Rice Serine Hydroxymethyltransferases: Evolution, Subcellular Localization, Function and Perspectives
Abstract
1. Introduction
2. Results
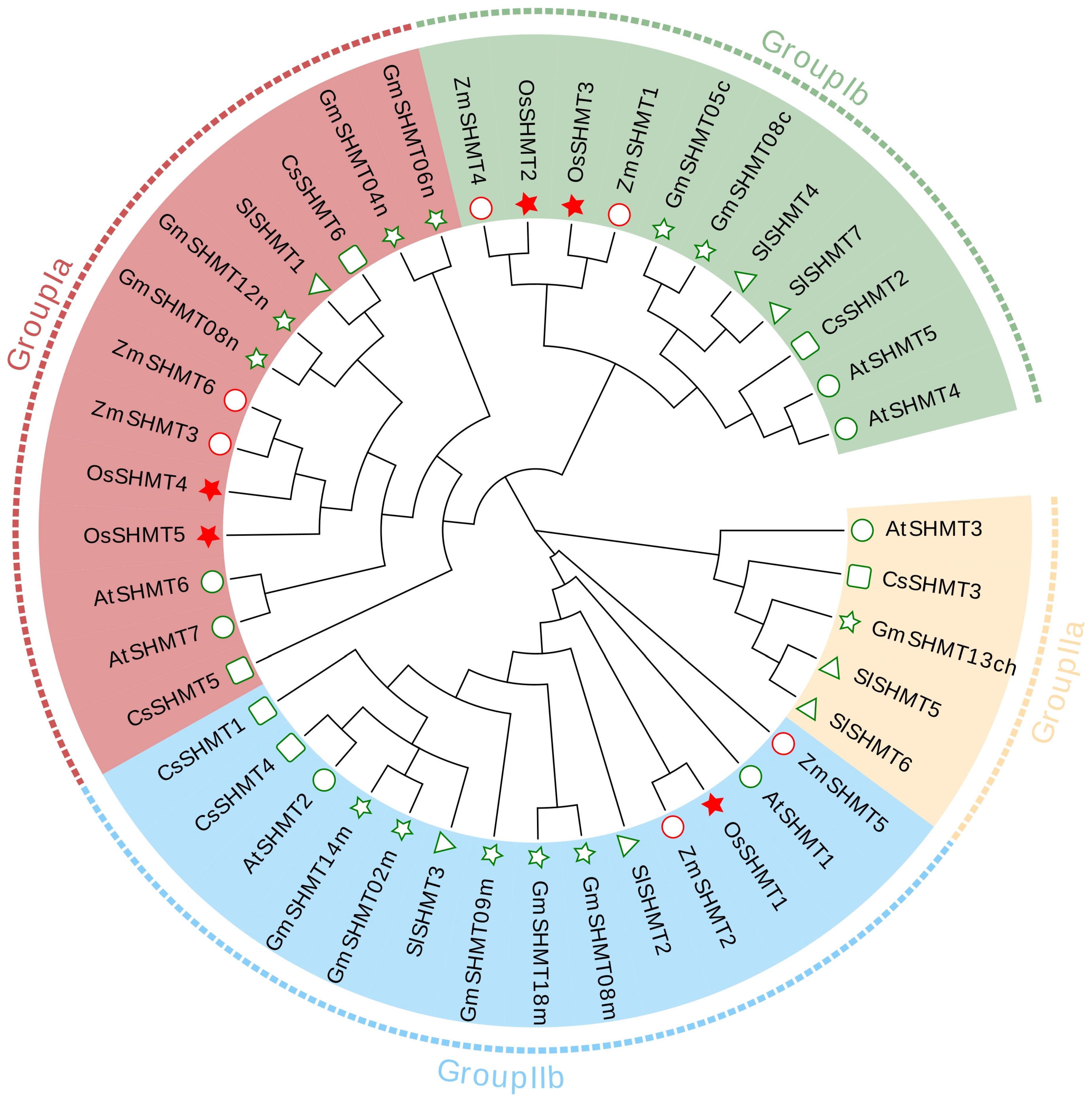
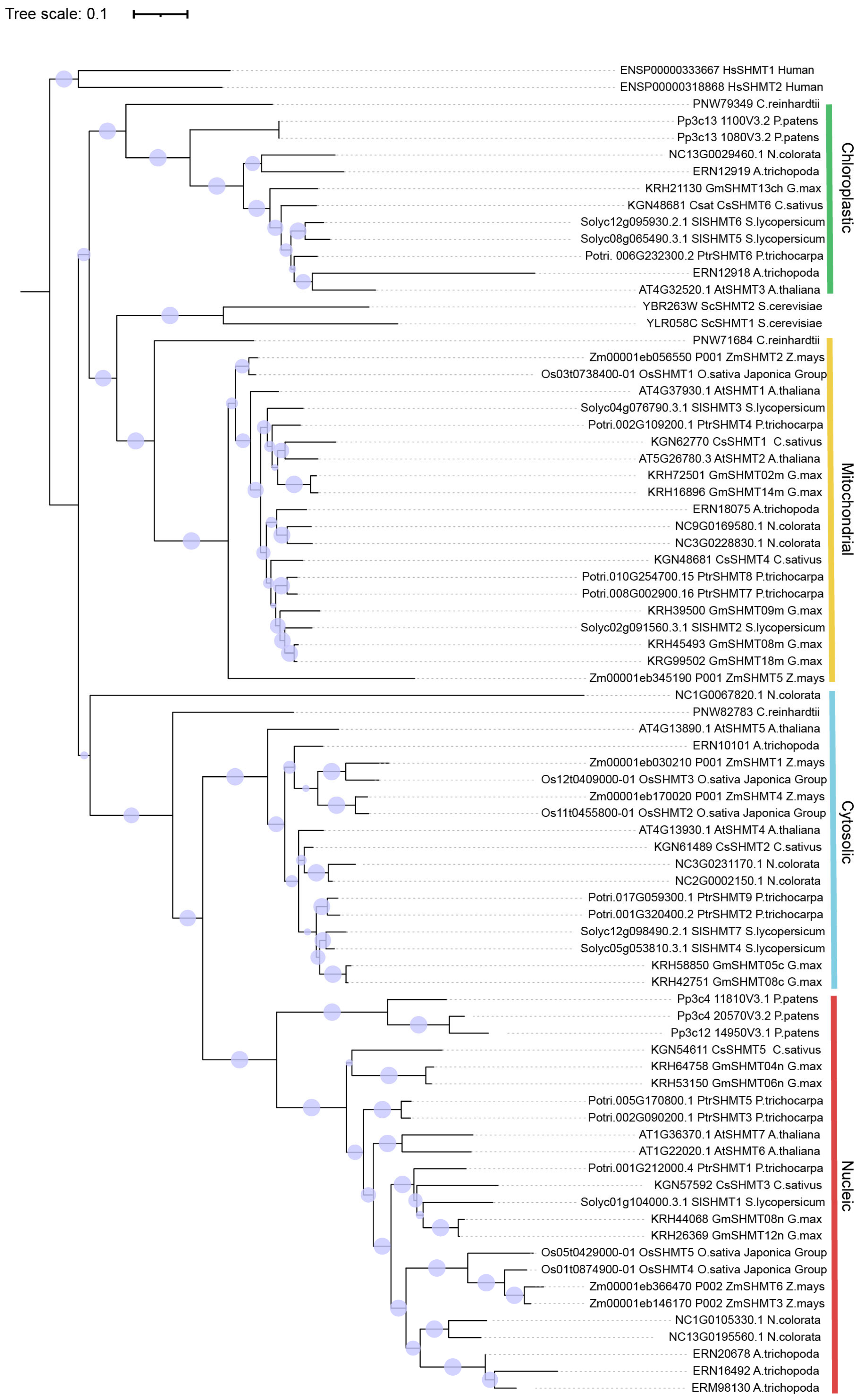
2.1. Identification and Evolutionary Analysis of SHMT Family Genes in the Rice Genome
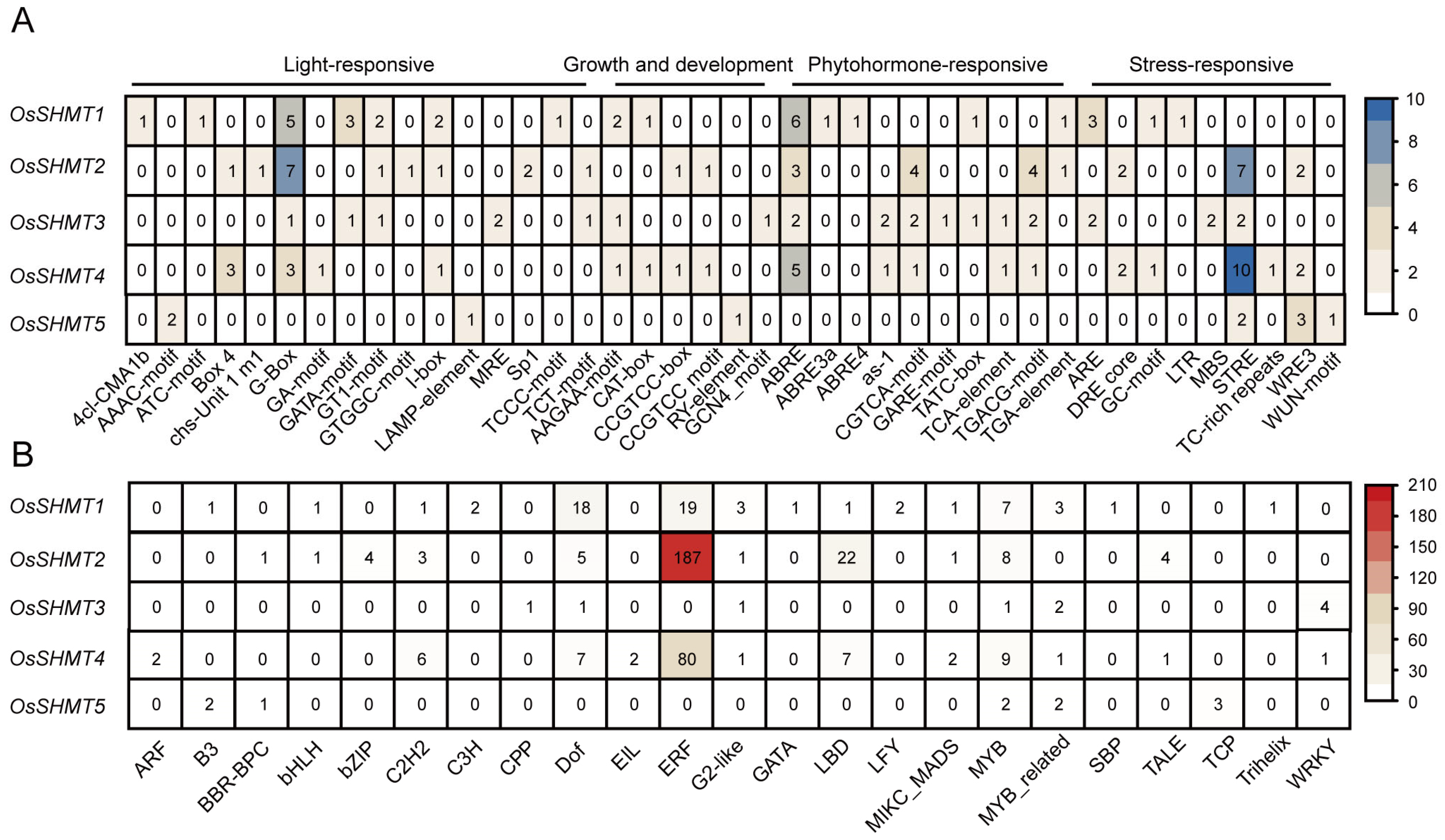
2.2. Gene Structure and Promoter Cis-Element Analysis of OsSHMTs
2.3. Expression Analysis of OsSHMTs in Various Tissues and in Response to Phytohormones
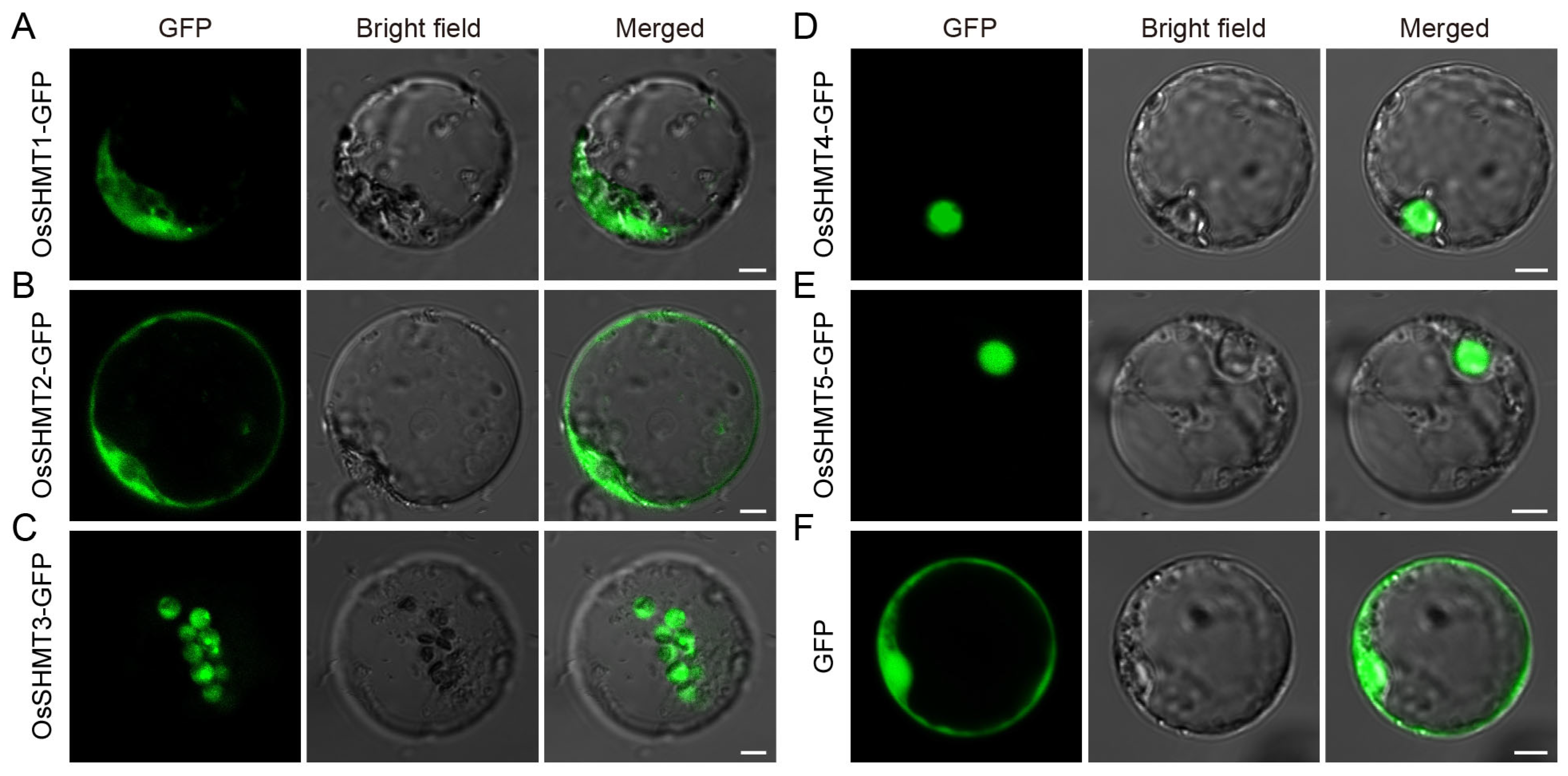
2.4. Subcellular Localization of OsSHMTs
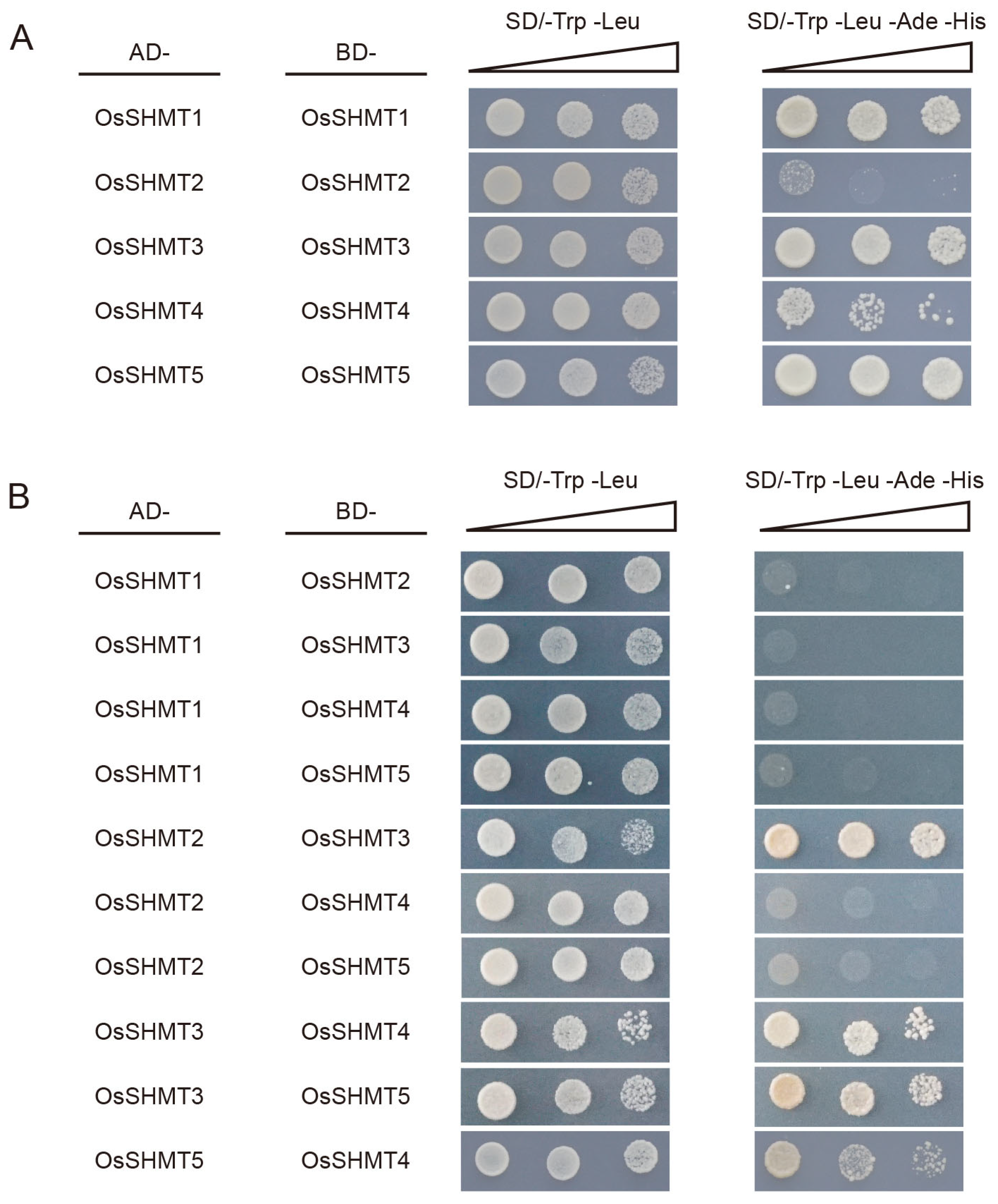
2.5. Protein Interaction Networks of OsSHMTs
2.6. OsSHMTs Form Polymers
3. Discussion
3.1. The Functions of OsSHMTs Show Differentiation
3.2. OsSHMTs Emerge as Promising Candidates for Bolstering Plant Resistance against Both Abiotic and Biotic Stressors
4. Materials and Methods
4.1. Identification and Phylogenetic Analysis of SHMT Family Genes in Rice Genome
4.2. Gene Structure Analysis of OsSHMTs
4.3. Predicted Cis-Element and Transcription Factor Binding Sites on OsSHMT Promoters
4.4. Expression Analysis
4.5. Protein–protein Interaction Prediction
4.6. Subcellular Localization Analysis
4.7. Yeast Two-Hybrid
4.8. Protein Structure Prediction
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Hanson, A.D.; Roje, S. One-Carbon Metabolism in Higher Plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 2001, 52, 119–137. [Google Scholar] [CrossRef] [PubMed]
- Mentch, S.J.; Locasale, J.W. One-Carbon Metabolism and Epigenetics: Understanding the Specificity. Ann. N. Y. Acad. Sci. 2016, 1363, 91–98. [Google Scholar] [CrossRef] [PubMed]
- Zrenner, R.; Stitt, M.; Sonnewald, U.; Boldt, R. Pyrimidine and Purine Biosynthesis and Degradation in Plants. Annu. Rev. Plant Biol. 2006, 57, 805–836. [Google Scholar] [CrossRef] [PubMed]
- Schulze, S.; Westhoff, P.; Gowik, U. Glycine Decarboxylase in C3, C4 and C3–C4 Intermediate Species. Curr. Opin. Plant Biol. 2016, 31, 29–35. [Google Scholar] [CrossRef]
- Hanson, A.D.; Gage, D.A.; Shachar-Hill, Y. Plant One-Carbon Metabolism and Its Engineering. Trends Plant Sci. 2000, 5, 206–213. [Google Scholar] [CrossRef]
- Huang, X.Y.; Chao, D.Y.; Koprivova, A.; Danku, J.; Wirtz, M.; Müller, S.; Sandoval, F.J.; Bauwe, H.; Roje, S.; Dilkes, B.; et al. Nuclear Localised More Sulphur Accumulation1 Epigenetically Regulates Sulphur Homeostasis in Arabidopsis Thaliana. PLoS Genet. 2016, 12, e1006298. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Sun, K.; Sandoval, F.J.; Santiago, K.; Roje, S. One-Carbon Metabolism in Plants: Characterization of a Plastid Serine Hydroxymethyltransferase. Biochem. J. 2010, 430, 97–105. [Google Scholar] [CrossRef] [PubMed]
- Wei, Z.; Sun, K.; Sandoval, F.J.; Cross, J.M.; Gordon, C.; Kang, C.; Roje, S. Folate Polyglutamylation Eliminates Dependence of Activity on Enzyme Concentration in Mitochondrial Serine Hydroxymethyltransferases from Arabidopsis Thaliana. Arch. Biochem. Biophys. 2013, 536, 87–96. [Google Scholar] [CrossRef] [PubMed]
- Douce, R.; Neuburger, M. Biochemical Dissection of Photorespiration. Curr. Opin. Plant Biol. 1999, 2, 214–222. [Google Scholar] [CrossRef] [PubMed]
- Rajinikanth, M.; Harding, S.A.; Tsai, C.J. The Glycine Decarboxylase Complex Multienzyme Family in Populus. J. Exp. Bot. 2007, 58, 1761–1770. [Google Scholar] [CrossRef]
- Somerville, C.R.; Ogren, W.L. Photorespiration-Deficient Mutants of Arabidopsis Thaliana Lacking Mitochondrial Serine Transhydroxymethylase Activity. Plant Physiol. 1981, 67, 666–671. [Google Scholar] [CrossRef] [PubMed]
- Voll, L.M.; Jamai, A.; Renné, P.; Voll, H.; McClung, C.R.; Weber, A.P.M. The Photorespiratory Arabidopsis Shm1 Mutant Is Deficient in SHM1. Plant Physiol. 2006, 140, 59–66. [Google Scholar] [CrossRef]
- Moreno, J.I.; Martín, R.; Castresana, C. Arabidopsis SHMT1, a Serine Hydroxymethyltransferase That Functions in the Photorespiratory Pathway Influences Resistance to Biotic and Abiotic Stress. Plant J. 2005, 41, 451–463. [Google Scholar] [CrossRef]
- Liu, Y.; Mauve, C.; Lamothe-Sibold, M.; Guérard, F.; Glab, N.; Hodges, M.; Jossier, M. Photorespiratory Serine Hydroxymethyltransferase 1 Activity Impacts Abiotic Stress Tolerance and Stomatal Closure. Plant Cell Environ. 2019, 42, 2567–2583. [Google Scholar] [CrossRef]
- Wu, J.; Zhang, Z.; Zhang, Q.; Han, X.; Gu, X.; Lu, T. The Molecular Cloning and Clarification of a Photorespiratory Mutant, Oscdm1, Using Enhancer Trapping. Front. Genet. 2015, 6, 226. [Google Scholar] [CrossRef][Green Version]
- Mishra, P.; Jain, A.; Takabe, T.; Tanaka, Y.; Negi, M.; Singh, N.; Jain, N.; Mishra, V.; Maniraj, R.; Krishnamurthy, S.L.; et al. Heterologous Expression of Serine Hydroxymethyltransferase-3 from Rice Confers Tolerance to Salinity Stress in E. coli and Arabidopsis. Front. Plant Sci. 2019, 10, 217. [Google Scholar] [CrossRef] [PubMed]
- Fang, C.; Zhang, P.; Li, L.; Yang, L.; Mu, D.; Yan, X.; Li, Z.; Lin, W. Serine Hydroxymethyltransferase Localised in the Endoplasmic Reticulum Plays a Role in Scavenging H2O2 to Enhance Rice Chilling Tolerance. BMC Plant Biol. 2020, 20, 236. [Google Scholar] [CrossRef]
- Besson, V.; Neuburger, M.; Rébeillé, F.; Douce, R. Evidence for Three Serine Hydroxymethyltransferases in Green Leaf Cells. Purification and Characterization of the Mitochondrial and Chloroplastic Isoforms. Plant Physiol. Biochem. 1995, 33, 665–673. [Google Scholar]
- Christensen, K.E.; MacKenzie, R.E. Mitochondrial One-Carbon Metabolism Is Adapted to the Specific Needs of Yeast, Plants and Mammals. Bioessays 2006, 28, 595–605. [Google Scholar] [CrossRef]
- Liu, S.; Kandoth, P.K.; Warren, S.D.; Yeckel, G.; Heinz, R.; Alden, J.; Yang, C.; Jamai, A.; El-Mellouki, T.; Juvale, P.S.; et al. A Soybean Cyst Nematode Resistance Gene Points to a New Mechanism of Plant Resistance to Pathogens. Nature 2012, 492, 256–260. [Google Scholar] [CrossRef]
- Rambani, A.; Pantalone, V.; Yang, S.; Rice, J.H.; Song, Q.; Mazarei, M.; Arelli, P.R.; Meksem, K.; Stewart, C.N.; Hewezi, T. Identification of Introduced and Stably Inherited DNA Methylation Variants in Soybean Associated with Soybean Cyst Nematode Parasitism. New Phytol. 2020, 227, 168–184. [Google Scholar] [CrossRef] [PubMed]
- Lakhssassi, N.; Piya, S.; Knizia, D.; El Baze, A.; Cullen, M.A.; Meksem, J.; Lakhssassi, A.; Hewezi, T.; Meksem, K. Mutations at the Serine Hydroxymethyltransferase Impact Its Interaction with a Soluble NSF Attachment Protein and a Pathogenesis-Related Protein in Soybean. Vaccines 2020, 8, 349. [Google Scholar] [CrossRef] [PubMed]
- Lakhssassi, N.; Piya, S.; Bekal, S.; Liu, S.; Zhou, Z.; Bergounioux, C.; Miao, L.; Meksem, J.; Lakhssassi, A.; Jones, K.; et al. A Pathogenesis-Related Protein GmPR08-Bet VI Promotes a Molecular Interaction between the GmSHMT08 and GmSNAP18 in Resistance to Heterodera Glycines. Plant Biotechnol. J. 2020, 18, 1810–1829. [Google Scholar] [CrossRef] [PubMed]
- Lakhssassi, N.; Knizia, D.; El Baze, A.; Lakhssassi, A.; Meksem, J.; Meksem, K. Proteomic, Transcriptomic, Mutational, and Functional Assays Reveal the Involvement of Both THF and PLP Sites at the GmSHMT08 in Resistance to Soybean Cyst Nematode. Int. J. Mol. Sci. 2022, 23, 11278. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Huang, X.Y.; Salt, D.E.; Zhao, F.J. Mutation in OsCADT1 Enhances Cadmium Tolerance and Enriches Selenium in Rice Grain. New Phytol. 2020, 226, 838–850. [Google Scholar] [CrossRef] [PubMed]
- Yan, M.; Pan, T.; Zhu, Y.; Jiang, X.; Yu, M.; Wang, R.; Zhang, F.; Luo, S.; Bao, X.; Chen, Y.; et al. FLOURY ENDOSPERM20 Encoding SHMT4 Is Required for Rice Endosperm Development. Plant Biotechnol. J. 2022, 20, 1438–1440. [Google Scholar] [CrossRef]
- Anderson, D.D.; Woeller, C.F.; Chiang, E.P.; Shane, B.; Stover, P.J. Serine Hydroxymethyltransferase Anchors de Novo Thymidylate Synthesis Pathway to Nuclear Lamina for DNA Synthesis. J. Biol. Chem. 2012, 287, 7051–7062. [Google Scholar] [CrossRef]
- Anderson, D.D.; Woeller, C.F.; Stover, P.J. Small Ubiquitin-like Modifier-1 (SUMO-1) Modification of Thymidylate Synthase and Dihydrofolate Reductase. Clin. Chem. Lab. Med. 2007, 45, 1760–1763. [Google Scholar] [CrossRef]
- Woeller, C.F.; Anderson, D.D.; Szebenyi, D.M.; Stover, P.J. Evidence for Small Ubiquitin-like Modifier-Dependent Nuclear Import of the Thymidylate Biosynthesis Pathway. J. Biol. Chem. 2007, 282, 17623–17631. [Google Scholar] [CrossRef]
- Timm, S.; Bauwe, H. The Variety of Photorespiratory Phenotypes—Employing the Current Status for Future Research Directions on Photorespiration. Plant Biol. 2013, 15, 737–747. [Google Scholar] [CrossRef]
- Zhou, Q.; Yu, Q.; Wang, Z.; Pan, Y.; Lv, W.; Zhu, L.; Chen, R.; He, G. Knockdown of GDCH Gene Reveals Reactive Oxygen Species-Induced Leaf Senescence in Rice. Plant Cell Environ. 2013, 36, 1476–1489. [Google Scholar] [CrossRef]
- Schirch, V.; Szebenyi, D.M. Serine Hydroxymethyltransferase Revisited. Curr. Opin. Chem. Biol. 2005, 9, 482–487. [Google Scholar] [CrossRef]
- Yan, Y.; Tao, H.; He, J.; Huang, S.Y. The HDOCK Server for Integrated Protein–Protein Docking. Nat. Protoc. 2020, 15, 1829–1852. [Google Scholar] [CrossRef]
- Mooers, B.H.M. Shortcuts for Faster Image Creation in PyMOL. Protein Sci. 2020, 29, 268–276. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly Accurate Protein Structure Prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef] [PubMed]
- Lakhssassi, N.; Patil, G.; Piya, S.; Zhou, Z.; Baharlouei, A.; Kassem, M.A.; Lightfoot, D.A.; Hewezi, T.; Barakat, A.; Nguyen, H.T.; et al. Genome Reorganization of the GmSHMT Gene Family in Soybean Showed a Lack of Functional Redundancy in Resistance to Soybean Cyst Nematode. Sci. Rep. 2019, 9, 1506. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Baxter, R.; Smith, B.S.; Partch, C.L.; Colbert, C.L.; Deisenhofer, J. Crystal Structure of Cryptochrome 3 from Arabidopsis Thaliana and Its Implications for Photolyase Activity. Proc. Natl. Acad. Sci. USA 2006, 103, 17701–17706. [Google Scholar] [CrossRef]
- Wang, Y.; Mostafa, S.; Zeng, W.; Jin, B. Function and Mechanism of Jasmonic Acid in Plant Responses to Abiotic and Biotic Stresses. Int. J. Mol. Sci. 2021, 22, 8568. [Google Scholar] [CrossRef] [PubMed]
- Vanitha, P.A.; Vijayaraghavareddy, P.; Uttarkar, A.; Dawane, A.; Sujitha, D.; Ashwin, V.; Babitha, K.C.; Niranjan, V.; Sheshshayee, M.S.; Anuradha, C.V.; et al. Novel Small Molecules Targeting BZIP23 TF Improve Stomatal Conductance and Photosynthesis under Mild Drought Stress by Regulating ABA. FEBS J. 2022, 289, 6058–6077. [Google Scholar]
- Ma, Z.; Hu, L.; Jiang, W. Understanding AP2/ERF Transcription Factor Responses and Tolerance to Various Abiotic Stresses in Plants: A Comprehensive Review. Int. J. Mol. Sci. 2024, 25, 893. [Google Scholar] [CrossRef]
- Wang, X.; Niu, Y.; Zheng, Y. Multiple Functions of Myb Transcription Factors in Abiotic Stress Responses. Int. J. Mol. Sci. 2021, 22, 6125. [Google Scholar] [CrossRef]
- Hu, P.; Song, P.; Xu, J.; Wei, Q.; Tao, Y.; Ren, Y.; Yu, Y.; Li, D.; Hu, H.; Li, C. Genome-Wide Analysis of Serine Hydroxymethyltransferase Genes in Triticeae Species Reveals That TaSHMT3A-1 Regulates Fusarium Head Blight Resistance in Wheat. Front. Plant Sci. 2022, 13, 847087. [Google Scholar] [CrossRef]
- Katoh, K.; Misawa, K.; Kuma, K.-I.; Miyata, T. MAFFT: A Novel Method for Rapid Multiple Sequence Alignment Based on Fast Fourier Transform. Nucleic Acids Res. 2002, 30, 3059–3066. [Google Scholar] [CrossRef] [PubMed]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. Fasttree: Computing Large Minimum Evolution Trees with Profiles Instead of a Distance Matrix. Mol. Biol. Evol. 2009, 26, 1641–1650. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Gao, S.; Lercher, M.J.; Hu, S.; Chen, W.H. EvolView, an Online Tool for Visualizing, Annotating and Managing Phylogenetic Trees. Nucleic Acids Res. 2012, 40, W569–W572. [Google Scholar] [CrossRef]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME Suite: Tools for Motif Discovery and Searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Wu, Y.; Li, J.; Wang, X.; Zeng, Z.; Xu, J.; Liu, Y.; Feng, J.; Chen, H.; He, Y.; et al. TBtools-II: A “One for All, All for One” Bioinformatics Platform for Biological Big-Data Mining. Mol. Plant 2023, 16, 1733–1742. [Google Scholar] [CrossRef]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van De Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a Database of Plant Cis-Acting Regulatory Elements and a Portal to Tools for in Silico Analysis of Promoter Sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Jin, J.; Tian, F.; Yang, D.C.; Meng, Y.Q.; Kong, L.; Luo, J.; Gao, G. PlantTFDB 4.0: Toward a Central Hub for Transcription Factors and Regulatory Interactions in Plants. Nucleic Acids Res. 2017, 45, D1040–D1045. [Google Scholar] [CrossRef]
- Chen, S.; Tao, L.; Zeng, L.; Vega-Sanchez, M.E.; Umemura, K.; Wang, G.L. A highly efficient transient protoplast system for analyzing defence gene expression and protein-protein interactions in rice. Mol. Plant Pathol. 2006, 7, 417–427. [Google Scholar] [CrossRef] [PubMed]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pan, T.; Jin, H.; Zhou, C.; Yan, M. Rice Serine Hydroxymethyltransferases: Evolution, Subcellular Localization, Function and Perspectives. Plants 2024, 13, 1116. https://doi.org/10.3390/plants13081116
Pan T, Jin H, Zhou C, Yan M. Rice Serine Hydroxymethyltransferases: Evolution, Subcellular Localization, Function and Perspectives. Plants. 2024; 13(8):1116. https://doi.org/10.3390/plants13081116
Chicago/Turabian StylePan, Tian, Hongmiao Jin, Chuanhui Zhou, and Mengyuan Yan. 2024. "Rice Serine Hydroxymethyltransferases: Evolution, Subcellular Localization, Function and Perspectives" Plants 13, no. 8: 1116. https://doi.org/10.3390/plants13081116
APA StylePan, T., Jin, H., Zhou, C., & Yan, M. (2024). Rice Serine Hydroxymethyltransferases: Evolution, Subcellular Localization, Function and Perspectives. Plants, 13(8), 1116. https://doi.org/10.3390/plants13081116





