Analysis of Ethylene Signal Regulating Sucrose Metabolism in Strawberry Fruits Based on RNA-seq
Abstract
1. Introduction
2. Results
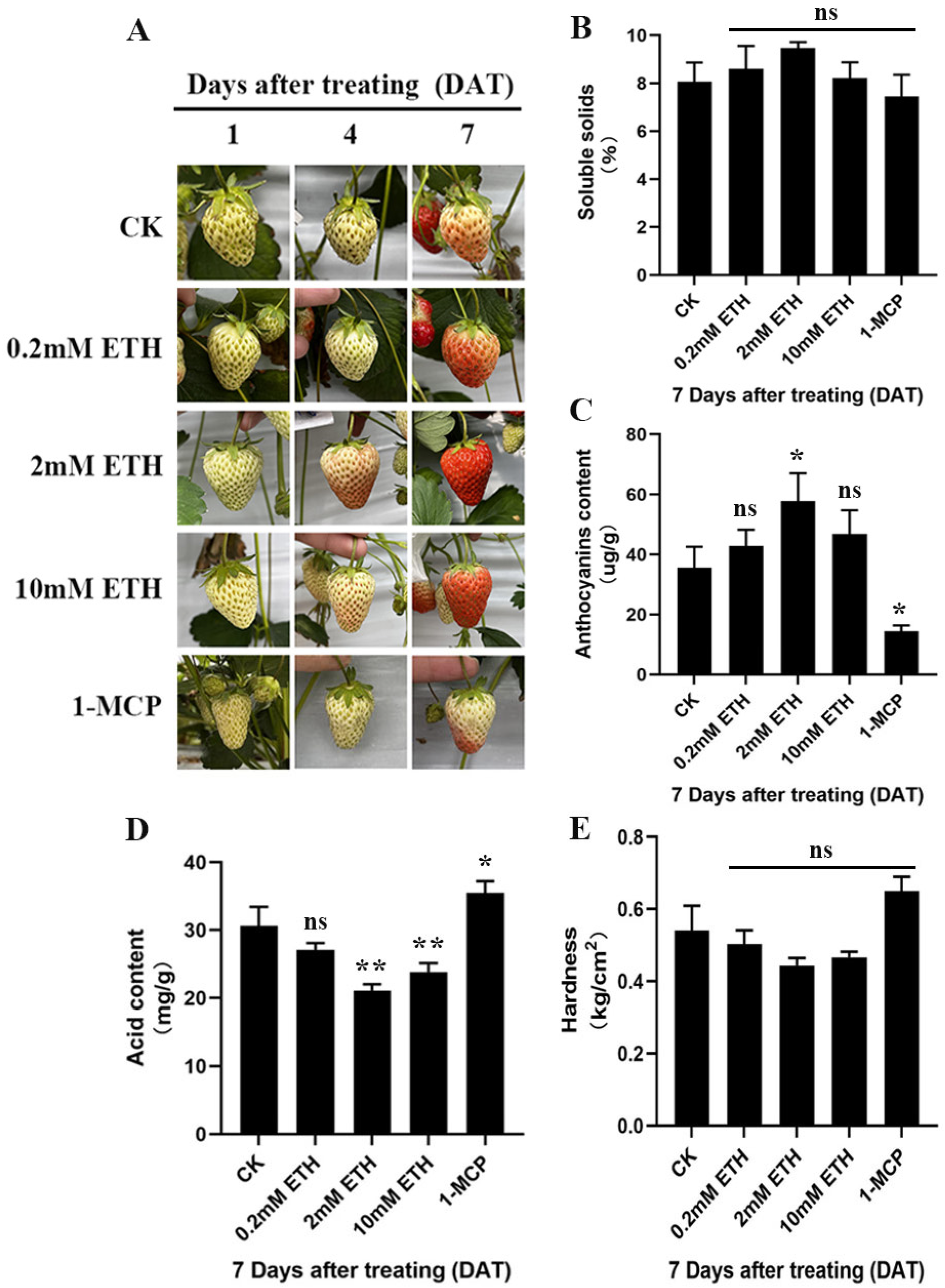
2.1. Ethylene Signal Can Affect the Ripening and Quality Formation of ‘Benihoppe’ Fruit
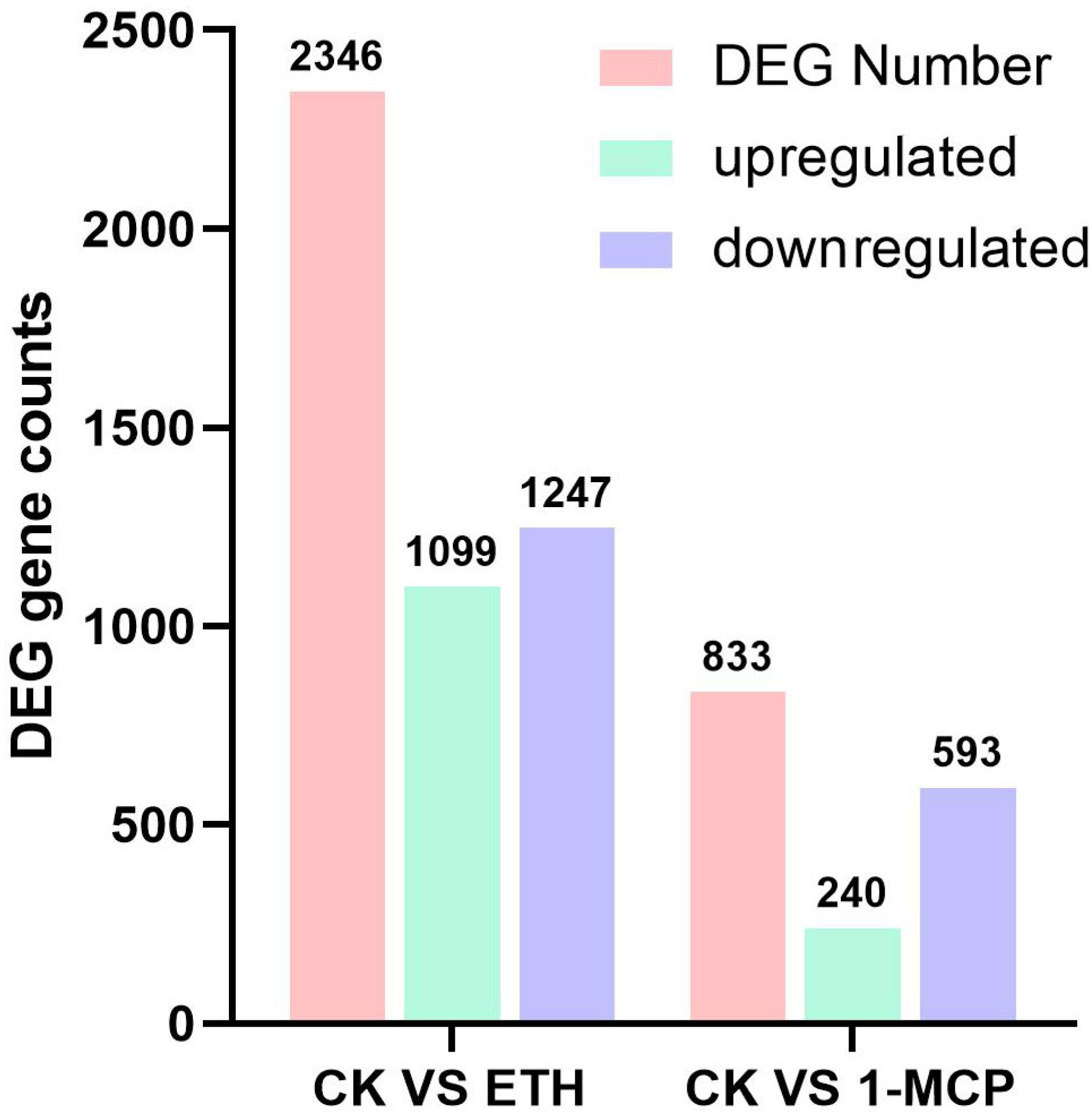
2.2. Transcriptome Analysis of Ethylene Treatment
2.2.1. Sequencing Statistics and Quality Control
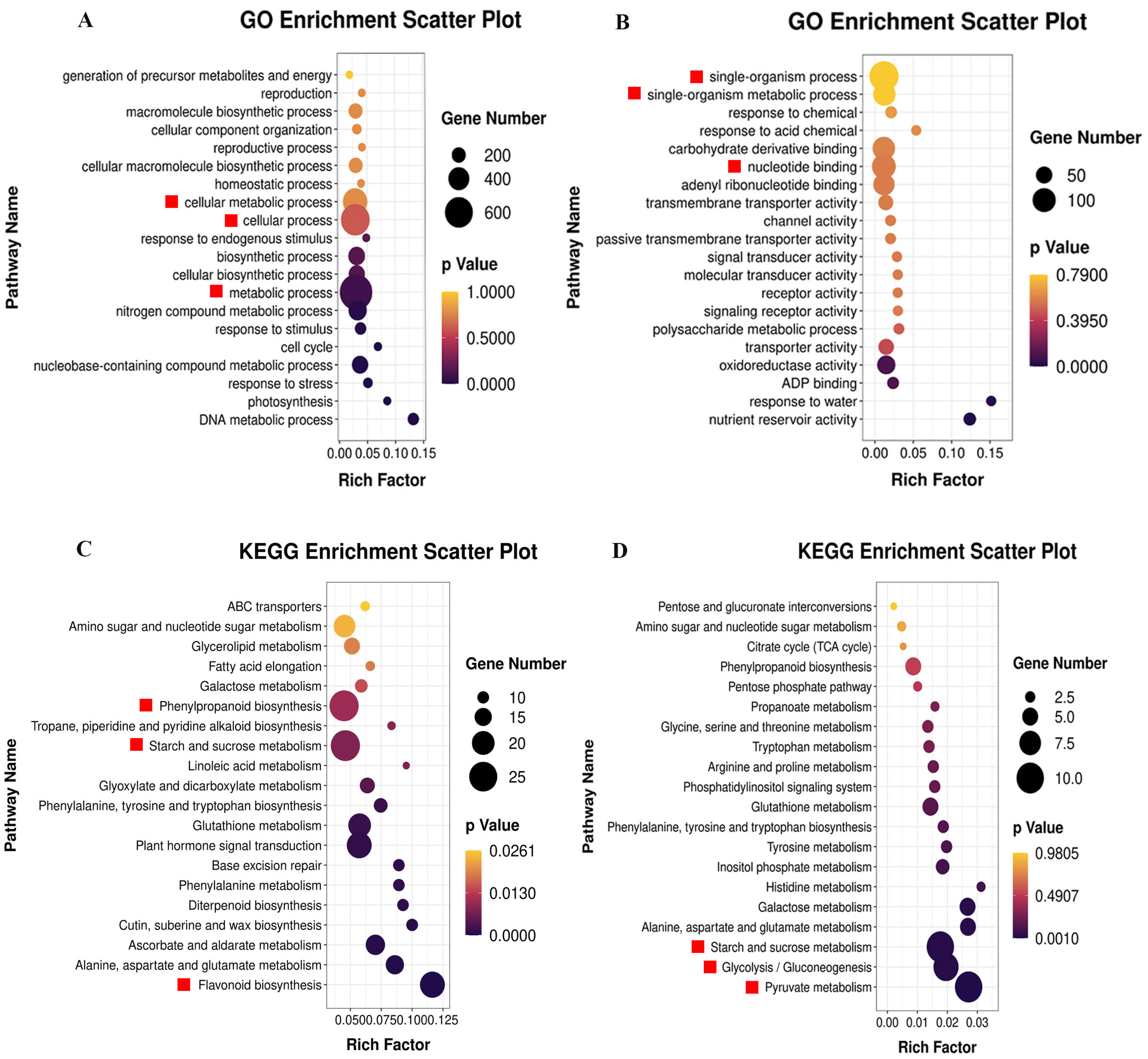
2.2.2. GO and KEGG Pathway Analysis
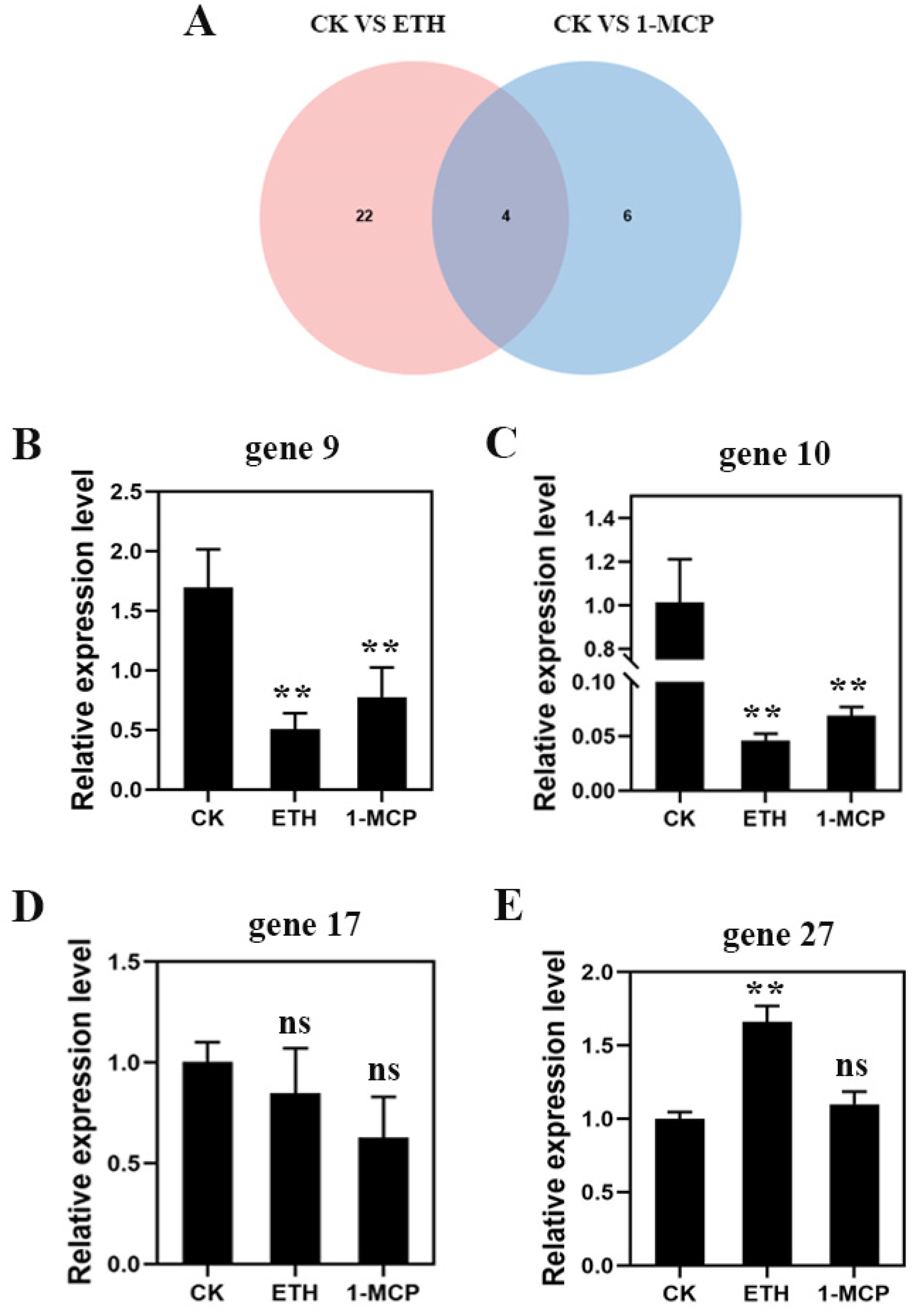
2.3. Differential Gene Analysis of Starch and Sucrose Metabolic Pathway
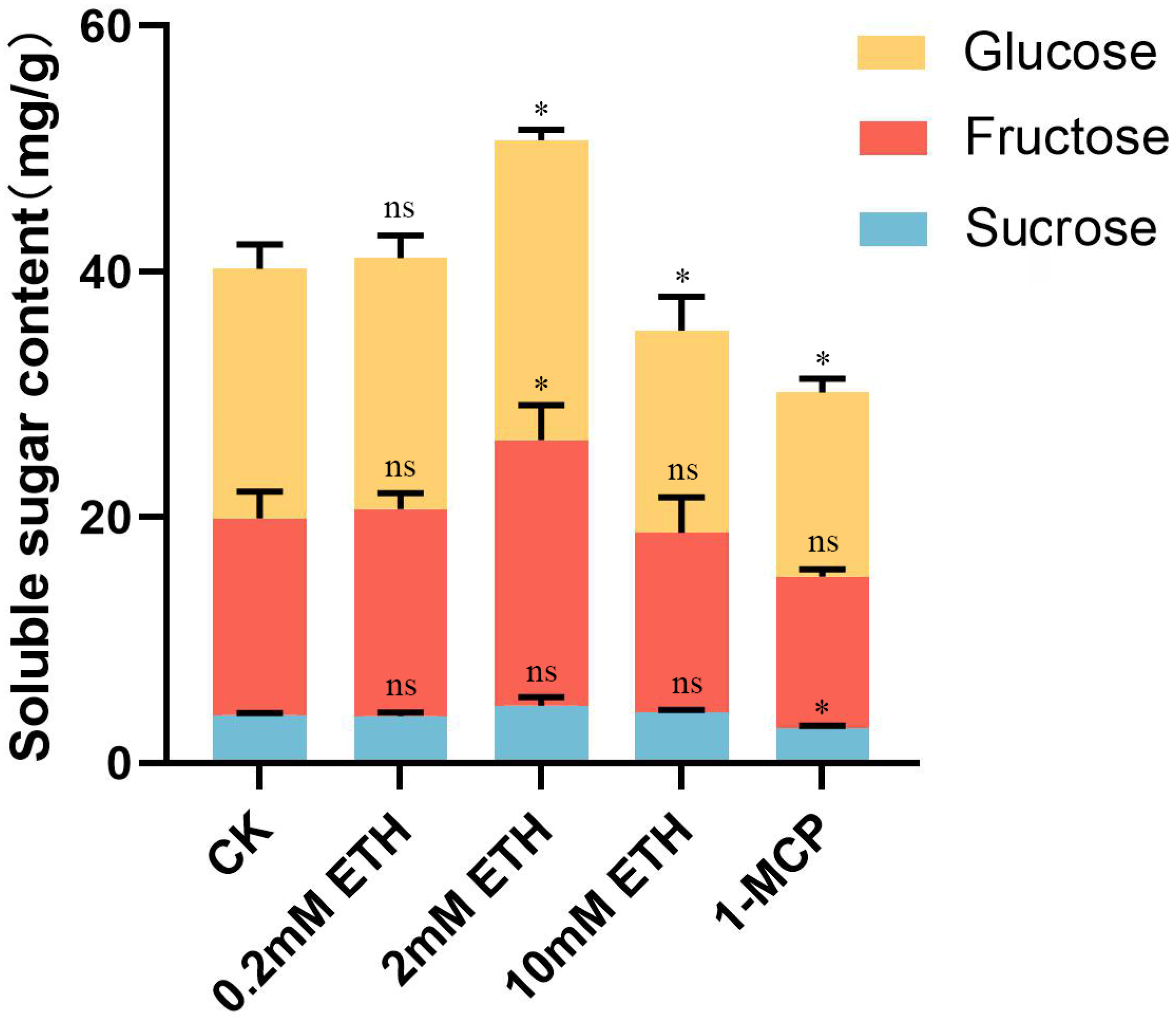
2.4. Effect of Ethylene Treatment on Soluble Sugar in Strawberry Fruit
3. Materials and Methods
3.1. Transcriptome Analysis of Ethylene Treatment
3.2. Physiological Index Determination
3.2.1. Anthocyanin Content Determination
3.2.2. Determination of Fruit Hardness and Soluble Solids
3.2.3. Determination of Total Acid Content
3.3. RNA-seq Analysis
3.3.1. Transcriptome Library Construction
3.3.2. Transcriptome Data Analysis
3.4. Determination of Soluble Sugar (Sucrose, Fructose, and Glucose) Content
3.5. qRT-PCR Analysis
3.6. Statistical Analysis
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Pang, Q.; Chen, X.; Lv, J.; Li, T.; Fang, J.; Jia, H. Triacontanol Promotes the Fruit Development and Retards Fruit Senescence in Strawberry: A Transcriptome Analysis. Plants 2020, 9, 488. [Google Scholar] [CrossRef] [PubMed]
- Carrasco-Orellana, C.; Stappung, Y.; Mendez-Yañez, A.; Allan, A.C.; Espley, R.V.; Plunkett, B.J.; Moya-Leon, M.A.; Herrera, R. Characterization of a ripening-related transcription factor FcNAC1 from Fragaria chiloensis fruit. Sci. Rep. 2018, 8, 10524. [Google Scholar] [CrossRef]
- Guan, J.; Liang, X.; Gao, G.; Yang, F.; Qi, H. The interaction between CmPIF8 and CmACO1 under postharvest red light treatment might affect fruit ripening and sucrose accumulation in oriental melon fruit. Postharvest Biol. Technol. 2024, 209, 112–717. [Google Scholar] [CrossRef]
- Hu, Y.; Han, Z.; Wang, T.; Li, H.; Li, Q.; Wang, S.; Tian, J.; Wang, Y.; Zhang, X.; Xu, X.; et al. Ethylene response factor MdERF4 and histone deacetylase MdHDA19 suppress apple fruit ripening through histone deacetylation of ripening-related genes. Plant Physiol. 2022, 188, 2166–2181. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Kuang, J.; Chen, J.; Liu, X.; Xiao, Y.; Fu, C.; Wang, J.; Wu, K.; Lu, W.J. Banana transcription factor MaERF11 recruits histone deacetylase MaHDA1 and represses the expression of MaACO1 and expansins during fruit ripening. Plant Physiol. 2016, 171, 1070–1084. [Google Scholar] [CrossRef] [PubMed]
- Lü, P.; Yu, S.; Zhu, N.; Chen, Y.R.; Zhou, B.; Pan, Y.; Tzeng, D.; Fabi, J.P.; Argyris, J.; Garcia-Mas, J.; et al. Genome encode analyses reveal the basis of convergent evolution of fleshy fruit ripening. Nat. Plants 2018, 4, 784–791. [Google Scholar] [CrossRef]
- Liu, M.; Julien, P.; Christian, C.; Jean-Paul, R.; Mondher, B. Ethylene Control of Fruit Ripening: Revisiting the Complex Network of Transcriptional Regulation. Plant Physiol. 2015, 4, 2380–2390. [Google Scholar] [CrossRef]
- Perkins-Veazie, P.M. Growth and ripening of strawberry fruit. Hortic. Rev. 1995, 17, 267–297. [Google Scholar]
- Perkins-Veazie, P.M.; Huber, D.J.; Brecht, J.K. In vitro growth and ripening of strawberry fruit in presence of ACC, STS or propylene. Ann. Appl. Biol. 1996, 128, 105–116. [Google Scholar] [CrossRef]
- Iannetta, P.P.M.; Laarhovenb, L.J.; Medina-Escobar, N.; James, E.K.; McManuse, M.T.; Davies, H.V.; Harren, F.J.M. Ethylene and carbon dioxide production by developing strawberries show a correlative pattern that is indicative of ripening climacteric fruit. Physiol. Plant. 2006, 127, 247–259. [Google Scholar] [CrossRef]
- Osorio, S.; Scossa, F.; Fernie, A. Molecular regulation of fruit ripening. Front. Plant Sci. 2013, 4, 198. [Google Scholar] [CrossRef] [PubMed]
- Park, J.I.; Lee, Y.K.; Chung, W.I.; Lee, I.H.; Choi, J.H.; Lee, W.M.; Ezur, H.; Lee, S.P.; Kim, I.J. Modification of sugar composition in strawberry fruit by antisense suppression of an ADP-glucose pyrophosphorylase. Mol. Breed. 2006, 17, 269–279. [Google Scholar] [CrossRef]
- Wei, L.; Mao, W.; Jia, M.; Xing, S.; Ali, U.; Zhao, Y.; Chen, Y.; Cao, M.; Dai, Z.; Zhang, K.; et al. FaMYB44.2, a transcriptional repressor, negatively regulates sucrose accumulation in strawberry receptacles through interplay with FaMYB10. J. Exp. Bot. 2018, 69, 4805–4820. [Google Scholar] [CrossRef] [PubMed]
- Li, B.-J.; Donald, G.; Shi, Y.; Chen, K.-S. Roles of abscisic acid in regulating ripening and quality of strawberry, a model non-climacteric fruit. Hortic. Res. 2022, 9, uhac089. [Google Scholar] [CrossRef] [PubMed]
- Symons, G.M.; Chua, Y.-J.; Ross, J.J.; Quittenden, L.J.; Davies, N.W.; Reid, J.B. Hormonal changes during non-climacteric ripening in strawberry. J. Exp. Bot. 2012, 63, 4741–4750. [Google Scholar] [CrossRef]
- Mou, W.S.; Li, D.D.; Luo, Z.S.; Li, L.; Mao, L.; Ying, T. SlAREB1 transcriptional activation of NOR is involved in abscisic acid-modulated ethylene biosynthesis during tomato fruit ripening. Plant Sci. 2018, 276, 239–249. [Google Scholar] [CrossRef]
- Liu, M.; Chen, Y.; Chen, Y.; Shin, J.H.; Mila, I.; Audran, C.; Zouine, M.; Pirrello, J.; Bouzayen, M. The tomato ethylene response factor Sl-ERF.B3 integrates ethylene and auxin signaling via direct regulation of Sl-Aux/IAA27. New Phytol. 2018, 219, 631–640. [Google Scholar] [CrossRef]
- Gambhir, P.; Singh, V.; Parida, A.; Raghuvanshi, U.; Kumar, R.; Sharma, A.K. Ethylene response factor ERF.D7 activates auxin response factor 2 paralogs to regulate tomato fruit ripening. Plant Physiol. 2022, 190, 2775–2796. [Google Scholar] [CrossRef]
- Li, S.J.; Yin, X.R.; Wang, W.L.; Liu, X.-F.; Zhang, B.; Chen, K.-S. Citrus CitNAC62 cooperates with CitWRKY1 to participate in citric acid degradation via up-regulation of CitAco3. J. Exp. Bot. 2017, 68, 3419–3426. [Google Scholar] [CrossRef]
- Ji, Y.L.; Xu, M.Y.; Liu, Z.; Yuan, H.; Lv, T.; Li, H.; Xu, Y.; Si, Y.; Wang, A. Nucleocytoplasmic shuttling of ethylene response factor 5 mediated by nitric oxide suppresses ethylene biosynthesis in apple fruit. New Phytol. 2022, 234, 1714–1734. [Google Scholar] [CrossRef]
- Chervin, C.; El-Kereamy, A.; Roustan, J.P.; Latché, A.; Lamon, J.; Bouzayen, M. Ethylene seems required for the berry development and ripening in grape, a non-climacteric fruit. Plant Sci. 2004, 167, 1301–1305. [Google Scholar] [CrossRef]
- Merchante, C.; Vallarino, J.G.; Aragüez, I.; Villarreal, N.; Ariza, M.T.; Martínez, G.A.; Medina-Escobar, N.; Civello, M.P.; Fernie, A.R.; Botella, M.A.; et al. Ethylene is involved in strawberry fruit ripening in an organ-specific manner. J. Exp. Bot. 2013, 64, 4421–4439. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.D.; Tang, M.F.; Liu, M.C.; Su, D.; Chen, J.; Gao, Y.; Bouzayen, M.; Li, Z. The molecular regulation of ethylene in fruit ripening. Small Methods 2020, 4, 1900485. [Google Scholar] [CrossRef]
- Beril, T.J.; Fanzone, M.; Piccill, P.; Botany, R. Solar UV-B and ABA are involved in phenol metabolism of Vitis vinfera L. increasing biosynthesis of berry skin polyphenols. J. Agric. Food Chem. 2011, 59, 4874–4884. [Google Scholar] [CrossRef] [PubMed]
- Shen, X.; Zhao, K.; Liu, L.; Zhang, K.; Yuan, H.; Liao, X. A role for PacMYBA in ABA-regulated anthocyanin biosynthesis in red-coloured sweet cherry cv. Hong Deng (Prunus avium L.). Plant Cell Physiol. 2014, 55, 862–880. [Google Scholar] [CrossRef]
- Oh, H.D. Abscisic Acid Stimulates Softening and Pigmentation of ‘Jersey’ Highbush Blueberry Fruits during Ripening. Master’s Thesis, Seoul National University, Seoul, Republic of Korea, 2017. [Google Scholar]
- Jia, H.; Chai, Y.; Li, C.; Lu, D.; Luo, J.; Qin, L. Abscisic acid plays an important role in the regulation of strawberry fruit ripening. Plant Physiol. 2011, 157, 183–199. [Google Scholar] [CrossRef] [PubMed]
- Oh, H.D.; Yu, D.; Chung, S.; Chea, S.; Lee, H. Abscisic acid stimulates anthocyanin accumulation in ‘Jersey’ highbush blueberry fruits during ripening. Food Chem. 2018, 244, 403–407. [Google Scholar] [CrossRef]
- Trainotti, L.; Pavanello, A.; Casadoro, G. Different ethylene receptors show an increased expression during the ripening of strawberries: Does such an increment imply a role for ethylene in the ripening of these non-climacteric fruits? J. Exp. Bot. 2005, 56, 2037–2046. [Google Scholar] [CrossRef]
- Zhang, X.R.; Luo, G.G.; Wang, R.H.; Wang, J.; Himelrick, D.G. Growth the developmental responses of seeded and seedless grape berries to shoot girdling. J. Am. Soc. Hortic. Sci. 2003, 128, 316–323. [Google Scholar] [CrossRef]
- Mukkun, L.; Singh, Z. Methyl jasmonate plays a role in fruit ripening of ‘Pajaro’ strawberry through stimulation of ethylene biosynthesis. Sci. Hortic. 2009, 123, 5–10. [Google Scholar] [CrossRef]
- Yamasaki, A.; Yamashita, M. Changes in endogenous cytokinins during flower induction of strawberry. Acta Hortic. 1993, 345, 93–99. [Google Scholar] [CrossRef]
- Eshghi, S.; Tafazoli, E. Possible role of cytokinins in flower induction in strawberry. Am. J. Plant Physiol. 2007, 2, 167–174. [Google Scholar] [CrossRef]
- Tosetti, R.; Elmi, F.; Pradas, I.; Cools, K.; Terry, L.A. Continuous exposure to ethylene differentially affects senescence in receptacle and achene tissues in strawberry fruit. Front. Plant Sci. 2020, 11, 174. [Google Scholar] [CrossRef] [PubMed]

| Sample ID | Read Sum | Base Sum | GC (%) | N (%) | Q20 (%) | Q30 (%) |
|---|---|---|---|---|---|---|
| CK-1 | 19,184,077 | 5,745,561,076 | 46.54 | 0 | 98.21 | 94.8 |
| CK-2 | 19,740,638 | 5,912,302,492 | 46.43 | 0 | 98.16 | 94.65 |
| CK-3 | 20,126,908 | 6,027,476,780 | 46.59 | 0 | 97.73 | 93.69 |
| M-1 | 21,968,697 | 6,578,189,556 | 46.46 | 0 | 98.43 | 95.42 |
| M-2 | 20,270,368 | 6,069,493,864 | 46.7 | 0 | 98.06 | 94.49 |
| M-3 | 20,371,609 | 6,101,097,966 | 46.42 | 0 | 98.08 | 94.42 |
| Y-1 | 21,581,232 | 6,464,065,172 | 46.27 | 0 | 98.11 | 94.37 |
| Y-2 | 20,591,704 | 6,166,415,648 | 46.33 | 0 | 98.29 | 94.93 |
| Y-3 | 20,438,566 | 6,120,413,870 | 46.48 | 0 | 97.86 | 93.85 |
| Enzyme | Gene ID | Gene ID |
|---|---|---|
| SPS | maker-Fvb2-2-augustus-gene-229.38 | gene 1 |
| glgC | maker-Fvb6-1-augustus-gene-76.45 | gene 2 |
| TPP | maker-Fvb7-3-snap-gene-42.66 | gene 3 |
| TPP | maker-Fvb7-1-augustus-gene-276.46 | gene 4 |
| beta-amylase | maker-Fvb7-1-augustus-gene-172.32 | gene 5 |
| beta-amylase | augustus_masked-Fvb7-3-processed-gene-118.5 | gene 6 |
| beta-amylase | maker-Fvb7-2-augustus-gene-115.58 | gene 7 |
| beta-amylase | maker-Fvb6-2-augustus-gene-329.38 | gene 8 |
| beta-amylase | maker-Fvb5-1-augustus-gene-130.60 | gene 9 |
| beta-amylase | maker-Fvb7-4-augustus-gene-113.46 | gene 10 |
| endoglucanase | augustus_masked-Fvb7-1-processed-gene-277.6 | gene 11 |
| endoglucanase | maker-Fvb2-1-augustus-gene-183.47 | gene 12 |
| endoglucanase | maker-Fvb2-3-augustus-gene-13.40 | gene 13 |
| MGAM | augustus_masked-Fvb3-3-processed-gene-44.10 | gene 14 |
| beta-glucosidase | maker-Fvb3-2-snap-gene-148.16 | gene 15 |
| beta-glucosidase | augustus_masked-Fvb3-4-processed-gene-18.7 | gene 16 |
| beta-glucosidase | maker-Fvb3-2-augustus-gene-265.38 | gene 17 |
| beta-glucosidase | maker-Fvb1-3-snap-gene-210.26 | gene 18 |
| beta-glucosidase | maker-Fvb1-3-snap-gene-210.24 | gene 19 |
| WAXY | maker-Fvb5-4-augustus-gene-52.55 | gene 20 |
| WAXY | snap_masked-Fvb5-3-processed-gene-221.46 | gene 21 |
| TPS | augustus_masked-Fvb4-3-processed-gene-14.14 | gene 22 |
| TPS | maker-Fvb5-4-augustus-gene-29.85 | gene 23 |
| TPS | maker-Fvb4-1-augustus-gene-191.38 | gene 24 |
| TPS | augustus_masked-Fvb4-4-processed-gene-21.7 | gene 25 |
| pgm | maker-Fvb7-3-augustus-gene-36.50 | gene 26 |
| pgm | maker-Fvb7-1-augustus-gene-282.41 | gene 27 |
| pgm | maker-Fvb7-3-augustus-gene-8.31 | gene 28 |
| EGLC | maker-Fvb1-4-augustus-gene-182.27 | gene 29 |
| INV | maker-Fvb7-2-snap-gene-260.73 | gene 30 |
| INV | maker-Fvb7-1-augustus-gene-276.47 | gene 31 |
| INV | maker-Fvb7-4-snap-gene-32.75 | gene 32 |
| Component | Retention Time/min | Regression Equations | Correlation Coefficient (R2) |
|---|---|---|---|
| sucrose | 7.6 | y = 55,641x + 2215.2 | 0.992 |
| fructose | 9.5 | y = 57,315x + 31,904 | 0.998 |
| glucose | 11.5 | y = 54,647x + 30,298 | 0.998 |
| Gene ID | Forward Primer Sequence (5′–3′) | Reverse Primer Sequence (5′–3′) |
|---|---|---|
| gene9 | ATGCGCTGCCGAGTATTGCT | ATGCCCTCCATGCGTCACTG |
| gene10 | TAGGAGGTTGGCGGTGGTGA | ATGACACCCAACTGCGGCAA |
| gene17 | ACGTCAAAGATGCGGGTCTGG | ACAGCCACACAGGGAGCAAA |
| gene27 | AGGAACCAGCGGTCTCCGAA | ACCATTCCCTGCGGCGATTT |
| FaActin | TGGGTTTGCTGGAGATGAT | CAGTTAGGAGAACTGGGTGC |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yu, J.-Q.; Li, Z.-T.; Chen, S.; Gao, H.-S.; Sheng, L.-X. Analysis of Ethylene Signal Regulating Sucrose Metabolism in Strawberry Fruits Based on RNA-seq. Plants 2024, 13, 1121. https://doi.org/10.3390/plants13081121
Yu J-Q, Li Z-T, Chen S, Gao H-S, Sheng L-X. Analysis of Ethylene Signal Regulating Sucrose Metabolism in Strawberry Fruits Based on RNA-seq. Plants. 2024; 13(8):1121. https://doi.org/10.3390/plants13081121
Chicago/Turabian StyleYu, Jian-Qiang, Zhao-Ting Li, Shen Chen, Hong-Sheng Gao, and Li-Xia Sheng. 2024. "Analysis of Ethylene Signal Regulating Sucrose Metabolism in Strawberry Fruits Based on RNA-seq" Plants 13, no. 8: 1121. https://doi.org/10.3390/plants13081121
APA StyleYu, J.-Q., Li, Z.-T., Chen, S., Gao, H.-S., & Sheng, L.-X. (2024). Analysis of Ethylene Signal Regulating Sucrose Metabolism in Strawberry Fruits Based on RNA-seq. Plants, 13(8), 1121. https://doi.org/10.3390/plants13081121





