flower bud development by jointly controlling the metabolism of plants. Yan et al. (2019) found that a higher IAA content was beneficial to the differentiation of licorice flower buds, eventually improving the seed yield of licorice [18]. Before floral bud growth, increasing the levels of ABA and GA3 in stem apical meristems may promote flower induction and floral bud growth [19]. Fang et al. (2018) explored the effect of growth regulators on cotton floral buds and concluded that the ratio of trans-Zeatin-riboside (ZR)/IAA and GA3/IAA was significantly elevated in the seeds after pretreatment with GA3 and N6-benzyladenine (6-BA), resulting in an increase in the number of floral buds [20]. During the process of grape floral bud differentiation, GAs inhibited the primordium differentiation of grape inflorescences, and CTK could promote grape flowering [21,22]. Understanding the dynamic change in endogenous phytohormones in the developmental stages of floral buds can establish a theoretical foundation for conducting cultivation management and improving the seed yield of alfalfa.
In recent years, with the development of high-throughput sequencing, omics technology has become an important method to explore the potential mechanism of floral bud growth. For example, Qu et al. demonstrated that GA3 played a crucial role in regulating the floral bud development of Cyclocarya paliurus based on transcriptome analysis [23]. Moreover, Xie et al. analyzed the gene expression and identified candidate genes associated with female and male floral bud development in Carya illinoensis by using RNA sequencing [24]. In this work, we investigated the gene expression and phytohormone accumulation in three developmental stages of floral bud development in alfalfa, which provided a theoretical basis for molecular breeding.
2. Results
2.1. Transcriptome Analysis
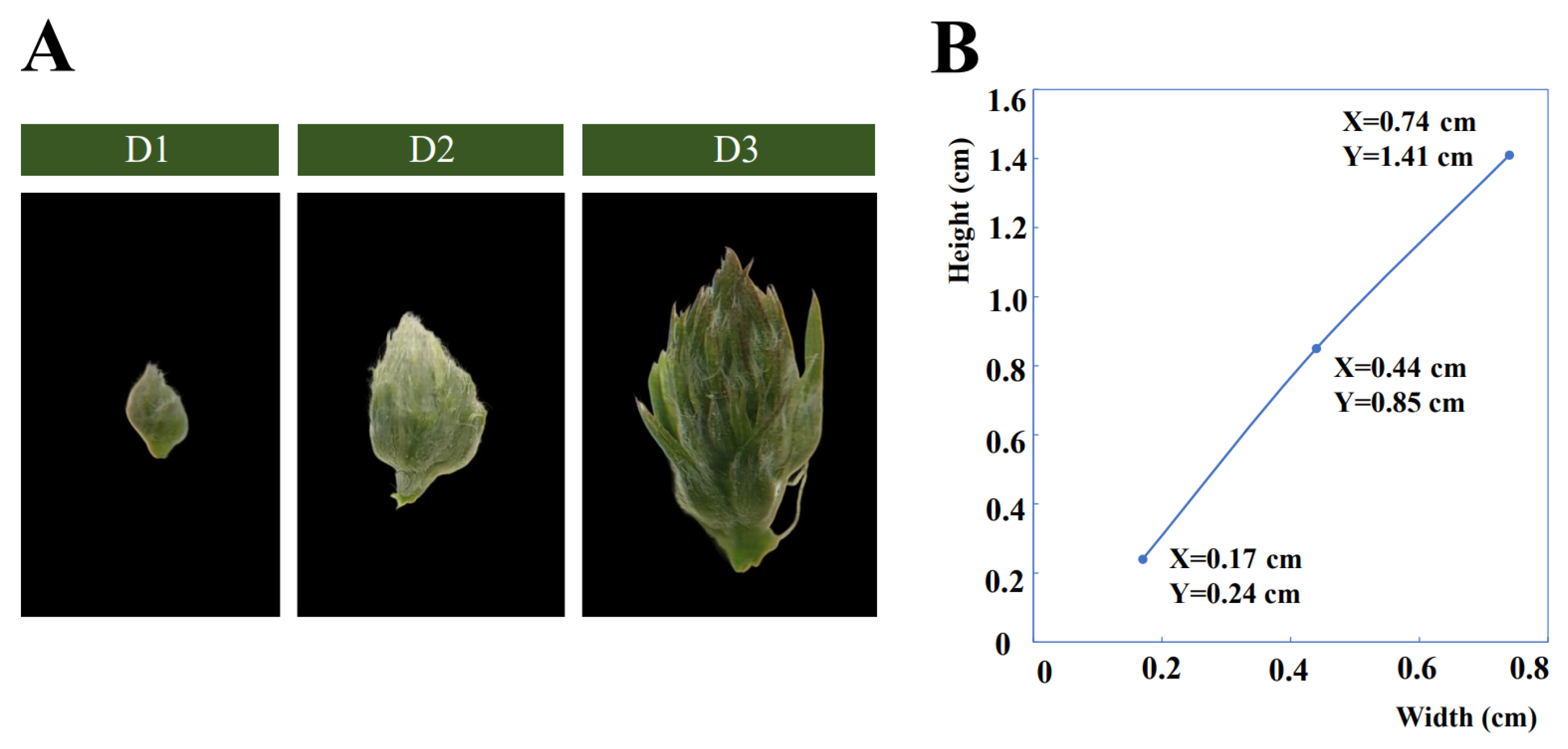
By observing the morphological changes in floral bud development of alfalfa, floral bud growth is divided into three developmental stages (D1, D2, and D3) (Figure 1A,B). At the early bud stage (D1), the floral buds are detectable as small swellings, surrounded by the leaf primordium. At the mid bud stage (D2), the floral bud is larger and easier to detect. Small swellings differentiate into multiple florets along with floral buds enlarging, and many white trichomes grow on the floret primordium. At the late bud stage (D3), each floret primordium becomes mature along with the bud expanding and lengthening rapidly. After the late bud stage, the florets will gradually bloom. These three stages are the most typical stages in the developmental process of floral buds.
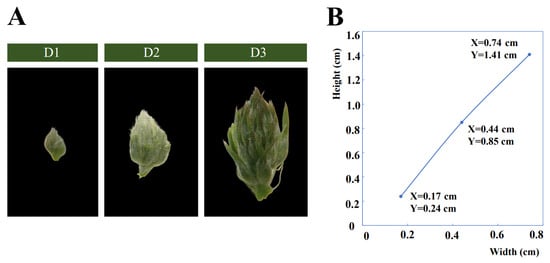
Figure 1.
(A) The phenotypes of the three developmental stages (D1, D2, and D3) of floral buds in alfalfa. (B) Width and height of D1, D2, and D3; X and Y represent values of width and height of floral bud, respectively.
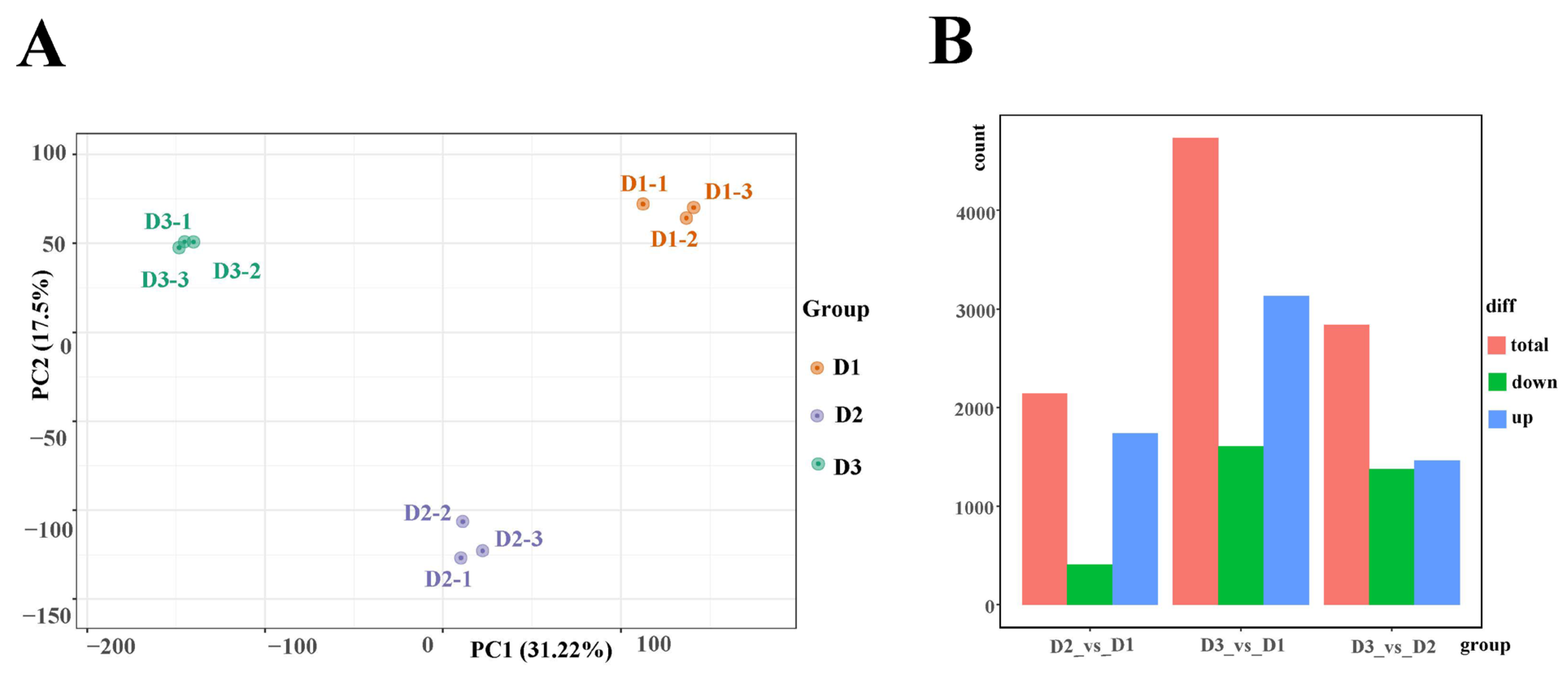
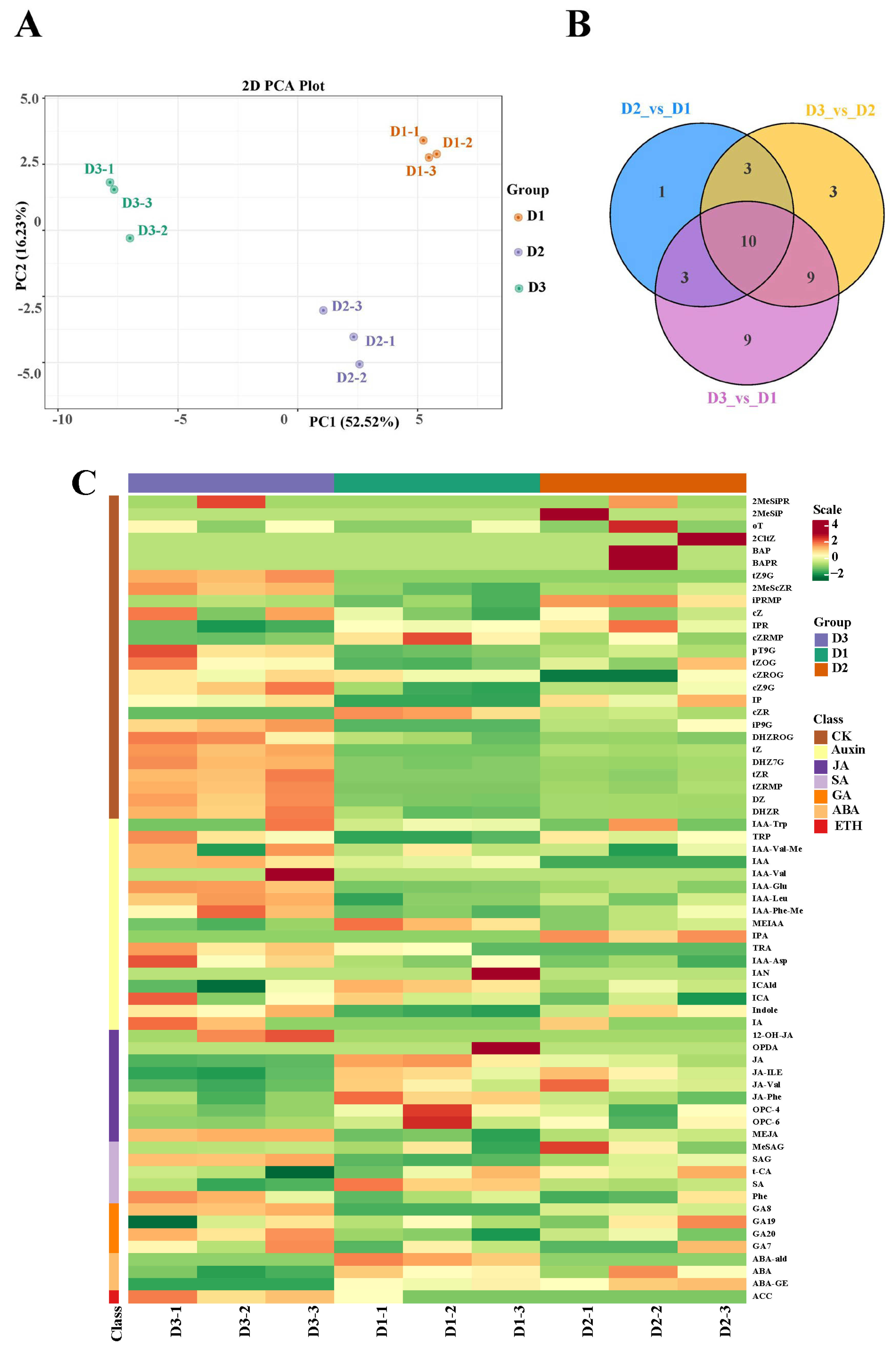
Floral buds at the three stages (D1, D2, and D3) were utilized in transcriptome sequencing to elucidate the potential mechanism of floral bud development in alfalfa. As a result, a total of 59.21 Gb clean data were identified, and the clean data of each sample reached 5 Gb. Moreover, the Q30 base percentage was above 94%, and more than 80% of the clean reads were mapped to the Medicago sativa reference genome (Table S1). Principal component analysis (PCA) suggested that the three samples were obviously separated (Figure 2A). These results indicated that the transcriptome datasets were reliable and accurate for further investigation. Furthermore, a total of 1736 up-regulated and 407 down-regulated genes, 3131 up-regulated and 1604 down-regulated genes, and 1465 up-regulated and 1373 down-regulated genes were identified in the D2 vs. D1, D3 vs. D1, and D3 vs. D2 comparisons, respectively (Figure 2B). In addition, nine genes were selected to identify the reliability of the transcriptome datasets. The results showed that the FPKM value of most genes had a similar tendency to the relative expression levels in the three groups, revealing that the transcriptome data in this work can be trusted for further investigation (Figure S1).
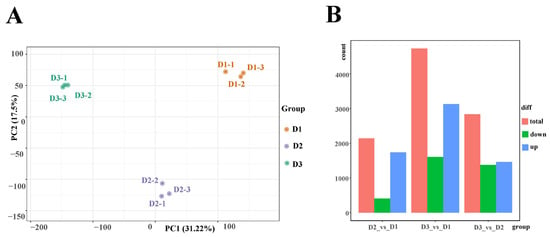
Figure 2.
(A) PCA plot analysis of transcriptome; the x-axis and y-axis represent principal component 1 (PC1) and principal component 2 (PC2), respectively. (B) Statistics of differentially expressed genes in the D2 vs. D1, D3 vs. D1, and D3 vs. D2 comparison.
2.2. GO and KEGG Enrichment Analysis
Subsequently, we conducted a GO and KEGG enrichment analysis of differentially expressed genes (DEGs) in the D2 vs. D1 and D3 vs. D2 comparisons. In the D2 vs. D1 comparison, the GO enrichment results revealed that most DEGs were mainly enriched in biological processes, cellular components, and molecular functions, including the anatomical structure formation involved in morphogenesis (GO:0048646), secondary metabolite biosynthetic process (GO:0044550), plant-type cell wall organization or biogenesis (GO:0071669), integral component of plasma membrane (GO:0005887), glucosyltransferase activity (GO:0046527), inorganic anion transmembrane transporter activity (GO:0015103), and some flower-development-related terms (Figure S2). In the D3 vs. D2 comparison, most DEGs were mainly enriched in biological processes and molecular functions, including anatomical structure formation involved in morphogenesis (GO:0048646), secondary metabolite biosynthetic process (GO:0044550), cell wall biogenesis (GO:0042546), glucosyltransferase activity (GO:0046527), UDP-glucosyltransferase activity (GO:0035251), dioxygenase activity (GO:0045543), and some flower-development-related terms (Figure S2).
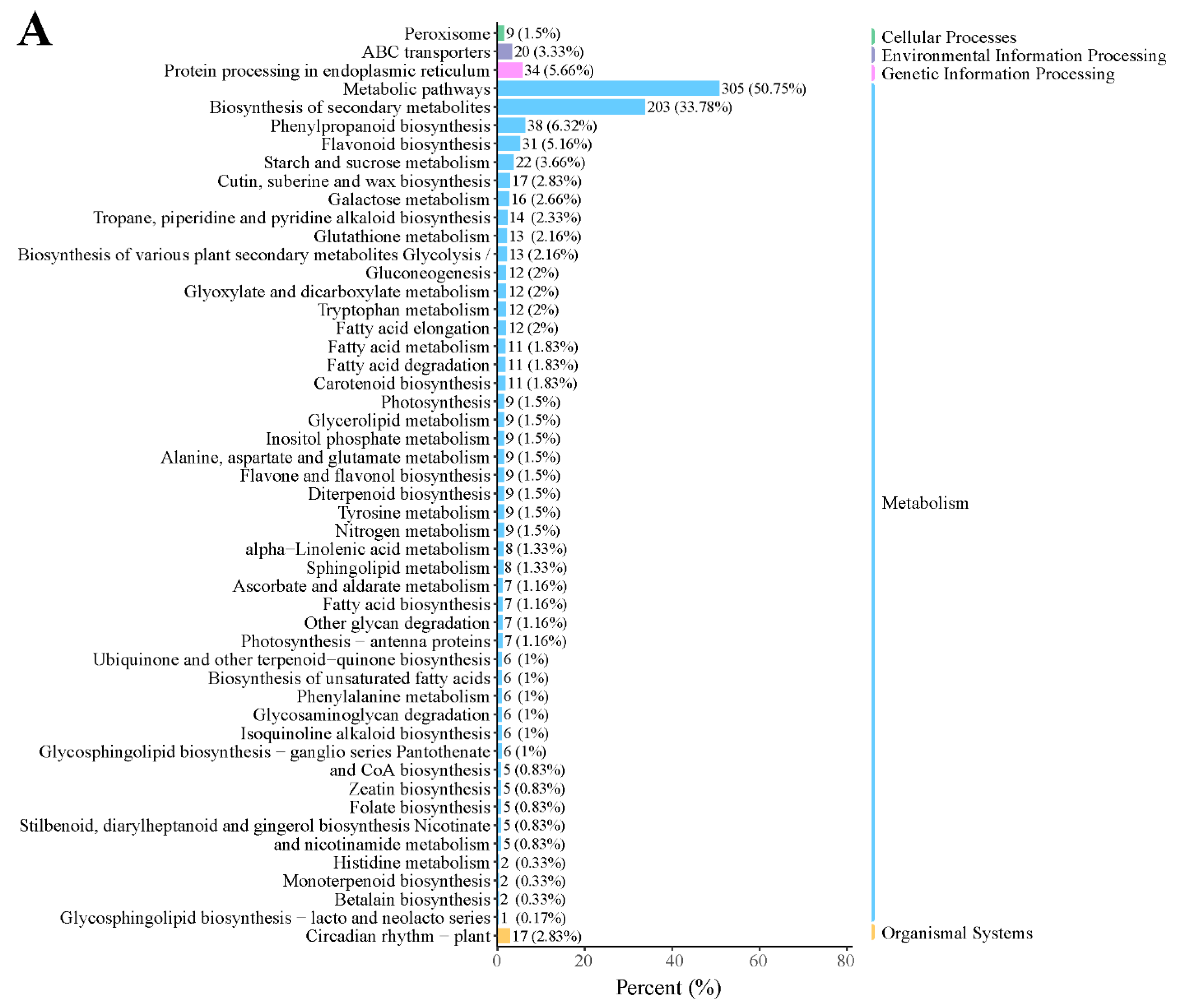
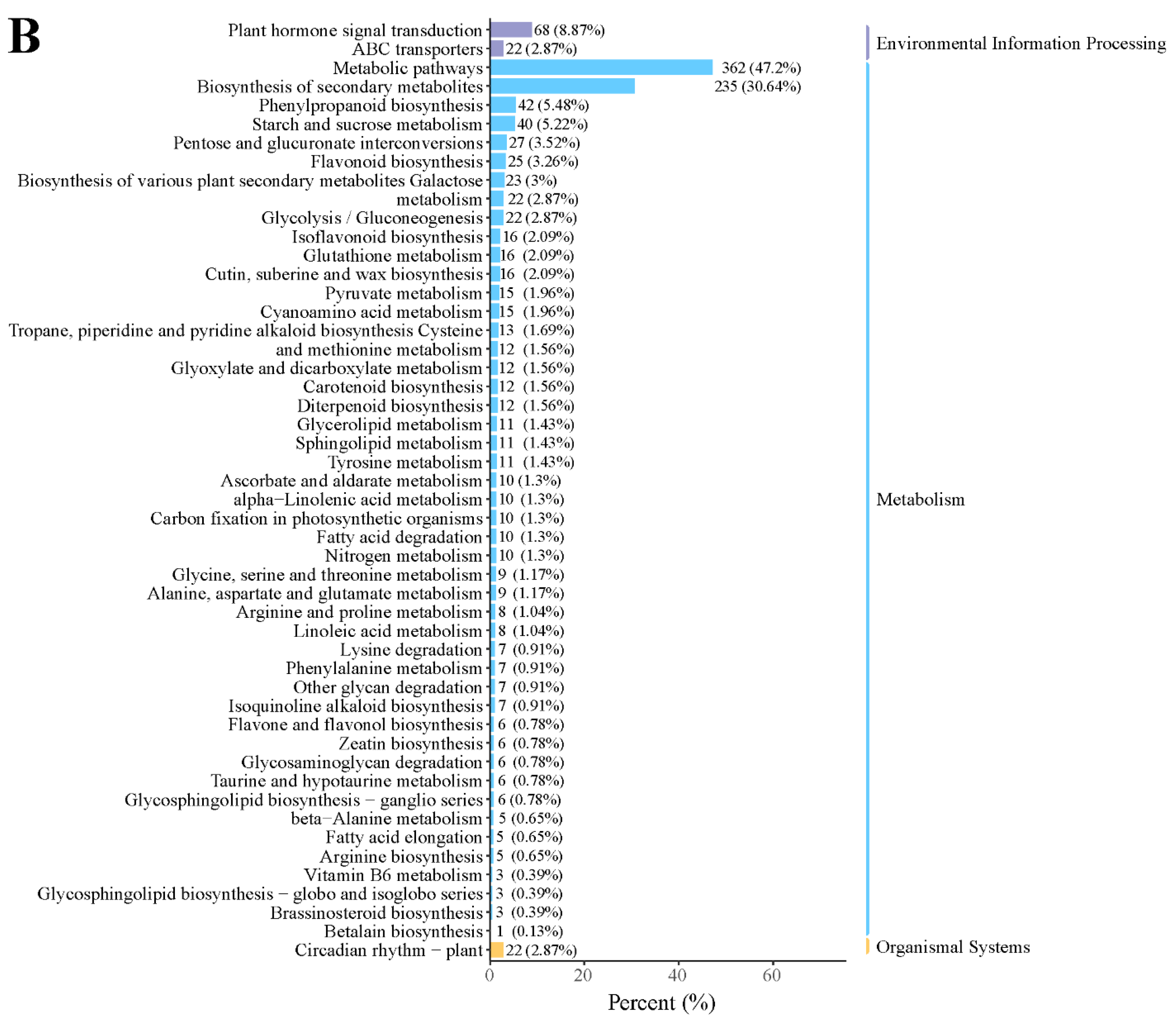
In addition, KEGG enrichment showed that many genes were enriched in metabolic pathways, biosynthesis of secondary metabolites, phenylpropanoid biosynthesis, protein processing in the endoplasmic reticulum, and flavonoid biosynthesis in the D2 vs. D1 comparison (Figure 3A). In the D3 vs. D2 comparison, most DEGs were enriched in metabolic pathways, biosynthesis of secondary metabolites, plant hormone signal transduction, and phenylpropanoid biosynthesis (Figure 3B). These results suggested that these pathways might play a critical role in floral bud development.
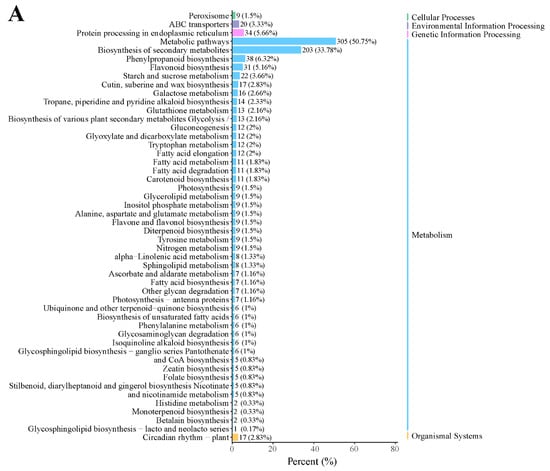

Figure 3.
KEGG classification in the (A) D2 vs. D1 and (B) D3 vs. D2 comparison.
2.3. Metabolomic Analysis
From the KEGG enrichment results, we discovered that many DEGs were enriched in phytohormone-biosynthesis-related pathways, such as tryptophan metabolism (ko00380), carotenoid biosynthesis (ko00906), diterpenoid biosynthesis (ko00904), alpha-Linolenic acid metabolism (ko00592), zeatin biosynthesis (ko00908), and cysteine and methionine metabolism (ko00270) (Figure 3A,B). These results indicated that phytohormones are closely connected to the floral bud development.
To detect the content of phytohormones in floral buds at the three stages (D1, D2, and D3), a targeted phytohormone metabolome analysis was conducted. The PCA results showed that the three groups had a good separation, suggesting that the experimental results were reliable for further study (Figure 4A). Subsequently, a total of 17, 31, and 25 differentially accumulated metabolites (DAMs) were identified in the D2 vs. D1, D3 vs. D1, and D3 vs. D2 comparisons, respectively (Figure 4B). Moreover, seven categories of plant hormones were differentially accumulated in the three groups, including cytokinin (CK), auxin (IAA), jasmonic acid (JA), salicylic acid (SA), gibberellin (GA), abscisic acid (ABA), and ethylene (ETH) (Figure 4C). These results suggested that these phytohormones might participate in the floral bud development.
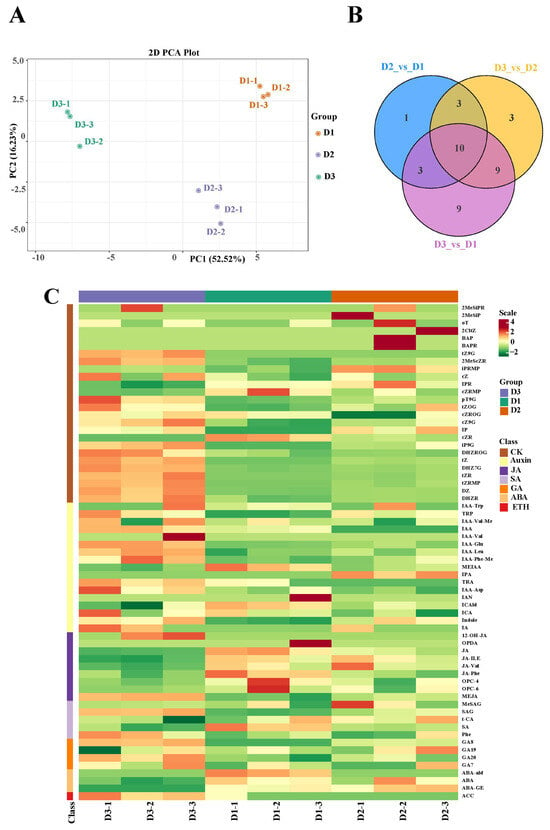
Figure 4.
(A) PCA plot analysis of metabolome; the x-axis and y-axis represent principal component 1 (PC1) and principal component 2 (PC2), respectively. (B) Venn map of the three comparisons. (C) Differentially accumulated phytohormones in the three groups.
2.4. Quantitative Analysis of Differentially Accumulated Phytohormones
Furthermore, we performed a quantitative analysis of DAMs in the D2 vs. D1 and D3 vs. D2 comparisons to investigate the dynamic changes in seven phytohormones in the developmental process of floral bud (Table 1 and Table 2). For GA, GA8 (Log2 fold-change value = Inf; Log2 fold-change value = 1.07) was up-accumulated in the D2 vs. D1 and D3 vs. D2 comparisons, and the content of GA7 (Log2 fold-change value = 1.13) was higher in the D3 group compared to the D2 group (Table 1 and Table 2).

Table 1.
Differentially accumulated phytohormones in the D2 vs. D1 comparison.

Table 2.
Differentially accumulated phytohormones in the D3 vs. D2 comparison.
For auxin, one up-accumulated (IPA, Log2 fold-change value = Inf) and two down-accumulated (IAA, Log2 fold-change value = −Inf; TRA, Log2 fold-change value = −Inf) auxins were detected in the D2 vs. D1 comparison. Additionally, IAA (Log2 fold-change value = Inf), IAA-Glu (Log2 fold-change value = 1.98), TRA (Log2 fold-change value = Inf), IAA-Asp (Log2 fold-change value = 1.05), and IA (Log2 fold-change value = 1.33) were up-accumulated in the D3 vs. D2 comparisons, and IPA did not accumulate in the D3 group (Table 1 and Table 2).
Furthermore, we found that nine up-accumulated and two down-accumulated CKs, and ten up-accumulated and two down-accumulated CKs, were detected in the D2 vs. D1 and D3 vs. D2 comparisons, respectively. Notably, we found that pT9G (Log2 fold-change value = 1.06; Log2 fold-change value = 1.23), iP9G (Log2 fold-change value = Inf; Log2 fold-change value = 1.20), tZ (Log2 fold-change value = Inf; Log2 fold-change value = 2.40), DHZ7G (Log2 fold-change value = 1.13; Log2 fold-change value = 2.59), tZR (Log2 fold-change value = Inf; Log2 fold-change value = 3.77), and tZRMP (Log2 fold-change value = Inf; Log2 fold-change value = 3.12) were significantly up-accumulated in the D2 vs. D1 and D3 vs. D2 comparisons (Table 1 and Table 2).
For JA, MEJA (Log2 fold-change value = 1.19; Log2 fold-change value = 1.04) was up-accumulated in the D2 vs. D1 and D3 vs. D2 comparisons, and 12-OH-JA (Log2 fold-change value = Inf) maintained a high accumulation in the D3 group compared to the D2 group. For ETH, we found that ACC (Log2 fold-change value = Inf) was up-accumulated in the D3 vs. D2 comparison. For ABA, ABA-ald (Log2 fold-change value = −Inf) and ABA-GE (Log2 fold-change value = −Inf) were down-accumulated in the D2 vs. D1 and D3 vs. D2 comparisons, respectively. For SA, t-CA (Log2 fold-change value = −1.21) was down-accumulated in the D3 vs. D2 comparison (Table 1 and Table 2).
Based on the results above, we concluded that the content of one GA (GA8), six CKs (pT9G, iP9G, tZ, DHZ7G, tZR, and tZRMP), and one JA (MEJA) continuously increased with development, and some auxins were differentially accumulated in the D3 vs. D2 comparison. Therefore, we speculated that GA8, pT9G, iP9G, tZ, DHZ7G, tZR, and tZRMP, and MEJA might be involved in floral bud growth, and auxins played a vital role in the mid and late bud stages.
2.5. Key Genes Involved in GA, IAA, CK, and JA Biosynthesis Pathways
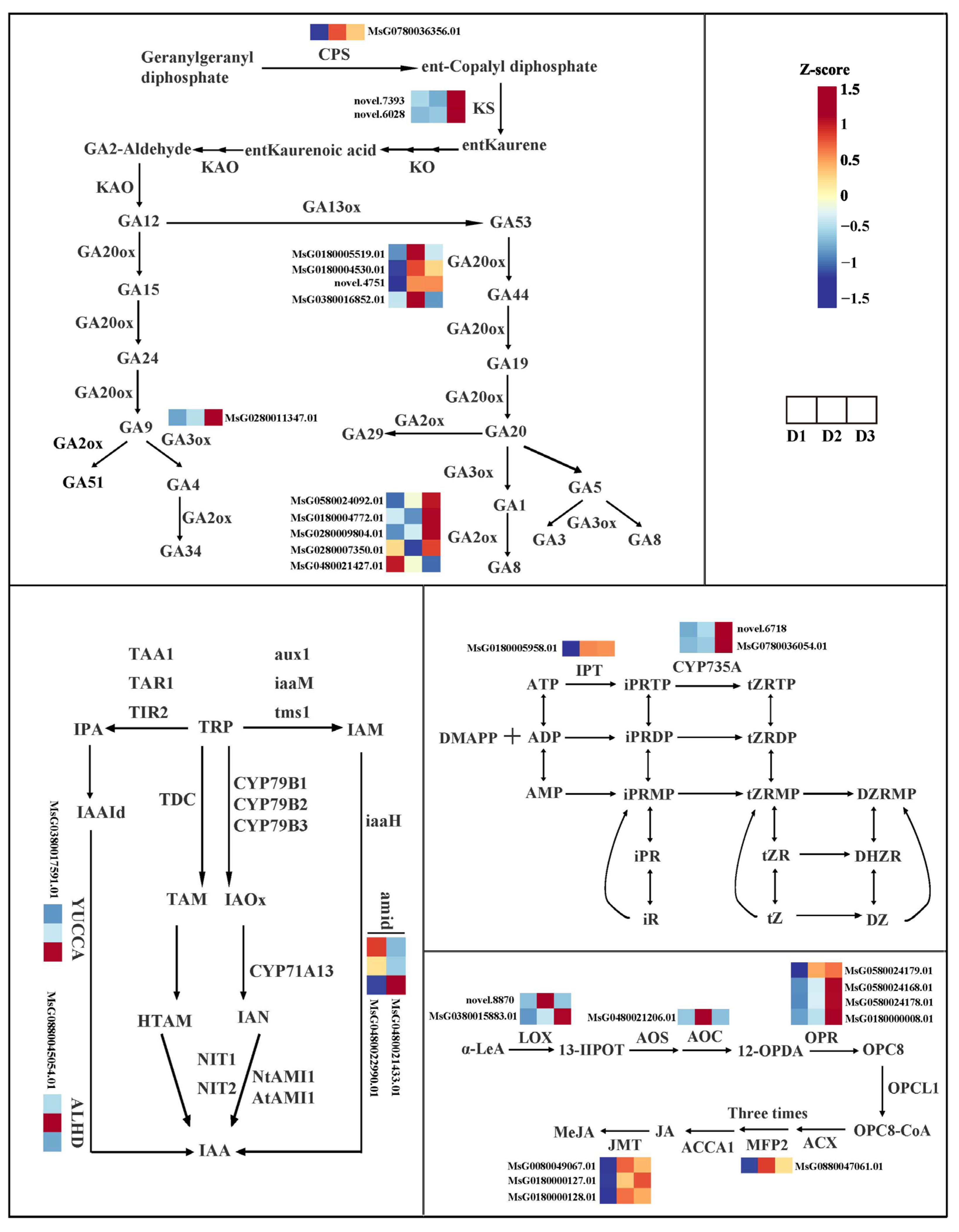
To screen the candidate genes involved in crucial phytohormone synthesis, the GA, IAA, CK, and JA biosynthesis pathways were examined, respectively (Figure 5). The FPKM value of DEGs in each group is shown in Table S2. In GA biosynthesis, we identified that one CPS (MsG0780036356.01), three GA20ox (MsG0180005519.01; MsG0180004530.01; novel.4751), and five GA2ox (MsG0780038278.01; MsG0880046136.01; MsG0580024092.01; MsG0280010379.01; MsG0280007031.01) genes were differentially expressed in the D2 vs. D1 comparison, and two KS (novel.7393, novel.6028), two GA20ox (MsG0180005519.01; MsG0380016852.01), GA3ox (MsG0280011347.01), and five GA2ox (MsG0580024092.01; MsG0180004772.01; MsG0280009804.01; MsG0280007350.01; MsG0480021427.01) genes were differentially expressed in the D3 vs. D2 comparison.

Figure 5.
Gene expression of GA, IAA, CK, and JA biosynthesis pathways.
In IAA biosynthesis, one YUCCA6 (MsG0380017591.01), two amid (MsG0480022990.01; MsG0480021433.01), and one ALDH (MsG0880045054.01) genes were differentially expressed in the D3 vs. D2 comparison.
In the CK biosynthesis, one IPT (MsG0180005958.01) and two CYP735A (novel.6718; MsG0780036054.01) genes were significantly up-regulated in the D2 vs. D1 and D3 vs. D2 comparisons, respectively.
In JA biosynthesis, one LOX (novel.8870), one AOC (MsG0480021206.01), two OPR (MsG0580024179.01; MsG0580024168.01), one MFP2 (MsG0880047061.01), and three JMT (MsG0080049067.01; MsG0180000127.01; MsG0180000128.01) genes were differentially expressed in the D2 vs. D1 comparison, and two LOX (MsG0380015883.01; novel.8870), one AOC (MsG0480021206.01), and three OPR (MsG0580024168.01; MsG0580024178.01; MsG0180000008.01) genes were differentially expressed in the D3 vs. D2 comparison. These DEGs might be involved in GA, IAA, CK, and JA accumulation in floral bud development.
2.6. Key Genes Involved in GA, IAA, CK, and JA Signaling Pathways
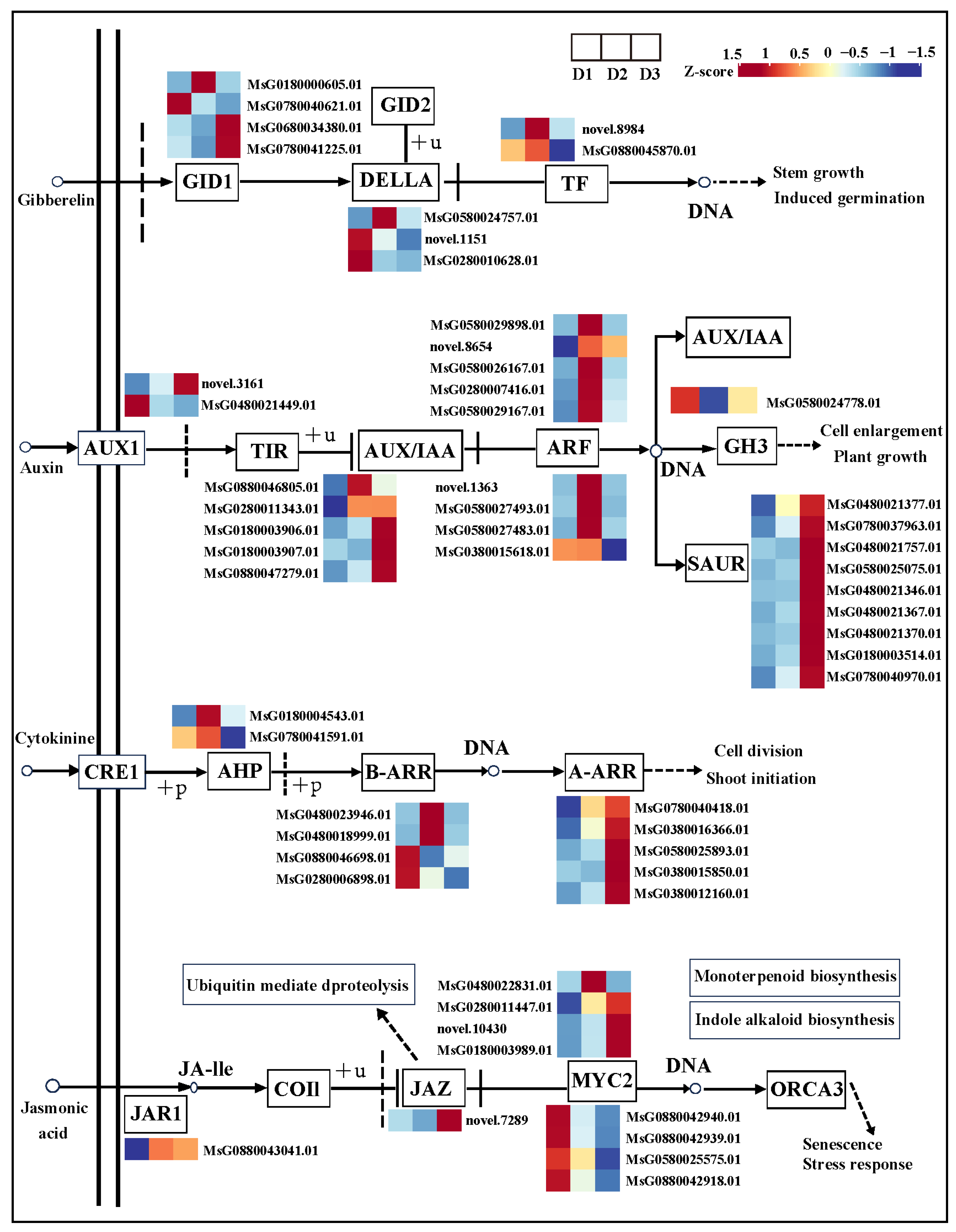
Simultaneously, some DEGs were identified in the GA, IAA, CK, and JA signaling pathways in the D2 vs. D1 and D3 vs. D2 comparisons (Figure 6). The FPKM value of DEGs in each group is shown in Table S3. In the GA signaling pathway, four GID1 (MsG0180000605.01; MsG0780040621.01; MsG0680034380.01; MsG0780041225.01), three DELLA (MsG0580024757.01; novel.1151; MsG0280010628.01), and two TF (novel.8984; MsG0880045870.01) genes were differentially expressed in the D2 vs. D1 and D3 vs. D2 comparisons.
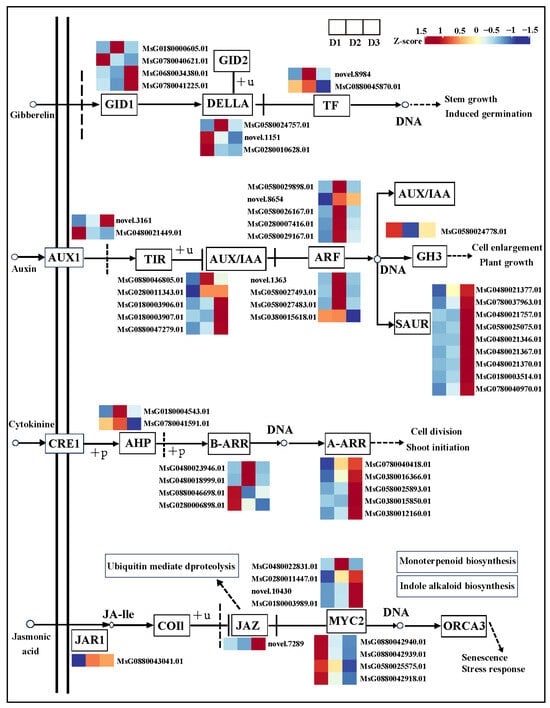
Figure 6.
Gene expression of GA, IAA, CK, and JA signaling pathways.
In the auxin signaling pathway, two AUX1 (novel.3161; MsG0480021449.01), five AUX/IAA (MsG0880046805.01; MsG0280011343.01; MsG0180003906.01; MsG0180003907.01; MsG0880047279.01), nine ARF (MsG0580029898.01; novel.8654; MsG0580026167.01; MsG0280007416.01; MsG0580029167.01; novel.1363; MsG0580027493.01; MsG0580027483.01; MsG0380015618.01), one GH3 (MsG0580024778.01), and nine SAUR (MsG0480021377.01; MsG0780037963.01; MsG0480021757.01; MsG0580025075.01; MsG0480021346.01; MsG0480021367.01; MsG0480021370.01; MsG0180003514.01; MsG0780040970.01) genes were differentially expressed in the D2 vs. D1 and D3 vs. D2 comparisons.
In the CK signaling pathway, two AHP (MsG0180004543.01; MsG0780041591.01), four B-ARR (MsG0480023946.01; MsG0480018999.01; MsG0880046698.01; MsG0280006898.01), and five A-ARR (MsG0780040418.01; MsG0380016366.01; MsG0580025893.01; MsG0380015850.01; MsG0380012160.01) genes were differentially expressed in the D2 vs. D1 and D3 vs. D2 comparisons.
In the JA signaling pathway, one JAR1 (MsG0880043041.01), one JAZ (novel.7289), and eight WYC2 (MsG0480022831.01; MsG0280011447.01; novel.10430; MsG0180003989.01; MsG0880042940.01; MsG0880042939.01; MsG0580025575.01; MsG0880042918.01) genes were differentially expressed in the D2 vs. D1 and D3 vs. D2 comparisons.
2.7. Transcription Factor Analysis
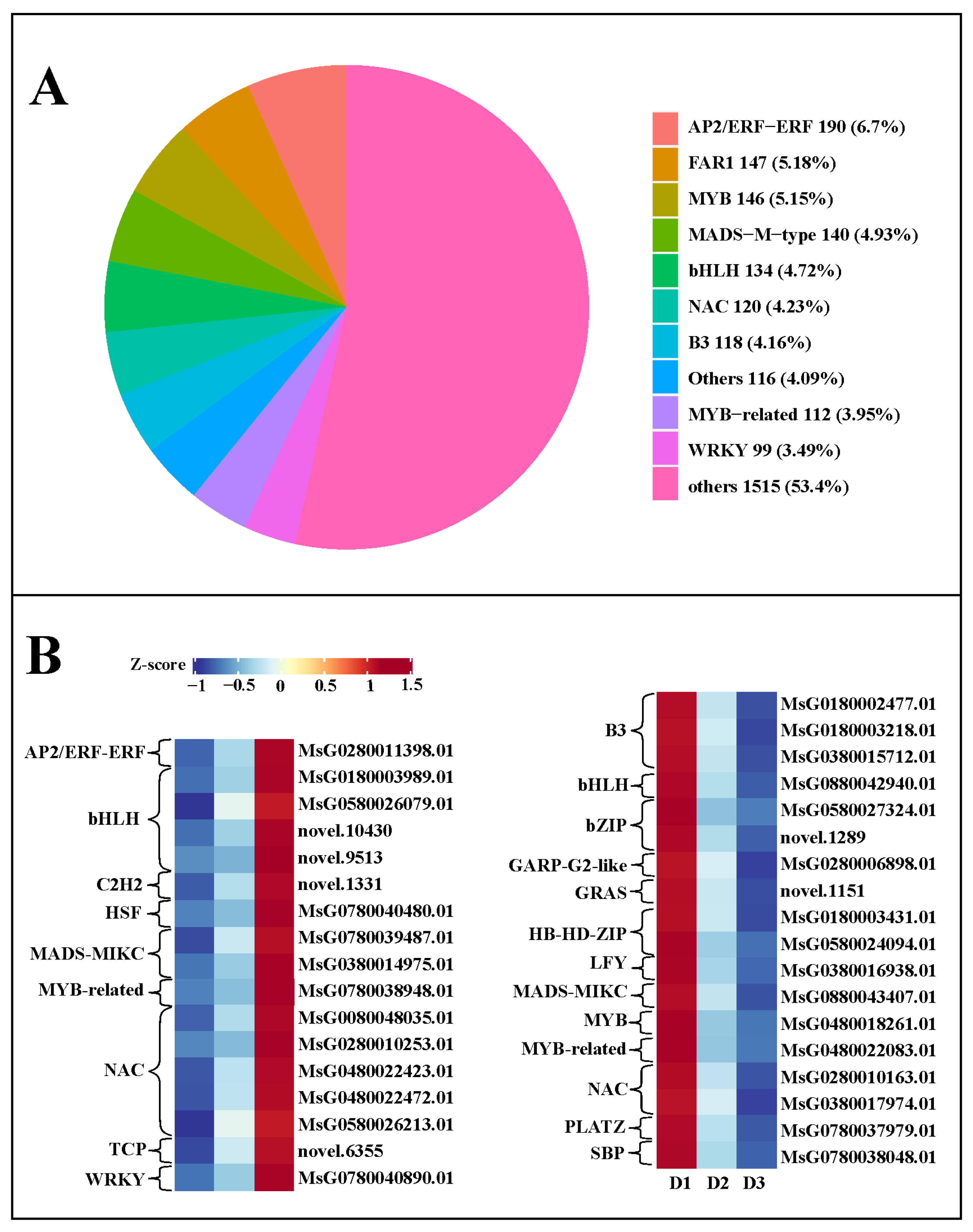
In this work, we identified a total of 2837 TFs (transcription factors) in the three groups, including 190 AP2/ERF-ERF (6.7%), 147 FAR1 (5.18%), 146 MYB (5.15%), 140 MADS-M-type (4.93%), 134 bHLH (4.72%), 120 NAC (4.23%), and 118 B3 (4.16%) families (Figure 7A), suggesting these TF families might play an essential role in regulating floral bud growth. Moreover, a total of 156 and 172 differentially expressed TFs were detected in the D2 vs. D1 and D3 vs. D2 comparisons, respectively. Among these differentially expressed TFs, 17 TFs with continuously increased expression levels were screened in the three developmental stages, including 1 AP2/ERF-ERF, 4 bHLH, 1 C2H2, 1 HSF, 2 MADS-MIKC, 1 7 1 TCP, and 1 WRKY (Figure 7B). Simultaneously, 18 TFs with persistently decreased expression levels were identified in the three developmental stages, including 3 B3, 1 bHLH, 2 bZIP, 1 GARP-G2-like, 1 GRAS, 2 HB-HD-ZIP, 1 LFY,1 MADS-MIKC, 1 MYB, 1 MYB-related, 2 NAC, 1 PLATZ, and 1 SBP (Figure 7B). The FPKM value of these differentially expressed TFs in each group is shown in Table S4. These TFs might be closely correlated with the floral bud development.
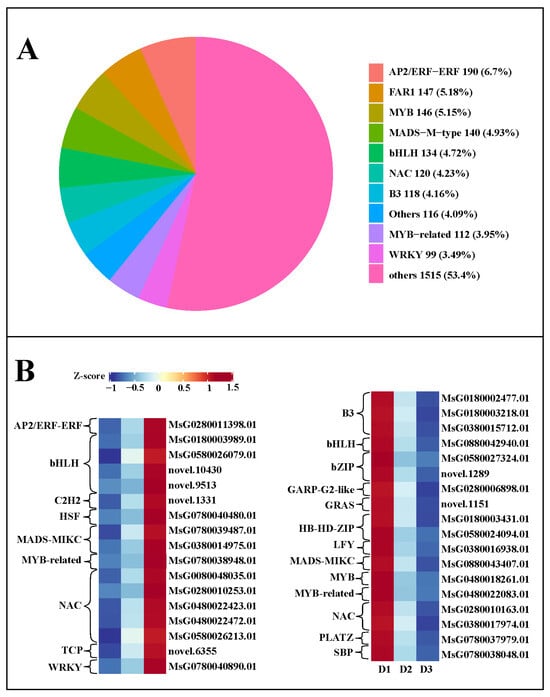
Figure 7.
(A) Transcription factors in the three groups. (B) TFs with continuously increased and decreased expression levels in the developmental process of floral buds.
3. Discussion
The development of floral buds determines the seed yield. To explore the gene expression patterns and phytohormone accumulation of the floral bud differentiation and development in alfalfa, we performed a comparative transcriptome and metabolome analysis for the three developmental stages of floral buds.
Plant hormones are the key regulatory factors during plant morphogenesis [16,17]. Spraying growth regulators on a plant is a conventional cultivation method to increase the crop yield during the flower bud developmental stage; therefore, exploring the accumulation of endogenous phytohormones in floral buds can provide a theoretical support for guiding the production and improving the seed yield. Cytokinin plays a key role in reproductive growth [25]. Gibberellin can affect flower bud differentiation by regulating the expression of downstream genes [26]. Auxin is a crucial plant hormone involved in the development of flower organs [27]. It is involved in regulating the gibberellin signaling pathway and eventually promoting flower formation [28]. Jasmonic acid can participate in floral organ development via promoting SlMYB21 expression in tomatoes [29]. In agreement with our findings, we found that the content of one GA (GA8), six CKs (pT9G, iP9G, tZ, DHZ7G, tZR, and tZRMP), and one JA (MEJA) continuously increased in the floral bud developmental process, and auxins were significantly differentially expressed in the D2 vs. D1 and D3 vs. D2 comparisons. These results indicated that these phytohormones might play a vital role in regulating the downstream genes related to flower organ development. Notably, previous studies have shown that auxin polar transport is closely related to plant morphological construction [30,31]; therefore, identifying the accumulation in specific cells of auxin will be important for exploring the mechanism of auxins involved in floral bud development in the future.
Moreover, some candidate genes related to GA, IAA, CK, and JA biosynthesis were identified based on the integrated transcriptome and metabolome results. CPS and KS are upstream enzymes of GA biosynthesis, catalyzing geranylgeranyl diphosphate to ent-Kaurene. The inhibition of CPS and KS enzymes influenced the GA biosynthesis and limited plant organ growth [32,33]. A previous study reported that the high expression levels of the SoGA20ox1 gene in shoot tips enhanced GA biosynthesis [34]. Additionally, overexpression of the GA2ox gene facilitated pollen growth in transgenic Arabidopsis plants [35]. In this work, we detected that the expression of one CPS, two KS, four GA20ox1, and five GA2ox genes maintained a high level in the D2 and D3 groups, suggesting that these genes might play an essential role in GA8 accumulation. Moreover, Liu et al. found FvYUCCCA6 played a critical role in vegetative and reproductive development in woodland strawberry [36]. Consistent with our results, one YUCCA6 gene maintained high expression levels in the D3 group compared to the D1 and D2 groups, and it might be involved in the IAA accumulation in the D3 group. IPT was the first enzyme participating in CK biosynthesis, catalyzing ATP, ADT, and AMP to iPRTP, iPRDP, and iPRMP, respectively [37,38]. Then, CYP735 converted iP-nucleotide to tZ-nucleotide [39]. In the present work, the expression levels of one IPT and two CYP735 genes were high in the D2 and D3 groups, suggesting that these genes may be closely related to CK accumulation in the floral bud developmental process. In addition, we identified that two LOX, one AOC, four OPR, one MFP2, and three JMT genes were highly expressed in the D2 and D3 groups. Previous studies have demonstrated that these genes have vital functions in JA biosynthesis [40,41]. An enhancement or inhibition of gene expression in these genes directly affects JA synthesis, eventually changing the plant physiological activities [42,43,44,45,46]. Therefore, these DEGs related to JA biosynthesis might play an essential role in regulating the floral bud growth.
The phytohormone signaling pathway was associated with plant organ development [47]. In accordance with our study, many DEGs were enriched in the plant hormone signal transduction pathway. The findings of a previous study revealed that GID1 and TF positively promote flower bud differentiation [48]. Consistent with our results, several GID1 and TF genes in the D2 or D3 group were highly expressed in the GA signaling pathway. Additionally, most studies demonstrated that the DELLA protein inhibits plant development and growth by binding to transcription factors [49,50,51]. In this work, the expression level of most DELLA genes was low in the D3 group, suggesting that the DELLA protein played a crucial role in participating in floral bud differentiation. In the auxin signaling pathway, the AUX1, TIR, AUX/IAA, ARF, GH3, and SAUR genes played a pivotal role in regulating plant growth [52]. In the present work, most differentially expressed AUX1, AUX/IAA, ARF, and SAUR genes in the auxin signaling pathway maintained high expression levels in the D2 and D3 groups compared to the D1 group, revealing that the auxin signaling pathway may participate in regulating floral bud development. A-ARR was a key gene involved in the CK signaling pathway and affected flower organ development and growth [53,54]. In this work, the expression level of four A-ARR genes was significantly higher in the D3 group than in the D1 and D2 groups, suggesting A-ARR genes might play a crucial role in regulating floral bud development in the D3 group. In the JA signaling pathway, JAR1 can induce the JA converted to the biologically active JA-Ile by responding to environmental stress [40], and the JAZ protein family usually regulates JA responses by interacting with the MYC family [55]. As we are currently aware, one JAR1, one JAZ, and eight MYC genes were differentially expressed in the three developmental stages, and these genes might be positively involved in floral bud development.
In this work, many AP2/ERF-ERF, MYB, MADS-M-type, bHLH, NAC, and WRKY genes were identified in the three developmental stages, which was consistent with previous studies [56,57]. AP2/ERF-ERF transcription factor family proteins are mainly involved in the development of sepals and petals in reproductive organs [58]. The overexpression of RcAP2 results in the transformation of stamens into petals, thereby increasing the number of petals in Arabidopsis, while silencing RcAP2 decreases the number of petals [59]. MYB transcription factors play a role in the flowering time, pollen development, flower color, and sex differentiation of flower organs [60]. During the developmental stage of Arabidopsis reproductive organs, AtMYB125 and AMYB98 are involved in the development of male and female gametes, respectively [61,62]. Most studies indicated that the MADS family is an important factor regulating flowering time and flower organ development in plants [63,64,65]. bHLH proteins CIB1, CIB2, CIB4, and CIB5 commonly regulate flowering initiation, which facilitates FT transcription by binding to the FT promoter in plants [66]. Additionally, AtWRKY75 may be a new member regulating flowering in the GA signaling pathway, and WRKY71 induces early flowering by activating FT and LFY in Arabidopsis [67,68]. The NAC transcription factor family can regulate flower growth by participating in the JA and GA signaling pathways [69,70]. Simultaneously, we also discovered that the expression level of some TFs, such as AP2/ERF-ERF, MYB, MADS-M-type, bHLH, NAC, WRKY, HSF, and LFY, is continuously increased or decreased in the three developmental stages, suggesting that these TFs might play a pivotal role in regulating floral bud development.
4. Materials and Methods
4.1. Plant Materials
Alfalfa seedlings were planted at the Institute of Grassland Research, Chinese Academy of Agricultural Sciences, Hohhot (40°58′ N, 111°78′ E). The samples of the three developmental stages were collected in the early flowering period. We observed the monological traits of floral buds and identified the developmental stages of floral buds by using a dissecting microscope (Dian Ying, Shanghai, China). The floral buds of the three developmental stages were named D1, D2, and D3. Three biological replicates were obtained for the three samples, and the mass of each biological replicate was more than 3 g. All samples were stored in liquid nitrogen.
4.2. Transcriptome Sequencing and Data Analysis
By utilizing ethanol precipitation and CTAB-PBIOZOL, the total RNA of floral buds (D1, D2, and D3) was obtained. Total RNA was analyzed by utilizing a Qubit fluorescence quantifier and a Qsep400 high-throughput biofragment analyzer (AUTO Q BIOSCIENCES, San Diego, CA, USA). Subsequently, all cDNA libraries were sequenced on the Illumina platform. After reads with adapters were removed by using fastp software (fastp v0.19.4), all non-redundant transcripts were mapped with Medicago sativa reference genome (https://figshare.com/articles/dataset/Medicago_sativa_genome_and_annotation_files/12623960 (accessed on 1 November 2023)). Novel genes were screened by using StringTie. FPKM (Fragments Per Kilobase Million) values were calculated in accordance with the gene length. Differentially expressed genes (DEGs) were identified between comparisons by using DESeq2. p-values, and log2 fold changes were set as criteria for obvious differential expression. In accordance with the hypergeometric test, with pathway-based hypergeometric distribution checking for Kyoto Encyclopedia of Genes and Genomes (KEGG) and Gene Ontology (GO) term-based profiles for GO, enrichment analysis was conducted. We thank Wuhan Metware Biotechnology Co., Ltd. (Wuhan, China) for assistance with sequencing.
4.3. Phytohormone Analysis
Endogenous auxin, cytokinin (CK), abscisic acid (ABA), jasmonate (Ja), salicylic acid (SA), gibberellin (Ga), ethylene (ETH), strigolactone (SL), and melatonin (MLT) were quantified by using LC–MS/MS. Firstly, the samples (15 mg) were dissolved in 1 mL of methanol/water/formic acid (15:4:1, v/v/v) and frozen in liquid nitrogen. Then, 10 mL of the internal standard mixed solution (100 ng/mL) was added to the extract as internal standards (IS) for further quantifying. After the liquid was vortexed for 10 min and centrifugated (12,000 r/min, 5 min, and 4 °C), the supernatant was poured into microtubes. Subsequently, the supernatant was evaporated, dissolved in 100 μL 80% methanol, and filtered for further analyses. The UPLC and ESI-MS/MS conditions were introduced by Niu et al. [71]. The detected metabolites were annotated based on the KEGG compound database (http://www.kegg.jp/kegg/compound/ (accessed on 10 November 2023)).
4.4. qRT-PCR Analysis
We extracted the total RNA by using the RNA pure plant kit (Tb Green® Premix Ex Taq™ II (TAKARA, Beijing, China)). Then, we obtained the first strand of the reverse-transcribed cDNA by using the specifications of the Monad first-strand cDNA Synthesis Kit. The primers were designed by utilizing PRIMER-BLAST (Table S5). The ABI7500 quantitative PCR instrument was adopted to perform real-time fluorescence quantitative PCR. The gene (MsG0180001288.01) was selected as an actin gene for high and stable levels of expression in nine samples according to the FPKM value, and each group was identified from three repetitions.
5. Conclusions
In this work, we elucidated the molecular mechanism of floral bud development in alfalfa based on the phenotypic, metabolome, and transcriptome in three stages (D1, D2, D3). The transcriptome results revealed that the phytohormone biosynthesis and signaling pathway were closely associated with floral bud growth. The metabolomic results indicated that GA, IAA, CK, and JA were the critical phytohormones involved in floral bud differentiation and development. Notably, many key genes participating in phytohormone biosynthesis and signaling pathways might play a crucial role in regulating floral bud growth. Finally, we uncovered that many TF family members were closely correlated with floral bud development, such as AP2/ERF-ERF, MYB, MADS-M-type, bHLH, NAC, and WRKY. This work established regulatory networks related to phytohormones regulating floral bud development, providing potential leads for the molecular breeding of alfalfa.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/plants13081078/s1, Table S1: Data filtering and comparison of reference tables; Table S2: FPKM of candidate genes related to phytohormones biosynthesis in each group; Table S3: FPKM of candidate genes related to phytohormones signaling pathway in each group; Table S4: FPKM of TFs with continuously increased or decreased expression levels in each group; Figure S1: primers used in qRT-PCR in this work (Table S5); verification of nine DEGs by qRT-PCR; Figure S2: GO enrichment analysis in the D2 vs. D1 and D3 vs. D2 comparison.
Author Contributions
X.H. is the first author. L.L. and X.Q. are the corresponding authors. X.H. conceived and designed the experiments. X.H. performed the experiments. X.H. analyzed the data and wrote the original manuscript. L.L., X.Q., Y.M., Z.L. and F.H. revised and approved the final version of the paper. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the projects from the Supplementary list and its assessment of the rare and endangered plant species in Inner Mongolia (Grant 2021MS03074); the secure conservation of forage germplasm resources in north of China (Grant 19230874); Study on Special Characteristics in Various Flower Color in Alfalfa (Medicago L.) (Grant 31402122).
Data Availability Statement
All data are open and available. The raw data are available in the NCBI (BioProject ID PRJNA1049519) (https://www.ncbi.nlm.nih.gov/sra/ (accessed on 6 December 2023)).
Acknowledgments
The authors are grateful to Linqing Yu from Inner Mongolia University for his suggestion for this article.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Ramírez, F.; Kallarackal, J. Phenological Growth Stages of Feijoa [Acca sellowiana (O. Berg) Burret] According to the BBCH Scale under Tropical Andean Conditions. Sci. Hortic. 2018, 232, 184–190. [Google Scholar] [CrossRef]
- Ramírez, F.; Kallarackal, J. Feijoa [Acca sellowiana (O. Berg) Burret] Pollination: A Review. Sci. Hortic. 2017, 226, 333–341. [Google Scholar] [CrossRef]
- Kozik, E.U.; Nowak, R.; Nowakowska, M.; Dyki, B. Level of Sterility and Morphological Flowers Differentiation of Petaloid Male-Sterile Plants of Carrot. J. Agric. Sci. 2012, 4, 187. [Google Scholar] [CrossRef]
- Lin, B.; Wang, D.; Li, Z.; Liu, J.; Xu, W.; Li, X.; Huang, J. Morphological Development and Nutritional Metabolism during Floral Bud Differentiation in Acca sellowiana (Feijoa). Int. J. Fruit Sci. 2023, 23, 102–115. [Google Scholar] [CrossRef]
- Pei, L.; Gao, Y.; Feng, L.; Zhang, Z.; Liu, N.; Yang, B.; Zhao, N. Phenolic Acids and Flavonoids Play Important Roles in Flower Bud Differentiation in Mikania Micrantha: Transcriptomics and Metabolomics. Int. J. Mol. Sci. 2023, 24, 16550. [Google Scholar] [CrossRef] [PubMed]
- Shang, C.; Cao, X.; Tian, T.; Hou, Q.; Wen, Z.; Qiao, G.; Wen, X. Cross-Talk between Transcriptome Analysis and Dynamic Changes of Carbohydrates Identifies Stage-Specific Genes during the Flower Bud Differentiation Process of Chinese Cherry (Prunus pseudocerasus L.). Int. J. Mol. Sci. 2022, 23, 15562. [Google Scholar] [CrossRef] [PubMed]
- Nagl, N.; Taski-Ajdukovic, K.; Barac, G.; Baburski, A.; Seccareccia, I.; Milic, D.; Katic, S. Estimation of the Genetic Diversity in Tetraploid Alfalfa Populations Based on RAPD Markers for Breeding Purposes. Int. J. Mol. Sci. 2011, 12, 5449–5460. [Google Scholar] [CrossRef] [PubMed]
- Atumo, T.T.; Kauffman, R.; Talore, D.G.; Abera, M.; Tesfaye, T.; Tunkala, B.Z.; Zeleke, M.; Kalsa, G.K. Adaptability, Forage Yield and Nutritional Quality of Alfalfa (Medicago sativa) Genotypes. Sustain. Environ. 2021, 7, 1895475. [Google Scholar] [CrossRef]
- Julier, B.; Huyghe, C.; Ecalle, C. Within- and Among-Cultivar Genetic Variation in Alfalfa: Forage Quality, Morphology, and Yield. Crop Sci. 2000, 40, 365–369. [Google Scholar] [CrossRef]
- Bolanos-Aguilar, E.D.; Huyghe, C.; Djukic, D.; Julier, B.; Ecalle, C. Genetic Control of Alfalfa Seed Yield and Its Components. Plant Breed. 2001, 120, 67–72. [Google Scholar] [CrossRef]
- Abadouz, G.; Hasanzadeh Gorttapeh, A.; Rahnema, A.A.; Behradfar, A. Effect of Row Spacing and Seeding Rate on Yield Component and Seed Yield of Alfalfa (Medicago sativa L.). Not. Sci. Biol. 2010, 2, 74–80. [Google Scholar] [CrossRef]
- Zik, M.; Irish, V.F. Flower Development: Initiation, Differentiation, and Diversification. Annu. Rev. Cell Dev. Biol. 2003, 19, 119–140. [Google Scholar] [CrossRef] [PubMed]
- Hidaka, K.; Dan, K.; Imamura, H.; Takayama, T. Crown-Cooling Treatment Induces Earlier Flower Bud Differentiation of Strawberry under High Air Temperatures. Environ. Control. Biol. 2017, 55, 21–27. [Google Scholar] [CrossRef][Green Version]
- Liao, Y.; Suzuki, K.; Yu, W.; Zhuang, D.; Takai, Y.; Ogasawara, R.; Shimazu, T.; Fukui, H. Night Break Effect of LED Light with Different Wavelengths on Floral Bud Differentiation of Chrysanthemum Morifolium Ramat ‘Jimba’ and Iwa No Hakusen. Environ. Control. Biol. 2014, 52, 45–50. [Google Scholar] [CrossRef]
- Fan, L.; Chen, M.; Dong, B.; Wang, N.; Yu, Q.; Wang, X.; Xuan, L.; Wang, Y.; Zhang, S.; Shen, Y. Transcriptomic Analysis of Flower Bud Differentiation in Magnolia Sinostellata. Genes 2018, 9, 212. [Google Scholar] [CrossRef] [PubMed]
- Koshita, Y.; Takahara, T.; Ogata, T.; Goto, A. Involvement of Endogenous Plant Hormones (IAA, ABA, GAs) in Leaves and Flower Bud Formation of Satsuma Mandarin (Citrus unshiu Marc.). Sci. Hortic. 1999, 79, 185–194. [Google Scholar] [CrossRef]
- Milyaev, A.; Kofler, J.; Moya, Y.A.T.; Lempe, J.; Stefanelli, D.; Hanke, M.-V.; Flachowsky, H.; Von Wirén, N.; Wünsche, J.-N. Profiling of Phytohormones in Apple Fruit and Buds Regarding Their Role as Potential Regulators of Flower Bud Formation. Tree Physiol. 2022, 42, 2319–2335. [Google Scholar] [CrossRef] [PubMed]
- Yan, B.; Hou, J.; Cui, J.; He, C.; Li, W.; Chen, X.; Li, M.; Wang, W. The Effects of Endogenous Hormones on the Flowering and Fruiting of Glycyrrhiza Uralensis. Plants 2019, 8, 519. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.J.; Li, A.; Liu, X.Q.; Sun, J.X.; Guo, W.J.; Zhang, J.W.; Lyu, Y.M. Changes in the Morphology of the Bud Meristem and the Levels of Endogenous Hormones after Low Temperature Treatment of Different Phalaenopsis Cultivars. S. Afr. J. Bot. 2019, 125, 499–504. [Google Scholar] [CrossRef]
- Fang, S.; Gao, K.; Hu, W.; Snider, J.L.; Wang, S.; Chen, B.; Zhou, Z. Chemical Priming of Seed Alters Cotton Floral Bud Differentiation by Inducing Changes in Hormones, Metabolites and Gene Expression. Plant Physiol. Biochem. 2018, 130, 633–640. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Yuan, M.; Dang, S.; Zhou, J.; Zhang, Y. Comparative Transcriptomic Analysis of Transcription Factors and Hormones during Flower Bud Differentiation in ‘Red Globe’ Grape under Red–blue Light. Sci. Rep. 2023, 13, 8932. [Google Scholar] [CrossRef]
- Vasconcelos, M.C.; Greven, M.; Winefield, C.S.; Trought, M.C.T.; Raw, V. The Flowering Process of Vitis vinifera: A Review. Am. J. Enol. Vitic. 2009, 60, 411–434. [Google Scholar] [CrossRef]
- Qu, Y.; Chen, X.; Mao, X.; Huang, P.; Fu, X. Transcriptome Analysis Reveals the Role of GA3 in Regulating the Asynchronism of Floral Bud Differentiation and Development in Heterodichogamous Cyclocarya paliurus (Batal.) Iljinskaja. Int. J. Mol. Sci. 2022, 23, 6763. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.; Hou, Z.; Shi, M.; Wang, Q.; Yang, Z.; Lim, K.-J.; Wang, Z. Transcriptional Regulation of Female and Male Flower Bud Initiation and Development in Pecan (Carya illinoensis). Plants 2023, 12, 1378. [Google Scholar] [CrossRef] [PubMed]
- Bartrina, I.; Otto, E.; Strnad, M.; Werner, T.; Schmülling, T. Cytokinin Regulates the Activity of Reproductive Meristems, Flower Organ Size, Ovule Formation, and Thus Seed Yield in Arabidopsis thaliana. Plant Cell 2011, 23, 69–80. [Google Scholar] [CrossRef]
- Proveniers, M. Sugars Speed up the Circle of Life. eLife 2013, 2, e00625. [Google Scholar] [CrossRef]
- Kućko, A.; Wilmowicz, E.; Ostrowski, M. Spatio-Temporal IAA Gradient Is Determined by Interactions with ET and Governs Flower Abscission. J. Plant Physiol. 2019, 236, 51–60. [Google Scholar] [CrossRef]
- Deng, W.; Ying, H.; Helliwell, C.A.; Taylor, J.M.; Peacock, W.J.; Dennis, E.S. FLOWERING LOCUS C (FLC) Regulates Development Pathways throughout the Life Cycle of Arabidopsis. Proc. Natl. Acad. Sci. USA 2011, 108, 6680–6685. [Google Scholar] [CrossRef]
- Niwa, T.; Suzuki, T.; Takebayashi, Y.; Ishiguro, R.; Higashiyama, T.; Sakakibara, H.; Ishiguro, S. Jasmonic Acid Facilitates Flower Opening and Floral Organ Development through the Upregulated Expression of SlMYB21 Transcription Factor in Tomato. Biosci. Biotechnol. Biochem. 2018, 82, 292–303. [Google Scholar] [CrossRef]
- Křeček, P.; Skůpa, P.; Libus, J.; Naramoto, S.; Tejos, R.; Friml, J.; Zažímalová, E. The PIN-FORMED (PIN) Protein Family of Auxin Transporters. Genome Biol. 2009, 10, 249. [Google Scholar] [CrossRef] [PubMed]
- Friml, J.; Wiśniewska, J.; Benková, E.; Mendgen, K.; Palme, K. Lateral Relocation of Auxin Efflux Regulator PIN3 Mediates Tropism in Arabidopsis. Nature 2002, 415, 806–809. [Google Scholar] [CrossRef] [PubMed]
- Sponsel, V.M.; Hedden, P. B2. Gibberellin Biosynthesis and Inactivation. In Plant Hormones; Springer: Dordrecht, The Netherlands, 2010. [Google Scholar]
- Bensen, R.J.; Johal, G.S.; Crane, V.C.; Tossberg, J.T.; Schnable, P.S.; Meeley, R.B.; Briggs, S.P. Cloning and Characterization of the Maize Anl Gene. Plant Cell 1995, 7, 75–84. [Google Scholar] [PubMed]
- Lee, D.J.; Zeevaart, J.A.D. Differential Regulation of RNA Levels of Gibberellin Dioxygenases by Photoperiod in Spinach. Plant Physiol. 2002, 130, 2085–2094. [Google Scholar] [CrossRef] [PubMed]
- Singh, D.P.; Jermakow, A.M.; Swain, S.M. Gibberellins Are Required for Seed Development and Pollen Tube Growth in Arabidopsis. Plant Cell 2002, 14, 3133–3147. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Xie, W.; Zhang, L.; Valpuesta, V.; Ye, Z.; Gao, Q.; Duan, K. Auxin Biosynthesis by the YUCCA6 Flavin Monooxygenase Gene in Woodland Strawberry. J. Integr. Plant Biol. 2014, 56, 350–363. [Google Scholar] [CrossRef] [PubMed]
- Sakamoto, T.; Sakakibara, H.; Kojima, M.; Yamamoto, Y.; Nagasaki, H.; Inukai, Y.; Sato, Y.; Matsuoka, M. Ectopic Expression of KNOTTED1-Like Homeobox Protein Induces Expression of Cytokinin Biosynthesis Genes in Rice. Plant Physiol. 2006, 142, 54–62. [Google Scholar] [CrossRef] [PubMed]
- Kakimoto, T. Identification of Plant Cytokinin Biosynthetic Enzymes as Dimethylallyl Diphosphate:ATP/ADP Isopentenyltransferases. Plant Cell Physiol. 2001, 42, 677–685. [Google Scholar] [CrossRef] [PubMed]
- Kamada-Nobusada, T.; Sakakibara, H. Molecular Basis for Cytokinin Biosynthesis. Phytochemistry 2009, 70, 444–449. [Google Scholar] [CrossRef] [PubMed]
- Ruan, J.; Zhou, Y.; Zhou, M.; Yan, J.; Khurshid, M.; Weng, W.; Cheng, J.; Zhang, K. Jasmonic Acid Signaling Pathway in Plants. Int. J. Mol. Sci. 2019, 20, 2479. [Google Scholar] [CrossRef] [PubMed]
- Wasternack, C.; Song, S. Jasmonates: Biosynthesis, Metabolism, and Signaling by Proteins Activating and Repressing Transciption. J. Exp. Bot. 2016, 68, 1303–1321. [Google Scholar] [CrossRef] [PubMed]
- Zhou, G.; Ren, N.; Qi, J.; Lu, J.; Xiang, C.; Ju, H.; Cheng, J.; Lou, Y. The 9-Lipoxygenase Osr9-LOX1 Interacts with the 13-Lipoxygenase-Mediated Pathway to Regulate Resistance to Chewing and Piercing-Sucking Herbivores in Rice. Physiol. Plant. 2014, 152, 59–69. [Google Scholar] [CrossRef] [PubMed]
- Riemann, M.; Haga, K.; Shimizu, T.; Okada, K.; Ando, S.; Mochizuki, S.; Nishizawa, Y.; Yamanouchi, U.; Nick, P.; Yano, M.; et al. Identification of Rice Allene Oxide Cyclase Mutants and the Function of Jasmonate for Defence against Magnaporthe oryzae. Plant J. 2013, 74, 226–238. [Google Scholar] [CrossRef] [PubMed]
- Scalschi, L.; Sanmartín, M.; Camañes, G.; Troncho, P.; Sánchez-Serrano, J.J.; García-Agustín, P.; Vicedo, B. Silencing of OPR3 in Tomato Reveals the Role of OPDA in Callose Deposition during the Activation of Defense Responses against Botrytis cinerea. Plant J. 2015, 81, 304–315. [Google Scholar] [CrossRef] [PubMed]
- Richmond, T.A.; Bleecker, A.B. A Defect in β-Oxidation Causes Abnormal Inflorescence Development in Arabidopsis. Plant Cell 1999, 11, 1911–1924. [Google Scholar] [PubMed]
- Stitz, M.; Gase, K.; Baldwin, I.T.; Gaquerel, E. Ectopic Expression of AtJMT in Nicotiana attenuata: Creating a Metabolic Sink Has Tissue-Specific Consequences for the Jasmonate Metabolic Network and Silences Downstream Gene Expression. Plant Physiol. 2011, 157, 341–354. [Google Scholar] [CrossRef]
- Waadt, R. Phytohormone Signaling Mechanisms and Genetic Methods for Their Modulation and Detection. Curr. Opin. Plant Biol. 2020, 57, 31–40. [Google Scholar] [CrossRef]
- Olszewski, N.; Sun, T.; Gubler, F. Gibberellin Signaling: Biosynthesis, Catabolism, and Response Pathways. Plant Cell 2002, 14, S61–S80. [Google Scholar] [CrossRef]
- Qi, T.; Huang, H.; Wu, D.; Yan, J.; Qi, Y.; Song, S.; Xie, D. Arabidopsis DELLA and JAZ Proteins Bind the WD-Repeat/bHLH/MYB Complex to Modulate Gibberellin and Jasmonate Signaling Synergy. Plant Cell 2014, 26, 1118–1133. [Google Scholar] [CrossRef] [PubMed]
- Yu, S.; Galvão, V.C.; Zhang, Y.-C.; Horrer, D.; Zhang, T.-Q.; Hao, Y.-H.; Feng, Y.-Q.; Wang, S.; Schmid, M.; Wang, J.-W. Gibberellin Regulates the Arabidopsis Floral Transition through miR156-Targeted SQUAMOSA PROMOTER BINDING–LIKE Transcription Factors. Plant Cell 2012, 24, 3320–3332. [Google Scholar] [CrossRef] [PubMed]
- Gubler, F.; Chandler, P.M.; White, R.G.; Llewellyn, D.J.; Jacobsen, J.V. Gibberellin Signaling in Barley Aleurone Cells. Control of SLN1 and GAMYB Expression. Plant Physiol. 2002, 129, 191–200. [Google Scholar] [CrossRef] [PubMed]
- Lavy, M.; Estelle, M. Mechanisms of Auxin Signaling. Development 2016, 143, 3226–3229. [Google Scholar] [CrossRef] [PubMed]
- D’Agostino, I.B.; Deruère, J.; Kieber, J.J. Characterization of the Response of the Arabidopsis Response Regulator Gene Family to Cytokinin. Plant Physiol. 2000, 124, 1706–1717. [Google Scholar] [CrossRef] [PubMed]
- D’Aloia, M.; Bonhomme, D.; Bouché, F.; Tamseddak, K.; Ormenese, S.; Torti, S.; Coupland, G.; Périlleux, C. Cytokinin Promotes Flowering of Arabidopsis via Transcriptional Activation of the FT Paralogue TSF. Plant J. 2011, 65, 972–979. [Google Scholar] [CrossRef] [PubMed]
- Fernández-Calvo, P.; Chini, A.; Fernández-Barbero, G.; Chico, J.-M.; Gimenez-Ibanez, S.; Geerinck, J.; Eeckhout, D.; Schweizer, F.; Godoy, M.; Franco-Zorrilla, J.M.; et al. The Arabidopsis bHLH Transcription Factors MYC3 and MYC4 Are Targets of JAZ Repressors and Act Additively with MYC2 in the Activation of Jasmonate Responses. Plant Cell 2011, 23, 701–715. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.; Yan, W.; Fu, L.-Y.; Kaufmann, K. Architecture of Gene Regulatory Networks Controlling Flower Development in Arabidopsis thaliana. Nat. Commun. 2018, 9, 4534. [Google Scholar] [CrossRef] [PubMed]
- Strader, L.; Weijers, D.; Wagner, D. Plant Transcription Factors—Being in the Right Place with the Right Company. Curr. Opin. Plant Biol. 2022, 65, 102136. [Google Scholar] [CrossRef] [PubMed]
- Bendahmane, M.; Dubois, A.; Raymond, O.; Bris, M.L. Genetics and Genomics of Flower Initiation and Development in Roses. J. Exp. Bot. 2013, 64, 847–857. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Tang, A.; Wan, H.; Zhang, T.; Cheng, T.; Wang, J.; Yang, W.; Pan, H.; Zhang, Q. An APETALA2 Homolog, RcAP2, Regulates the Number of Rose Petals Derived From Stamens and Response to Temperature Fluctuations. Front. Plant Sci. 2018, 9, 481. [Google Scholar] [CrossRef] [PubMed]
- Dubos, C.; Stracke, R.; Grotewold, E.; Weisshaar, B.; Martin, C.; Lepiniec, L. MYB Transcription Factors in Arabidopsis. Trends Plant Sci. 2010, 15, 573–581. [Google Scholar] [CrossRef]
- Brownfield, L.; Hafidh, S.; Borg, M.; Sidorova, A.; Mori, T.; Twell, D. A Plant Germline-Specific Integrator of Sperm Specification and Cell Cycle Progression. PLoS Genet. 2009, 5, e1000430. [Google Scholar] [CrossRef] [PubMed]
- Punwani, J.A.; Rabiger, D.S.; Lloyd, A.; Drews, G.N. The MYB98 Subcircuit of the Synergid Gene Regulatory Network Includes Genes Directly and Indirectly Regulated by MYB98. Plant J. 2008, 55, 406–414. [Google Scholar] [CrossRef] [PubMed]
- Sung, S.-K.; Yu, G.-H.; An, G. Characterization of MdMADS2, a Member of the SQUAMOSA Subfamily of Genes, in Apple. Plant Physiol. 1999, 120, 969–978. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.-K.; Lin, I.-C.; Yang, C.-H. Functional Analysis of Three Lily (Lilium longiflorum) APETALA1-like MADS Box Genes in Regulating Floral Transition and Formation. Plant Cell Physiol. 2008, 49, 704–717. [Google Scholar] [CrossRef] [PubMed]
- Hsu, H.-F.; Huang, C.-H.; Chou, L.-T.; Yang, C.-H. Ectopic Expression of an Orchid (Oncidium Gower Ramsey) AGL6-like Gene Promotes Flowering by Activating Flowering Time Genes in Arabidopsis thaliana. Plant Cell Physiol. 2003, 44, 783–794. [Google Scholar] [CrossRef]
- Liu, Y.; Li, X.; Li, K.; Liu, H.; Lin, C. Multiple bHLH Proteins Form Heterodimers to Mediate CRY2-Dependent Regulation of Flowering-Time in Arabidopsis. PLoS Genet. 2013, 9, e1003861. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Chen, L.; Yu, D. Transcription Factor WRKY75 Interacts with DELLA Proteins to Affect Flowering. Plant Physiol. 2018, 176, 790–803. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Liu, Z.; Wang, L.; Kim, S.; Seo, P.J.; Qiao, M.; Wang, N.; Li, S.; Cao, X.; Park, C.; et al. WRKY 71 Accelerates Flowering via the Direct Activation of FLOWERING LOCUS T and LEAFY in Arabidopsis thaliana. Plant J. 2016, 85, 96–106. [Google Scholar] [CrossRef] [PubMed]
- OsNAC2 Encoding a NAC Transcription Factor That Affects Plant Height through Mediating the Gibberellic Acid Pathway in Rice. Available online: https://onlinelibrary.wiley.com/doi/epdf/10.1111/tpj.12819 (accessed on 29 February 2024).
- Shih, C.-F.; Hsu, W.-H.; Peng, Y.-J.; Yang, C.-H. The NAC-like Gene ANTHER INDEHISCENCE FACTOR Acts as a Repressor That Controls Anther Dehiscence by Regulating Genes in the Jasmonate Biosynthesis Pathway in Arabidopsis. J. Exp. Bot. 2014, 65, 621–639. [Google Scholar] [CrossRef] [PubMed]
- Niu, Q.; Zong, Y.; Qian, M.; Yang, F.; Teng, Y. Simultaneous Quantitative Determination of Major Plant Hormones in Pear Flowers and Fruit by UPLC/ESI-MS/MS. Anal. Methods 2014, 6, 1766–1773. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).