Genome-Wide Identification of BrCMF Genes in Brassica rapa and Their Expression Analysis under Abiotic Stresses
Abstract
1. Introduction
2. Results
2.1. Identification and Analysis of BrCMFs
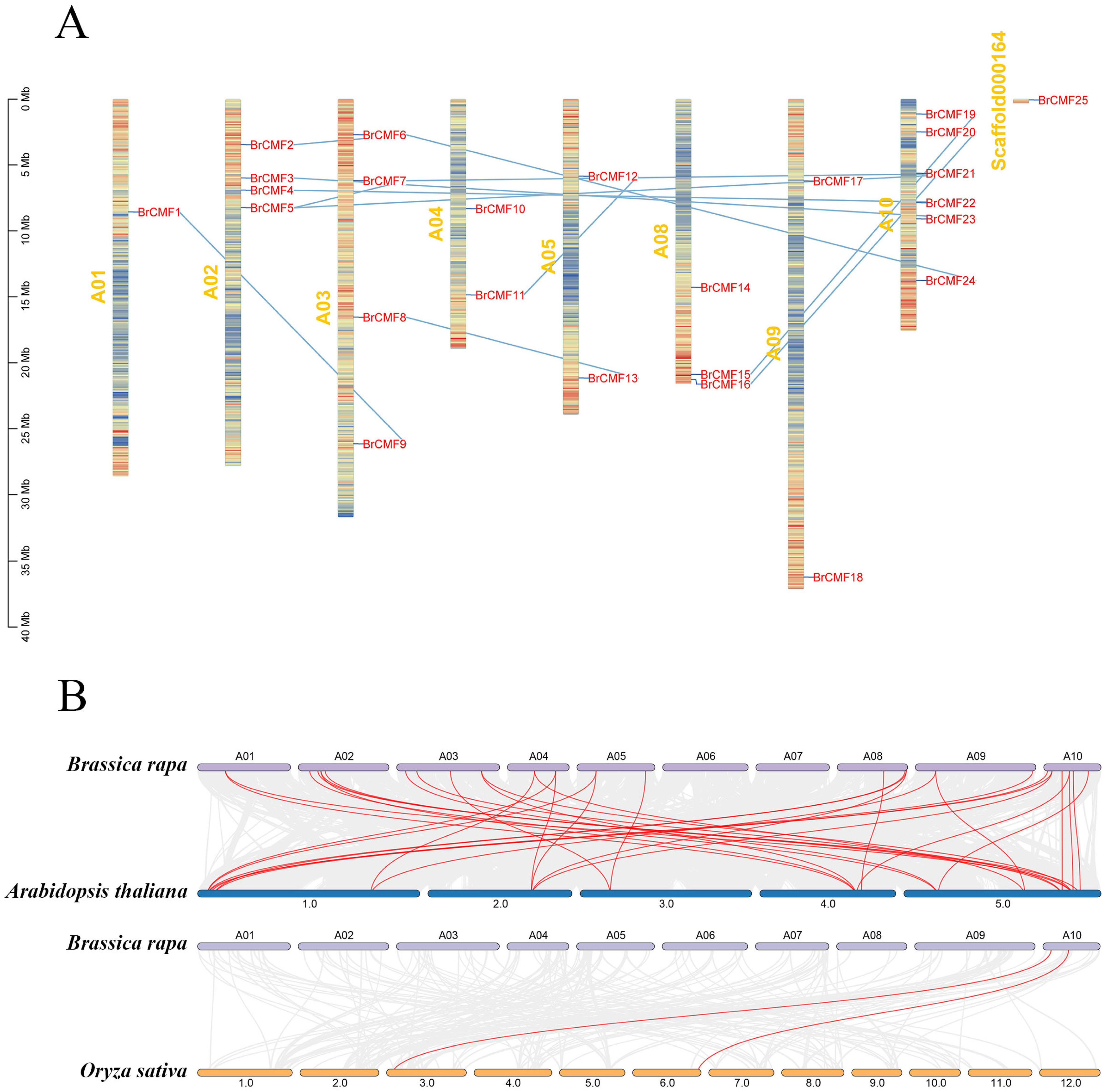
2.2. Chromosomal Localization, Synteny, and Phylogenetic Analysis of BrCMFs
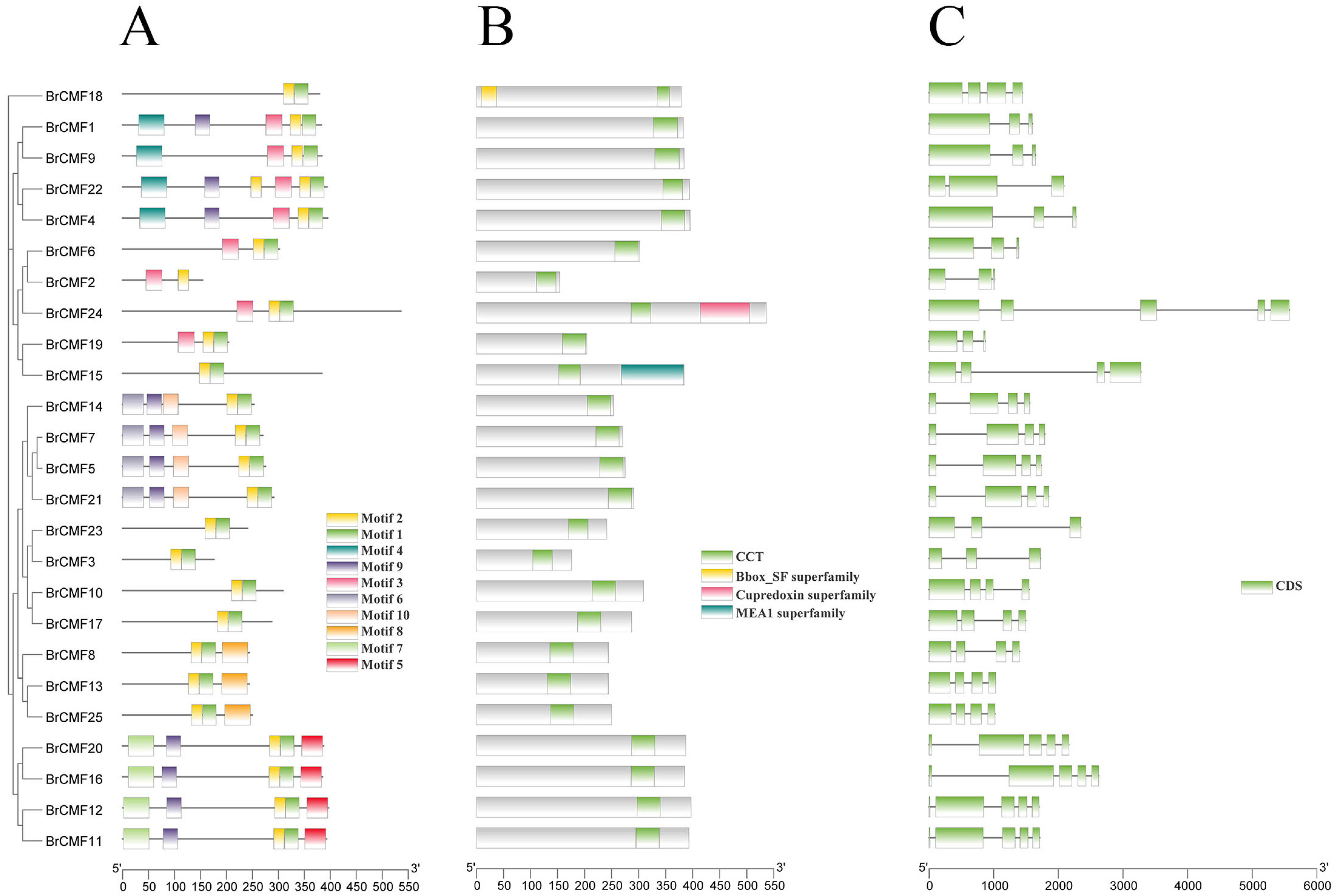
2.3. Motif Analysis, Gene Structure, and Conserved Domain Analysis of BrCMFs
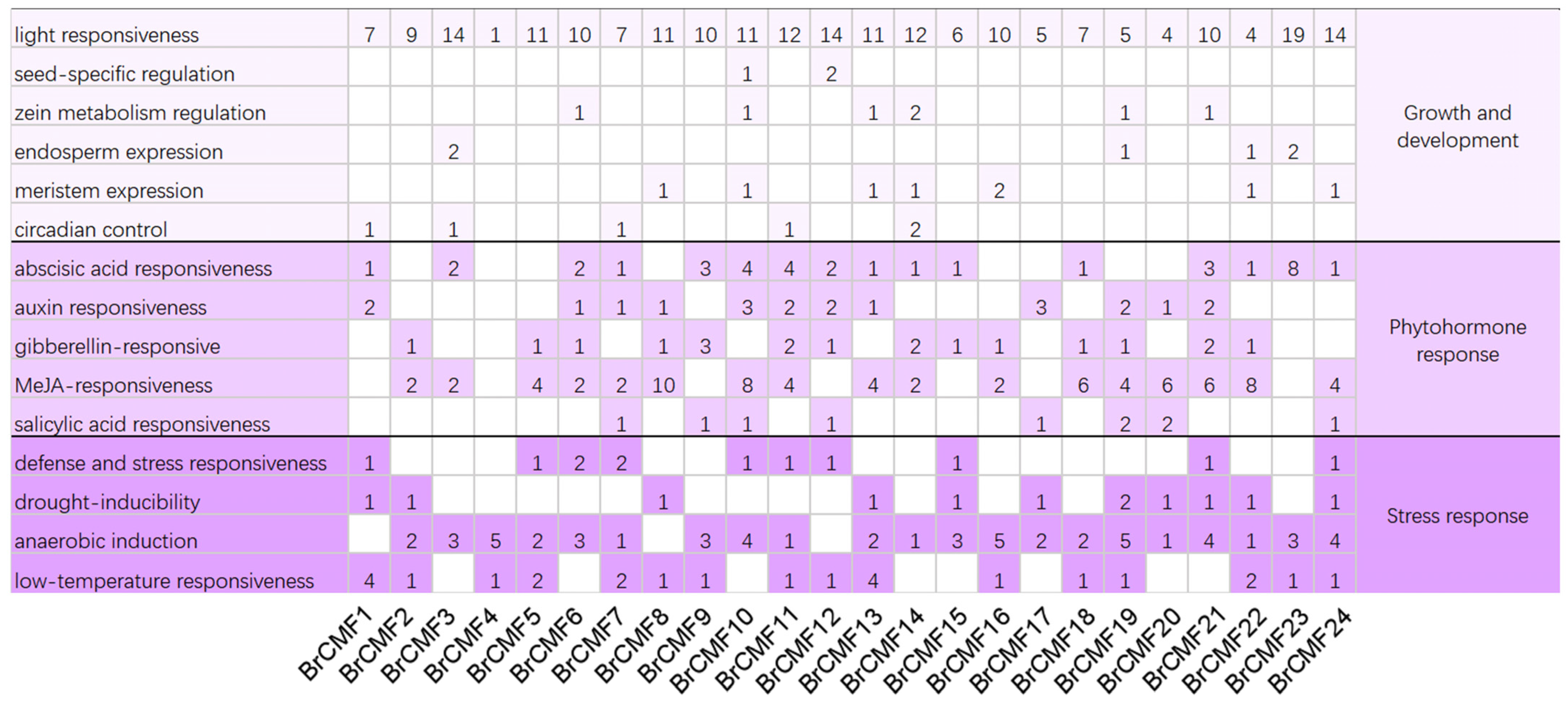
2.4. Cis-Acting Element Analysis of BrCMFs
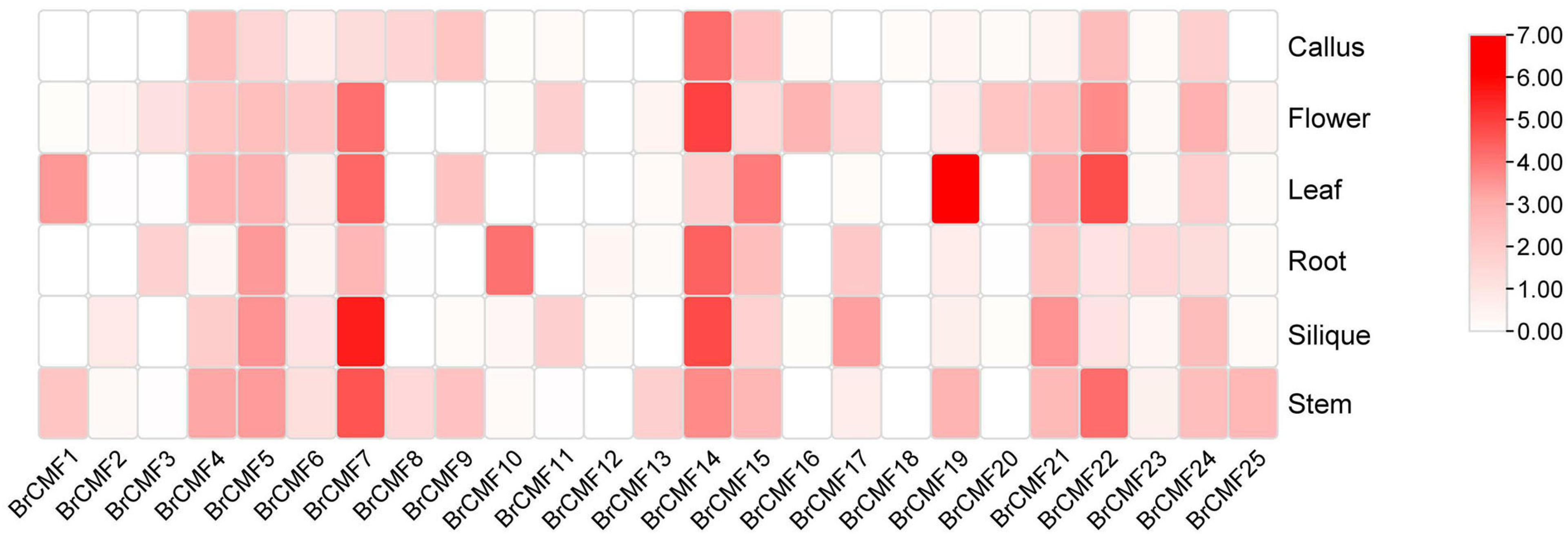
2.5. Tissue Specific Expression of BrCMFs
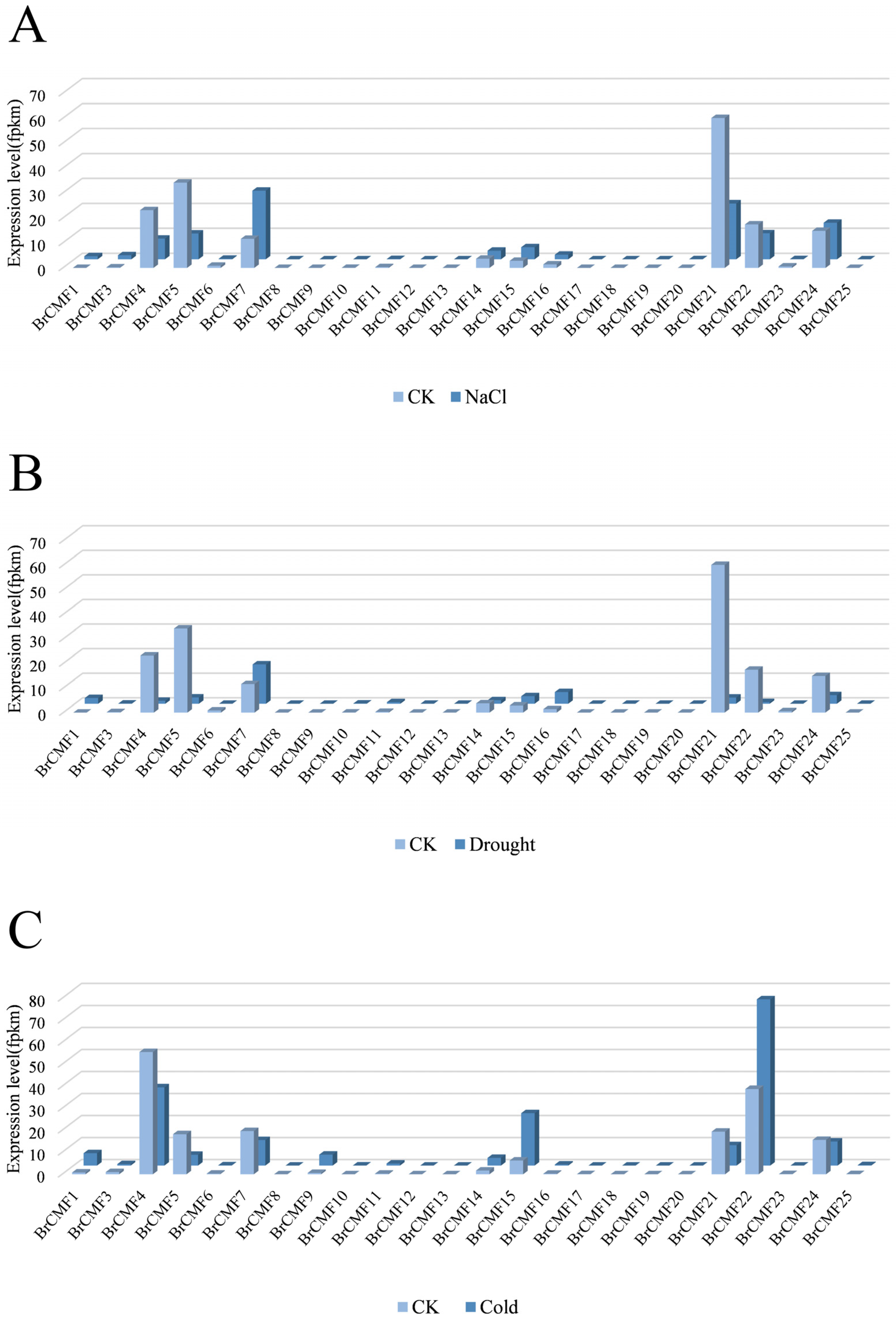
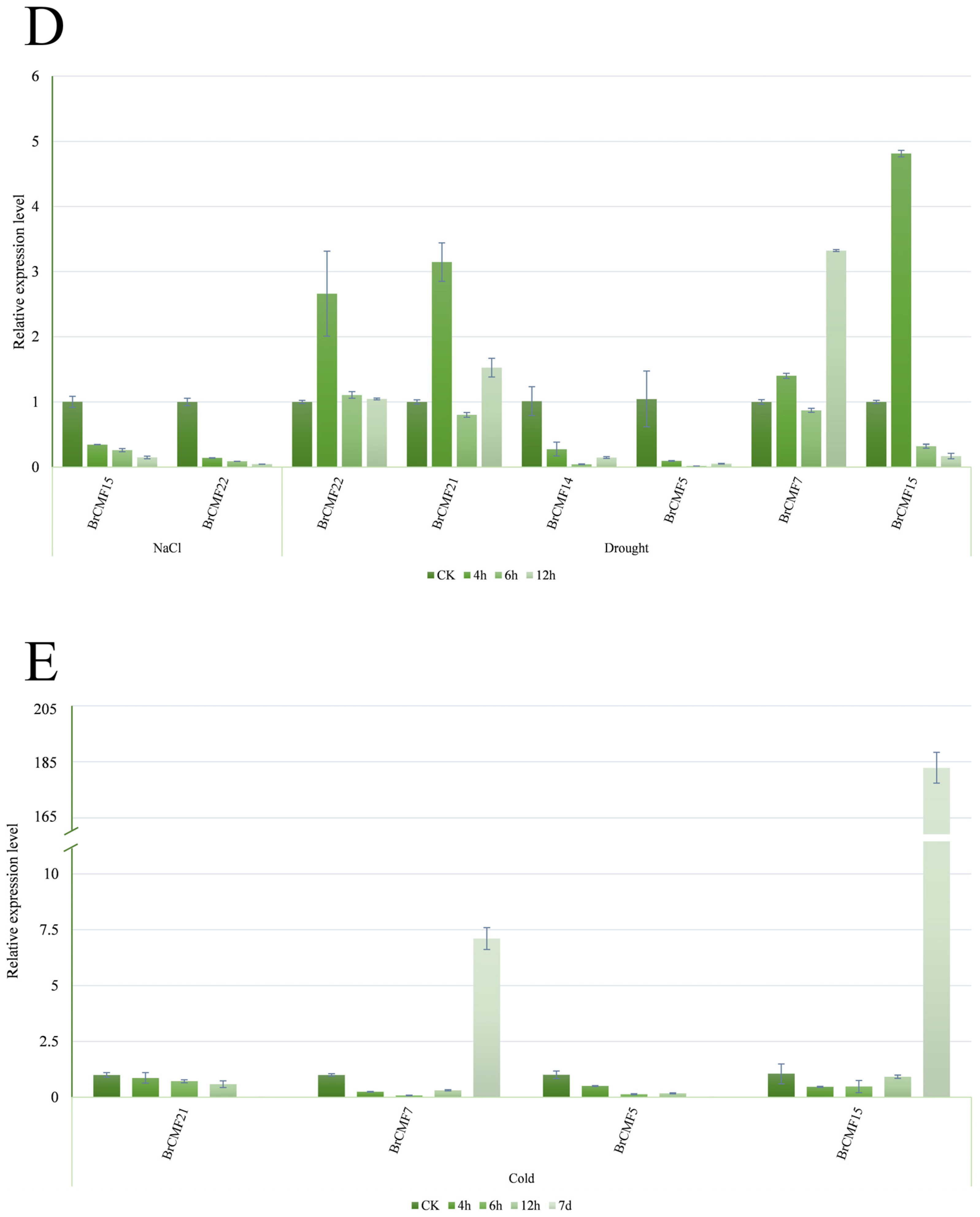
2.6. Transcriptome Analysis of BrCMFs
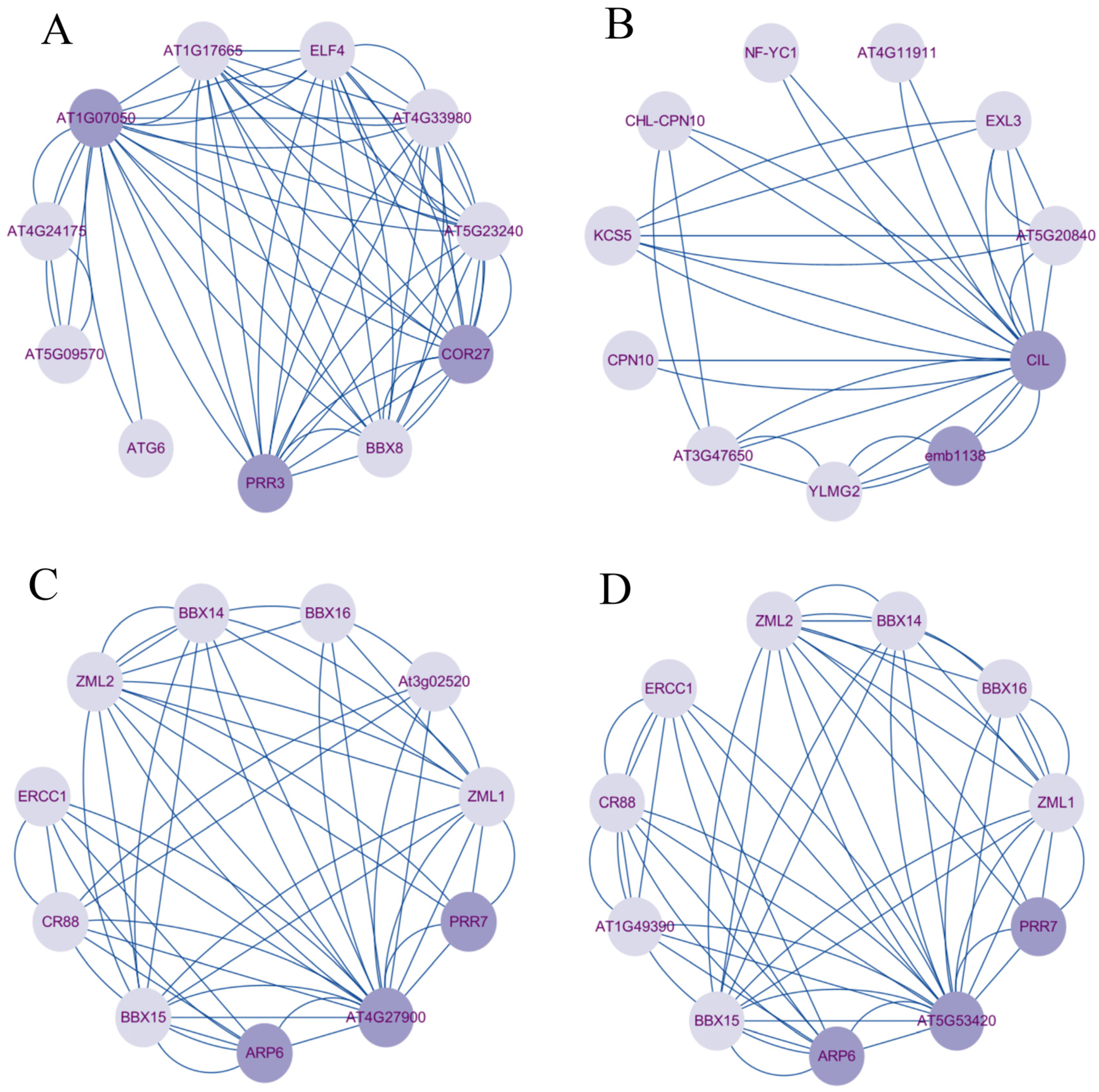
2.7. Protein–Protein Interaction (PPI) Network Analysis of BrCMFs
3. Discussion
4. Materials and Methods
4.1. Identification of CMF Genes in the B. rapa Genome
4.2. Chromosomal Localization, Synteny, and Phylogenetic Analysis
4.3. Motif analysis, Gene Structure, and Conservative Domain Analysis
4.4. Cis-Acting Element Analysis
4.5. Tissue Specific Expression
4.6. Stress Treatments, Total RNA Extraction, Transcriptome and RT-qPCR Analysis
4.7. PPI Network Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Putterill, J.; Robson, F.; Lee, K.; Simon, R.; Coupland, G. The CONSTANS gene of Arabidopsis promotes flowering and encodes a protein showing similarities to zinc finger transcription factors. Cell 1995, 80, 847–857. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Zhou, X.; Li, Q.; Wang, L.; Xing, Y. CCT domain-containing genes in cereal crops: Flowering time and beyond. Theor. Appl. Genet. 2020, 133, 1385–1396. [Google Scholar] [CrossRef] [PubMed]
- Satbhai, S.B.; Yamashino, T.; Okada, R.; Nomoto, Y.; Mizuno, T.; Tezuka, Y.; Itoh, T.; Tomita, M.; Otsuki, S.; Aoki, S. Pseudo-response regulator (PRR) homologues of the moss Physcomitrella patens: Insights into the evolution of the PRR family in land plants. DNA Res. 2011, 18, 39–52. [Google Scholar] [CrossRef] [PubMed]
- Cockram, J.; Thiel, T.; Steuernagel, B.; Stein, N.; Taudien, S.; Bailey, P.C.; O’Sullivan, D.M. Genome dynamics explain the evolution of flowering time CCT domain gene families in the Poaceae. PLoS ONE 2012, 7, e45307. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Li, T.; Wang, Y.; Zheng, J.; Li, H.; Hao, C.; Zhang, X. TaZIM-A1 negatively regulates flowering time in common wheat (Triticum aestivum L.). J. Integr. Plant Biol. 2019, 61, 359–376. [Google Scholar] [CrossRef] [PubMed]
- Shikata, M.; Matsuda, Y.; Ando, K.; Nishii, A.; Takemura, M.; Yokota, A.; Kohchi, T. Characterization of Arabidopsis ZIM, a member of a novel plant-specific GATA factor gene family. J. Exp. Bot. 2004, 55, 631–639. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Li, Q.; Dong, H.; He, Q.; Liang, L.; Tan, C.; Han, Z.; Yao, W.; Li, G.; Zhao, H.; et al. Three CCT domain-containing genes were identified to regulate heading date by candidate gene-based association mapping and transformation in rice. Sci. Rep. 2015, 5, 7663. [Google Scholar] [CrossRef]
- Zhang, H.; Jiao, B.; Dong, F.; Liang, X.; Zhou, S.; Wang, H. Genome-wide identification of CCT genes in wheat (Triticum aestivum L.) and their expression analysis during vernalization. PLoS ONE 2022, 17, e0262147. [Google Scholar] [CrossRef] [PubMed]
- Peng, X.; Tun, W.; Dai, S.F.; Li, J.Y.; Zhang, Q.J.; Yin, G.Y.; Yoon, J.; Cho, L.H.; An, G.; Gao, L.Z. Genome-Wide Analysis of CCT Transcript Factors to Identify Genes Contributing to Photoperiodic Flowering in Oryza rufipogon. Front. Plant Sci. 2021, 12, 736419. [Google Scholar] [CrossRef]
- Ma, L.; Yi, D.; Yang, J.; Liu, X.; Pang, Y. Genome-Wide Identification, Expression Analysis and Functional Study of CCT Gene Family in Medicago truncatula. Plants 2020, 9, 513. [Google Scholar] [CrossRef]
- Li, Y.; Yu, S.; Zhang, Q.; Wang, Z.; Liu, M.; Zhang, A.; Dong, X.; Fan, J.; Zhu, Y.; Ruan, Y.; et al. Genome-Wide Identification and Characterization of the CCT Gene Family in Foxtail Millet (Setaria italica) Response to Diurnal Rhythm and Abiotic Stress. Genes 2022, 13, 1829. [Google Scholar] [CrossRef] [PubMed]
- Mengarelli, D.A.; Zanor, M.I. Genome-wide characterization and analysis of the CCT motif family genes in soybean (Glycine max). Planta 2021, 253, 15. [Google Scholar] [CrossRef] [PubMed]
- Dong, M.Y.; Lei, L.; Fan, X.W.; Li, Y.Z. Analyses of open-access multi-omics data sets reveal genetic and expression characteristics of maize ZmCCT family genes. AoB Plants 2021, 13, plab048. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.; Li, X.; Ge, C.; Chang, J.; Shi, M.; Chen, J.; Qiao, L.; Chang, Z.; Zheng, J.; Zhang, J. Characterization of the CCT family and analysis of gene expression in Aegilops tauschii. PLoS ONE 2017, 12, e0189333. [Google Scholar] [CrossRef] [PubMed]
- Tribhuvan, K.U.; Kaila, T.; Srivastava, H.; Das, A.; Kumar, K.; Durgesh, K.; Joshi, R.; Singh, B.K.; Singh, N.K.; Gaikwad, K. Structural and functional analysis of CCT family genes in pigeonpea. Mol. Biol. Rep. 2022, 49, 217–226. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Zhang, S.; Du, K.; Kang, X. Genome-wide identification, characterization, and expression analysis of CCT transcription factors in poplar. Plant Physiol. Biochem. 2023, 204, 108101. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.Y.; Yan, W.Y.; Chang, H.Y.; Sun, C.W. Arabidopsis CIA2 and CIL have distinct and overlapping functions in regulating chloroplast and flower development. Plant Direct 2022, 6, e380. [Google Scholar] [CrossRef] [PubMed]
- Masaki, T.; Tsukagoshi, H.; Mitsui, N.; Nishii, T.; Hattori, T.; Morikami, A.; Nakamura, K. Activation tagging of a gene for a protein with novel class of CCT-domain activates expression of a subset of sugar-inducible genes in Arabidopsis thaliana. Plant J. 2005, 43, 142–152. [Google Scholar] [CrossRef] [PubMed]
- Osella, A.V.; Mengarelli, D.A.; Mateos, J.; Dong, S.; Yanovsky, M.J.; Balazadeh, S.; Valle, E.M.; Zanor, M.I. FITNESS, a CCT domain-containing protein, deregulates reactive oxygen species levels and leads to fine-tuning trade-offs between reproductive success and defence responses in Arabidopsis. Plant Cell Environ. 2018, 41, 2328–2341. [Google Scholar] [CrossRef]
- Weng, X.; Wang, L.; Wang, J.; Hu, Y.; Du, H.; Xu, C.; Xing, Y.; Li, X.; Xiao, J.; Zhang, Q. Grain number, plant height, and heading date7 is a central regulator of growth, development, and stress response. Plant Physiol. 2014, 164, 735–747. [Google Scholar] [CrossRef]
- Fedoroff, N.V.; Battisti, D.S.; Beachy, R.N.; Cooper, P.J.; Fischhoff, D.A.; Hodges, C.N.; Knauf, V.C.; Lobell, D.; Mazur, B.J.; Molden, D.; et al. Radically rethinking agriculture for the 21st century. Science 2010, 327, 833–834. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Lu, G.; Long, W.; Zou, X.; Li, F.; Nishio, T. Recent progress in drought and salt tolerance studies in Brassica crops. Breed. Sci. 2014, 64, 60–73. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.C.H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Zhang, B.; Gu, G.; Yuan, J.; Shen, S.; Jin, L.; Lin, Z.; Lin, J.; Xie, X. Genome-wide identification and expression analysis of the R2R3-MYB gene family in tobacco (Nicotiana tabacum L.). BMC Genom. 2022, 23, 432. [Google Scholar] [CrossRef] [PubMed]
- Ding, F.R.L.; Xie, F.; Wang, M.; Zhang, S. Jasmonate and Melatonin Act Synergistically to Potentiate Cold Tolerance in Tomato Plants. Front. Plant Sci. 2022, 12, 763284. [Google Scholar] [CrossRef]
- Han, Z.; Zhang, C.; Zhang, H.; Duan, Y.; Zou, Z.; Zhou, L.; Zhu, X.; Fang, W.; Ma, Y. CsMYB Transcription Factors Participate in Jasmonic Acid Signal Transduction in Response to Cold Stress in Tea Plant (Camellia sinensis). Plants 2022, 11, 2869. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Cui, X.; Zhao, C.; Shi, L.; Zhang, G.; Sun, F.; Cao, X.; Yuan, L.; Xie, Q.; Xu, X. COR27 and COR28 encode nighttime repressors integrating Arabidopsis circadian clock and cold response. J. Integr. Plant Biol. 2017, 59, 78–85. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Carlsson, J.; Takeuchi, T.; Newton, L.; Farre, E.M. Direct regulation of abiotic responses by the Arabidopsis circadian clock component PRR7. Plant J. 2013, 76, 101–114. [Google Scholar] [CrossRef] [PubMed]
- Nemhauser, J.L.; Hong, F.; Chory, J. Different plant hormones regulate similar processes through largely nonoverlapping transcriptional responses. Cell 2006, 126, 467–475. [Google Scholar] [CrossRef] [PubMed]
- Song, X.; Duan, W.; Huang, Z.; Liu, G.; Wu, P.; Liu, T.; Li, Y.; Hou, X. Comprehensive analysis of the flowering genes in Chinese cabbage and examination of evolutionary pattern of CO-like genes in plant kingdom. Sci. Rep. 2015, 5, 14631. [Google Scholar] [CrossRef]
- Kim, J.A.; Kim, J.S.; Hong, J.K.; Lee, Y.H.; Choi, B.S.; Seol, Y.J.; Jeon, C.H. Comparative mapping, genomic structure, and expression analysis of eight pseudo-response regulator genes in Brassica rapa. Mol. Genet. Genom. 2012, 287, 373–388. [Google Scholar] [CrossRef] [PubMed]
- Cheng, F.; Wu, J.; Wang, X. Genome triplication drove the diversification of Brassica plants. Hortic. Res. 2014, 1, 14024. [Google Scholar] [CrossRef] [PubMed]
- Waadt, R.; Seller, C.A.; Hsu, P.K.; Takahashi, Y.; Munemasa, S.; Schroeder, J.I. Plant hormone regulation of abiotic stress responses. Nat. Rev. Mol. Cell Biol. 2022, 23, 680–694. [Google Scholar] [CrossRef] [PubMed]
- Hong, J.H.; Seah, S.W.; Xu, J. The root of ABA action in environmental stress response. Plant Cell Rep. 2013, 32, 971–983. [Google Scholar] [CrossRef] [PubMed]
- Sah, S.K.; Reddy, K.R.; Li, J. Abscisic Acid and Abiotic Stress Tolerance in Crop Plants. Front. Plant Sci. 2016, 7, 571. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Jia, W.; Yang, J.; Ismail, A.M. Role of ABA in integrating plant responses to drought and salt stresses. Field Crops Res. 2006, 97, 111–119. [Google Scholar] [CrossRef]
- Zhu, J.K. Salt and drought stress signal transduction in plants. Annu. Rev. Plant Biol. 2002, 53, 247–273. [Google Scholar] [CrossRef] [PubMed]
- Ikegami, K.; Okamoto, M.; Seo, M.; Koshiba, T. Activation of abscisic acid biosynthesis in the leaves of Arabidopsis thaliana in response to water deficit. J. Plant Res. 2009, 122, 235–243. [Google Scholar] [CrossRef]
- Yoshida, T.; Mogami, J.; Yamaguchi-Shinozaki, K. ABA-dependent and ABA-independent signaling in response to osmotic stress in plants. Curr. Opin. Plant Biol. 2014, 21, 133–139. [Google Scholar] [CrossRef]
- Kalinina, E.B.; Keith, B.K.; Kern, A.J.; Dyer, W.E. Salt- and osmotic stress-induced choline monooxygenase expression in Kochia scoparia is ABA-independent. Biol. Plant. 2012, 56, 699–704. [Google Scholar] [CrossRef]
- Munemasa, S.; Mori, I.C.; Murata, Y. Methyl jasmonate signaling and signal crosstalk between methyl jasmonate and abscisic acid in guard cells. Plant Signal Behav. 2011, 6, 939–941. [Google Scholar] [CrossRef]
- Yu, X.; Zhang, W.; Zhang, Y.; Zhang, X.; Lang, D.; Zhang, X. The roles of methyl jasmonate to stress in plants. Funct. Plant Biol. 2019, 46, 197–212. [Google Scholar] [CrossRef]
- Tayyab, N.; Naz, R.; Yasmin, H.; Nosheen, A.; Keyani, R.; Sajjad, M.; Hassan, M.N.; Roberts, T.H. Combined seed and foliar pre-treatments with exogenous methyl jasmonate and salicylic acid mitigate drought-induced stress in maize. PLoS ONE 2020, 15, e0232269. [Google Scholar] [CrossRef] [PubMed]
- Xiong, B.; Wang, Y.; Zhang, Y.; Ma, M.; Gao, Y.; Zhou, Z.; Wang, B.; Wang, T.; Lv, X.; Wang, X.; et al. Alleviation of drought stress and the physiological mechanisms in Citrus cultivar (Huangguogan) treated with methyl jasmonate. Biosci. Biotechnol. Biochem. 2020, 84, 1958–1965. [Google Scholar] [CrossRef]
- Jin, Y.; Ding, X.; Li, J.; Guo, Z. Isolation and characterization of wheat ice recrystallisation inhibition gene promoter involved in low temperature and methyl jasmonate responses. Physiol. Mol. Biol. Plants 2022, 28, 1969–1979. [Google Scholar] [CrossRef] [PubMed]
- Lantzouni, O.; Alkofer, A.; Falter-Braun, P.; Schwechheimer, C. GROWTH-REGULATING FACTORS Interact with DELLAs and Regulate Growth in Cold Stress. Plant Cell 2020, 32, 1018–1034. [Google Scholar] [CrossRef]
- Achard, P.; Renou, J.P.; Berthome, R.; Harberd, N.P.; Genschik, P. Plant DELLAs restrain growth and promote survival of adversity by reducing the levels of reactive oxygen species. Curr. Biol. 2008, 18, 656–660. [Google Scholar] [CrossRef] [PubMed]
- Achard, P.; Cheng, H.; De Grauwe, L.; Decat, J.; Schoutteten, H.; Moritz, T.; Van Der Straeten, D.; Peng, J.; Harberd, N.P. Integration of plant responses to environmentally activated phytohormonal signals. Science 2006, 311, 91–94. [Google Scholar] [CrossRef]
- Artimo, P.; Jonnalagedda, M.; Arnold, K.; Baratin, D.; Csardi, G.; de Castro, E.; Duvaud, S.; Flegel, V.; Fortier, A.; Gasteiger, E.; et al. ExPASy: SIB bioinformatics resource portal. Nucleic Acids Res. 2012, 40, W597–W603. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Rombauts, S.D.P.; Van Montagu, M.; Rouzé, P. PlantCARE, a plant cis-acting regulatory element database. Nucleic Acids Res. 1999, 27, 295–296. [Google Scholar] [CrossRef] [PubMed]
- Livak KJ, S.T. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]

| Gene Name | Gene ID | Chromosome | pI | MW (Da) | Protein (aa) | Subcellular | A. thaliana ID | A. thaliana Name |
|---|---|---|---|---|---|---|---|---|
| BrCMF1 | Bra013939 | A01:8554984-8556579 | 6.37 | 42,602.83 | 382 | Nuclear | AT4G25990 | AtCMF9(CIL) |
| BrCMF2 | Bra023461 | A02:3453187-3454201 | 7.79 | 17,412.59 | 153 | Nuclear, Cytosol | AT5G14370 | AtCMF11 |
| BrCMF3 | Bra020271 | A02:5974545-5976267 | 8.81 | 19,452.58 | 175 | Nuclear | AT5G59990 | AtCMF15 |
| BrCMF4 | Bra020461 | A02:6894679-6896952 | 7.73 | 43,853.06 | 394 | Nuclear | AT5G57180 | AtCMF14(CIA2) |
| BrCMF5 | Bra022655 | A02:8231286-8233020 | 5.16 | 30,843.57 | 274 | Nuclear | AT5G53420 | AtCMF13 |
| BrCMF6 | Bra006252 | A03:2698486-2699873 | 9.34 | 33,962.85 | 301 | Nuclear | AT5G14370 | AtCMF11 |
| BrCMF7 | Bra029069 | A03:6182682-6184469 | 4.77 | 30,419.14 | 269 | Nuclear | AT5G53420 | AtCMF13 |
| BrCMF8 | Bra001475 | A03:16536050-16537448 | 5.9 | 28,039.74 | 243 | Nuclear | AT3G12890 | AtCMF8 (ASML2) |
| BrCMF9 | Bra019134 | A03:26145861-26147506 | 6.22 | 42,959.09 | 383 | Nuclear, Chloroplast, Mitochondrion | AT4G25990 | AtCMF9(CIL) |
| BrCMF10 | Bra025502 | A04:8024942-8029130 | 5.33 | 34,785.22 | 308 | Nuclear | AT5G41380 | AtCMF12 |
| BrCMF11 | Bra021846 | A04:14851491-14853202 | 4.45 | 43,463.76 | 392 | Nuclear | AT2G33350 | AtCMF6 |
| BrCMF12 | Bra005503 | A05:5839671-5841373 | 4.68 | 44,285.42 | 396 | Nuclear | AT2G33350 | AtCMF6 |
| BrCMF13 | Bra034721 | A05:21150216-21151247 | 5.09 | 27,909.4 | 243 | Nuclear | AT3G12890 | AtCMF8 (ASML2) |
| BrCMF14 | Bra010391 | A08:14276724-14278279 | 5.06 | 28,599.34 | 252 | Nuclear | AT4G27900 | AtCMF10 |
| BrCMF15 | Bra030669 | A08:20873312-20876588 | 4.56 | 43,689.41 | 383 | Nuclear, Cytosol | AT1G07050 | AtCMF3(FITNESS) |
| BrCMF16 | Bra030574 | A08:21264649-21267272 | 4.61 | 42,706.8 | 384 | Nuclear | AT1G04500 | AtCMF1 |
| BrCMF17 | Bra027785 | A09:6208655-6210149 | 5.05 | 32,671.79 | 286 | Nuclear | AT1G63820 | AtCMF4 |
| BrCMF18 | Bra032471 | A09:36209798-36211242 | 5.83 | 43,399.58 | 378 | Nuclear | AT1G05290 | AtCMF2 |
| BrCMF19 | Bra015548 | A10:1127207-1128077 | 5.06 | 24,336.74 | 203 | Nuclear | AT1G07050 | AtCMF3(FITNESS) |
| BrCMF20 | Bra015321 | A10:2486571-2488734 | 4.48 | 43,148.13 | 386 | Nuclear | AT1G04500 | AtCMF1 |
| BrCMF21 | Bra003057 | A10:5619206-5621057 | 4.68 | 32,644.78 | 290 | Nuclear, Cytosol, Mitochondrion | AT5G53420 | AtCMF13 |
| BrCMF22 | Bra002752 | A10:7861017-7863103 | 8.95 | 43,930.57 | 393 | Nuclear | AT4G25990 | AtCMF9(CIL) |
| BrCMF23 | Bra002516 | A10:9083709-9086058 | 6.3 | 26,425.3 | 240 | Nuclear | AT5G59990 | AtCMF15 |
| BrCMF24 | Bra008763 | A10:13747298-13752870 | 9.67 | 60,150.03 | 535 | Nuclear, Chloroplast | AT5G14370 | AtCMF11 |
| BrCMF25 | Bra039351 | Scaffold000164 | 5.61 | 28,406.09 | 249 | Nuclear | AT3G12890 | AtCMF8 (ASML2) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, L.; Wu, X.; Zhang, M.; Yang, L.; Ji, Z.; Chen, R.; Cao, Y.; Huang, J.; Duan, Q. Genome-Wide Identification of BrCMF Genes in Brassica rapa and Their Expression Analysis under Abiotic Stresses. Plants 2024, 13, 1118. https://doi.org/10.3390/plants13081118
Chen L, Wu X, Zhang M, Yang L, Ji Z, Chen R, Cao Y, Huang J, Duan Q. Genome-Wide Identification of BrCMF Genes in Brassica rapa and Their Expression Analysis under Abiotic Stresses. Plants. 2024; 13(8):1118. https://doi.org/10.3390/plants13081118
Chicago/Turabian StyleChen, Luhan, Xiaoyu Wu, Meiqi Zhang, Lin Yang, Zhaojing Ji, Rui Chen, Yunyun Cao, Jiabao Huang, and Qiaohong Duan. 2024. "Genome-Wide Identification of BrCMF Genes in Brassica rapa and Their Expression Analysis under Abiotic Stresses" Plants 13, no. 8: 1118. https://doi.org/10.3390/plants13081118
APA StyleChen, L., Wu, X., Zhang, M., Yang, L., Ji, Z., Chen, R., Cao, Y., Huang, J., & Duan, Q. (2024). Genome-Wide Identification of BrCMF Genes in Brassica rapa and Their Expression Analysis under Abiotic Stresses. Plants, 13(8), 1118. https://doi.org/10.3390/plants13081118





