The First Records of the In Silico Antiviral and Antibacterial Actions of Molecules Detected in Extracts of Algerian Fir (Abies numidica De Lannoy) Using LC-MS/MS Analysis
Abstract
:1. Introduction
2. Results
2.1. Molecular Docking
2.1.1. Antiviral Ability: Anti-SARS-CoV-2
2.1.2. In Silico Antibacterial Effect Evaluation
Escherichia coli
Staphylococcus aureus
3. Discussion
4. Materials and Methods
4.1. Ligand Preparation
4.2. Programs and Methods
4.3. Antiviral Effect
Main Protease of SARS-CoV-2 Virus (Mpro)
4.4. Antibacterial Effect
4.4.1. PRV for Escherichia coli
4.4.2. URO for Staphylococcus aureus
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mazid, M.; Khan, T.A.; Mohammad, F. Role of secondary metabolites in defense mechanisms of plants. Biol. Med. 2011, 3, 232–249. [Google Scholar]
- Quezel, P. L’endémisme dans la flore de l’Algérie. Editions Du Centre National De LA Recherche Scientifique, Quai 15 Anatole-France-Paris 7. 1964, pp. 138–149. Available online: https://www.persee.fr/doc/revec_0040-3865_1964_num_18_2_4410_t1_0238_0000_2 (accessed on 5 March 2022).
- Bennadja, S.; TliliAit-Kaki, Y. The Fir of Numidia: A Threatened Kind. Kastamonu Üniversitesi Orman Fakültesi Derg. 2012, 12, 283–286. [Google Scholar]
- Tlili-Ait Kaki, Y.; Bennadja, S.; Chefrour, A. Révalorisation d’une essence endémique: Le sapin de Numidie (Abies numidica). Flora Mediterr. 2013, 23, 123–129. [Google Scholar] [CrossRef]
- Ramdani, M.; Lograda, T.; Chalard, P.; Figueredo, G. Chemical and antimicrobial properties of essential. Int. J. Phytopharm. 2014, 5, 432–440. [Google Scholar]
- Ghadbane, M.; Bounar, R.; Khellaf, R.; Medjekal, S.; Belhadj, H.; Benderradji, L.; Smaili, T.; Harzallah, D. Antioxidant and antimicrobial activities of endemic tree Abies numidica growing in babor mountains from Algeria. Glob. J. Res. Med. Plants Indig. Med. 2016, 5, 19–28. [Google Scholar]
- Belhadj Mostefa, M.; Abedini, A.; Voutquenne-Nazabadioko, L.; Gangloff, S.C.; AhmedKabouche, K.; Kabouche, K. Abi-etane Diterpenes from the Cones of Abies numidica De Lannoy Ex Carrière (Pinaceae) and in Vitro Evaluation of Their Antimicrobial Properties. Nat. Prod. Res. 2017, 31, 568–571. [Google Scholar] [CrossRef]
- Benouchenne, D.; Khelifi, D.; Bellil, I. Phenanthroline antioxidant activity and α-glucosidase inhibition of essential oil extracted from Abies numidica leaves. NAJFNR 2019, 03, 26–27. [Google Scholar] [CrossRef]
- Benouchenne, D.; Bellil, I.; Akkal, S.; Bensouici, C.; Khelifi, D. LC–MS/MS analysis, antioxidant and antibacterial activities of Algerian fir (Abies numidica de LANNOY ex CARRIÈRE) ethyl acetate fraction extracted from needles. J. King Saud Univ.-Sci. 2020, 32, 3321–3327. [Google Scholar] [CrossRef]
- Benouchenne, D.; Bellil, I.; Bensouici, C.; Yilmaz, M.A.; Akkal, S.; Keskinkaya, H.B.; Khelifi, D. GC-MS Chemical Profile, Antioxidant Ability, Antibacterial Effect, A-Glucosidase, A-Amylase and Acetylcholinesterase Inhibitory Activity of Algerian Fir Essential Oil. Jordan J. Biol. Sci. 2022, 15, 303–310. [Google Scholar]
- Benouchenne, D.; Bellil, I.; Tachour, S.H.; Akkal, S.; Djeghim, H.; Kebaili, F.F.; Nieto, G.; Khelifi, D. Tyrosinase Inhibitory Ability and In Vitro, In Vivo Acute Oral and In Silico Toxicity Evaluation of Extracts Obtained from Algerian Fir (Abies numidica de Lannoy ex CARRIERE) Needles. Plants 2022, 11, 2389. [Google Scholar] [CrossRef]
- Yi, F.; Li, L.; Xu, L.; Meng, H.; Dong, Y.; Liu, H.; Xiao, P. In silico approach in reveal traditional medicine plants pharma-cological material basis. Chin. Med. 2018, 13, 33. [Google Scholar] [CrossRef]
- Shin, H.K.; Kang, Y.M.; No, K.T. Predicting ADME Properties of Chemicals. In Handbook of Computational Chemistry; Leszczynski, J., Kaczmarek-Kedziera, A., Puzyn, T., Papadopoulos, M.G., Reis, H., Shukla, M.H., Eds.; Springer International Publishing: Berlin/Heidelberg, Germany, 2017; pp. 2265–2301. [Google Scholar] [CrossRef]
- Dassamiour, S.; Bensaad, M.S.; Hambaba, L.; Melakhessou, M.A.; Sami, R.; Al-Mushin, A.A.M.; Aljahani, A.H.; Masoudi, M.A. In silico investigation of some compounds from N-Butanol extract of Centaurea tougourensis Boiss & Reut. Crystals 2022, 12, 355. [Google Scholar] [CrossRef]
- Ceylan, Y.; Usta, K.; Usta, A.; Maltas, E.; Yildiz, S. Evaluation of Antioxidant Activity, Phytochemicals and ESR Analysis of Lavandula stoechas. Acta Phys. Pol. 2015, 128, 483–488. [Google Scholar] [CrossRef]
- Ceylan, S.; Cetin, S.; Camadan, Y.; Saral, O.; Ozsen, O.; Tutus, A. Antibacterial and antioxidant activities of traditional medicinal plants from the Erzurum region of Turkey. Ir. J. Med. Sci. 2019, 188, 1303–1309. [Google Scholar] [CrossRef]
- Bucao, X.E.; Solidum, J. In-Silico Evaluation of Antidiabetic Activity and ADMET Prediction of Compounds from Musa acuminata Colla Peel. Philipp. J. Sci. 2021, 151, 171–192. [Google Scholar] [CrossRef]
- Ntie-Kang, F.; Lifongo, L.L.; Mbah, J.A.; Owono Owono, L.C.; Megnassan, E.; Mbaze, L.M.; Judson, P.N.; Sippl, W.; Efange, S.M.N. In silico drug metabolism and pharmacokinetic profiles of natural products from medicinal plants in the Congo basin. Silico Pharmacol. 2013, 1, 12. [Google Scholar] [CrossRef]
- Benouchenne, D.; Bellil, I.; Akkal, S.; Khelifi, D. Investigation of Phytochemical and Evaluation of Antioxidant and Anti-bacterial Activities from Abies Extract. Sci. J. King Faisal. Univ. 2021, 22, 26–32. [Google Scholar]
- Kumar, A.; Mishra, D.C.; Angadi, U.B.; Yadav, R.; Rai, A.; Kumar, D. Inhibition Potencies of Phytochemicals Derived from Sesame Against SARS-CoV-2 Main Protease: A Molecular Docking and Simulation Study. Front. Chem. 2021, 9, 744376. [Google Scholar] [CrossRef]
- Liang, J.; Zheng, Y.; Tong, X.; Yang, N.; Dai, S. In Silico Identification of Anti-SARS-CoV-2 Medicinal Plants Using Cheminformatics and Machine Learning. Molecules 2023, 28, 208. [Google Scholar] [CrossRef] [PubMed]
- Kaul, T.N.; Middleton, E., Jr.; Ogra, P.L. Antiviral effect of flavonoids on human viruses. J. Med. Virol. 1985, 15, 71–79. [Google Scholar] [CrossRef] [PubMed]
- Cataneo, A.H.D.; Avila, E.P.; Mendes, L.A.D.; de Oliveira, V.G.; Ferraz, C.R.; de Almeida, M.V.; Frabasile, S.; dos Santos, C.N.D.; Verri, W.A.; Bordignon, J. Flavonoids as Molecules with Anti-Zika virus Activity. Front. Microbiol. 2021, 12, 710359. [Google Scholar] [CrossRef] [PubMed]
- Abian, O.; Ortega-Alarcon, D.; Jimenez-Alesanco, A.; Ceballos-Laita, L.; Vega, S.; Reyburn, H.T.; Rizzuti, B.; Velazquez-Campoy, A. Structural stability of SARS-CoV-2 3CLpro and identification of quercetin as an inhibitor by experimental screening. Int. J. Biol. Macromol. 2020, 164, 1693–1703. [Google Scholar] [CrossRef] [PubMed]
- Xiao, T.; Cui, M.; Zheng, C.; Wang, M.; Sun, R.; Gao, D.; Bao, J.; Ren, S.; Yang, B.; Lin, J. Myricetin Inhibits SARS-CoV-2 Viral Replication by Targeting M(pro) and Ameliorates Pulmonary Inflammation. Front. Pharmacol. 2021, 12, 669642. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.; Zhang, M.; Liu, H.; Wei, K.; He, M.; Li, X.; Hu, D.; Yang, S.; Zheng, Y. Antiviral activity of aconite alkaloids from Aconitum carmichaelii Debx. Nat. Prod. Res. 2019, 33, 1486–1490. [Google Scholar] [CrossRef] [PubMed]
- Matos, M.J. Coumarin and Its Derivatives-Editorial. Molecules 2021, 26, 6320. [Google Scholar] [CrossRef]
- Al-Maharik, N. Isolation of naturally occurring novel isoflavonoids: An update. Nat. Prod. Rep. 2019, 36, 1156–1195. [Google Scholar] [CrossRef]
- Maaroufi, I.; Jamsransuren, D.; Hashida, K.; Matsuda, S.; Ogawa, H.; Takeda, Y. An Abies Extract Containing Nonvolatile Polyphenols Shows Virucidal Activity against SARS-CoV-2 That Is Enhanced in Increased pH Conditions. Pathogens 2023, 12, 1093. [Google Scholar] [CrossRef]
- Murata, T.; Jamsransuren, D.; Matsuda, S.; Ogawa, H.; Takeda, Y. Rapid Virucidal Activity of Japanese Saxifraga Species-Derived Condensed Tannins against SARS-CoV-2, Influenza A Virus, and Human Norovirus Surrogate Viruses. Appl. Environ. Microbiol. 2023, 89, e0023723. [Google Scholar] [CrossRef]
- Belščak-Cvitanović, A.; Durgo, K.; Hudek, A.; Bačun-Družina, V.; Komes, D. Overview of polyphenols and their properties. In Polyphenols: Properties, Recovery, and Applications; Elsevier: Amsterdam, The Netherlands, 2018; pp. 3–44. ISBN 978-0-12-813572-3. [Google Scholar]
- Chen, R.; Wang, J.B.; Zhang, X.Q.; Ren, J.; Zeng, C.M. Green tea polyphenol epigallocatechin-3-gallate (EGCG) induced intermolecular cross-linking of membrane proteins. Arch. Biochem. Biophys. 2011, 507, 343–349. [Google Scholar] [CrossRef]
- Bolton, J.L.; Dunlap, T. Formation and biological targets of quinones: Cytotoxic versus cytoprotective effects. Chem. Res. Toxicol. 2017, 30, 13–37. [Google Scholar] [CrossRef]
- Ozdal, T.; Capanoglu, E.; Altay, F. A review on protein-phenolic interactions and associated changes. Food Res. Int. 2013, 51, 954–970. [Google Scholar] [CrossRef]
- Phan, H.T.T.; Yoda, T.; Chahal, B.; Morita, M.; Takagi, M.; Vestergaard, M.C. Structure-dependent interactions of polyphenols with a biomimetic membrane system. Biochim. Biophys. Acta BBA—Biomembr. 2014, 1838, 2670–2677. [Google Scholar] [CrossRef]
- Sydiskis, R.J.; Owen, D.G.; Lohr, J.L.; Rosler, K.H.A.; Blomster, A.R.N. Inactivation of Enveloped Viruses by Anthraquinones Extracted from Plants. Antimicrob. Agents Chemother. 1991, 35, 2463–2466. [Google Scholar] [CrossRef] [PubMed]
- Alves, D.S.; Pérez-Fons, L.; Estepa, A.; Micol, V. Membrane-related effects underlying the biological activity of the anthraquinones emodin and barbaloin. Biochem. Pharmacol. 2004, 68, 549–561. [Google Scholar] [CrossRef] [PubMed]
- Konappa, N.; Udayashankar, A.C.; Krishnamurthy, S.; Pradeep, C.K.; Chowdappa, S.; Jogaiah, S. GC–MS analysis of phytoconstituents from Amomum nilgiricum and molecular docking interactions of bioactive serverogenin acetate with target proteins. Sci. Rep. 2020, 10, 1–23. [Google Scholar] [CrossRef] [PubMed]
- Vanitha, V.; Vijayakumar, S.; Nilavukkarasi, M.; Punitha, V.; Vidhya, E.; Praseetha, P. Heneicosane—A novel microbicidal bioactive alkane identified from Plumbago zeylanica L. Ind. Crops Prod. 2020, 154, 112748. [Google Scholar] [CrossRef]
- Ralte, L.; Khiangte, L.; Thangjam, N.M.; Kumar, A.; Singh, Y.T. GC–MS and molecular docking analyses of phytochemicals from the underutilized plant, Parkia timoriana revealed candidate anti-cancerous and anti-inflammatory agents. Sci. Rep. 2022, 12, 1–21. [Google Scholar] [CrossRef] [PubMed]
- Brantner, A.; Males, Z.; Pepeljnjak, S.; Antolic, A. Antimicrobial activity of Paliurus spina-christi Mill (Christ’s thorn). J. Ethnopharmacol. 1996, 52, 119–122. [Google Scholar] [CrossRef] [PubMed]
- Askun, T.; Tumen, G.; Satil, F.; Ates, M. In vitro activity of methanol extracts of plants used as spices against Mycobacterium tuberculosis and other bacteria. Food Chem. 2009, 116, 289–294. [Google Scholar] [CrossRef]
- Bendjedid, S.; Lekmine, S.; Tadjine, A.; Djelloul, R.; Bensouici, C. Analysis of Phytochemical Constituents, Antibacterial, Antioxidant, Photoprotective Activities, and cytotoxic effect of Leaves Extracts and Fractions of Aloe vera. Biocatal. Agric. Biotechnol. 2021, 33, 101991. [Google Scholar] [CrossRef]
- Dabai, Y.U.; Muhammad, S.; Aliyu, B.S. Antibacterial activity of anthraquinone fraction of Vitex doniana. PJBS 2007, 1–3. [Google Scholar]
- Reynolds, T.; Dweck, A.C. Aloe vera leaf gel: A review update. J. Ethnopharmacol. 1999, 68, 3–37. [Google Scholar] [CrossRef] [PubMed]
- Abdelli, I.; Hassani, F.; Brikci, S.B.; Ghalem, S. In silico study the inhibition of angiotensin converting enzyme 2 receptor of COVID-19 by Ammoides verticillata components harvested from Western Algeria. J. Biomol. Struct. Dyn. 2021, 39, 3263–3276. [Google Scholar] [CrossRef] [PubMed]
- Stewart, J.J.P. MOPAC2012, Version 13.159 M. Stewart computational chemistry 28. Allen, FH.; Kennard, O.; Taylor, R 1983 Systematic analysis of structural data as a research technique in organic chemistry. Acc. Chem. Res. 2012, 16, 146–153. [Google Scholar] [CrossRef]
- Kong, R.; Yang, G.B.; Xue, R.; Liu, M.; Wang, F.; Hu, J.P.; Guo, X.Q.; Chang, S. COVID-19 Docking Server: An interactive server for docking small molecules, peptides and antibodies against potential targets of COVID-19. Bioinformatics 2020, 36, 5109–5111. [Google Scholar] [CrossRef] [PubMed]
- Guedes, I.A.; Costa, L.S.C.; Dos Santos, K.b.; Karl, A.L.M.; Rocha, G.K.; Teixeira, L.M. Drug design and repurposing with DockThor-VS web server focusing on SARS-CoV-2 therapeutic targets and their non-synonym variants. Sci. Rep. 2021, 11, 5543. [Google Scholar] [CrossRef] [PubMed]
- Wrapp, D.; Wang, N.; Corbett, K.S.; Goldsmith, J.A.; Hsieh, C.L.; Abiona, O.; Graham, B.S.; McLellan, J.S. Cryo-EM structure ofthe 2019-nCoV spike in the prefusion conformation. Science 2020, 367, 1260–1263. [Google Scholar] [CrossRef]
- Bansal, P.; Kumar, R.; Singh, J.; Dhanda, S. In silico molecular docking of SARS CoV 2 surface proteins with microbial non ribosomal peptides: Identification of potential drugs. JPP 2021, 12, 177–184. [Google Scholar] [CrossRef]
- Biovia, D.S. Discovery Studio Visualizer; Dassoult Systemes: San Diego, CA, USA, 2019. [Google Scholar]
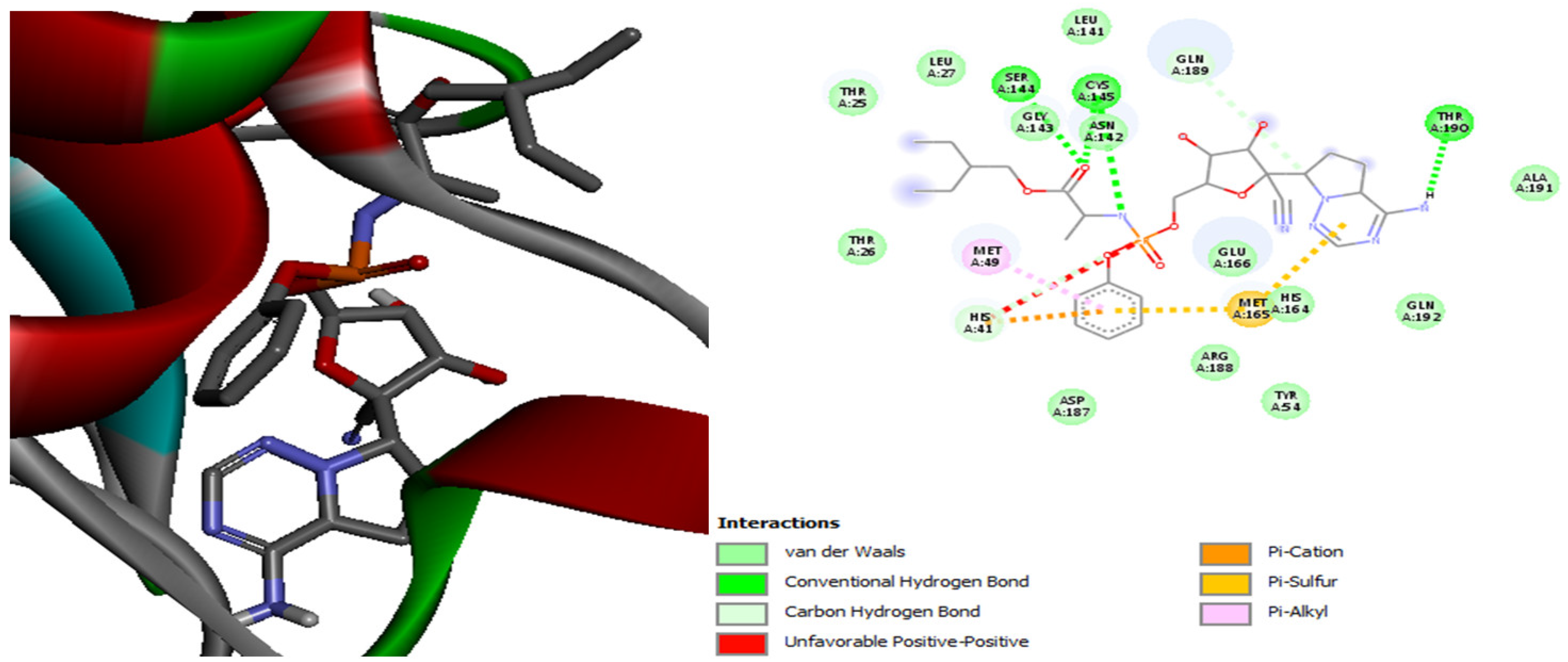
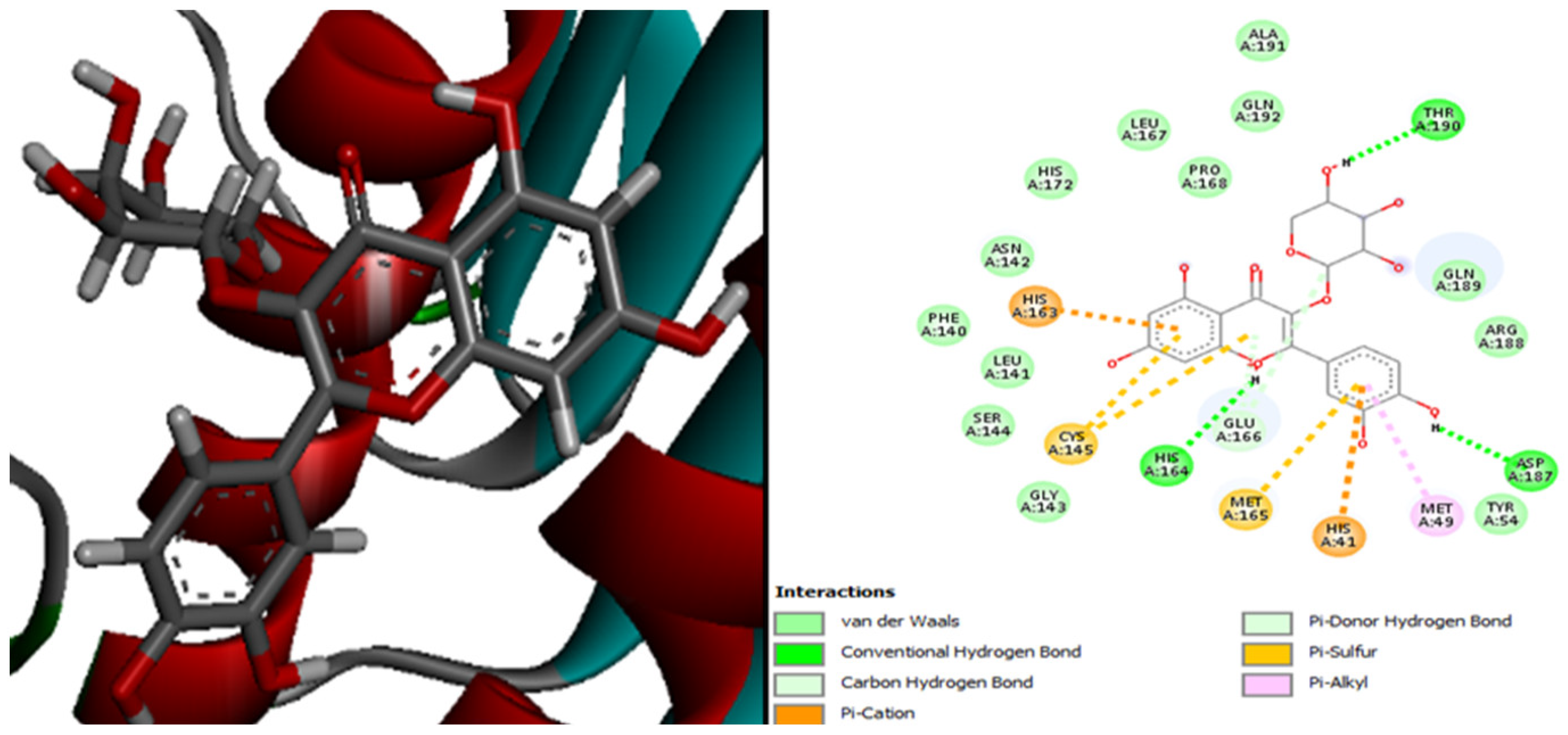
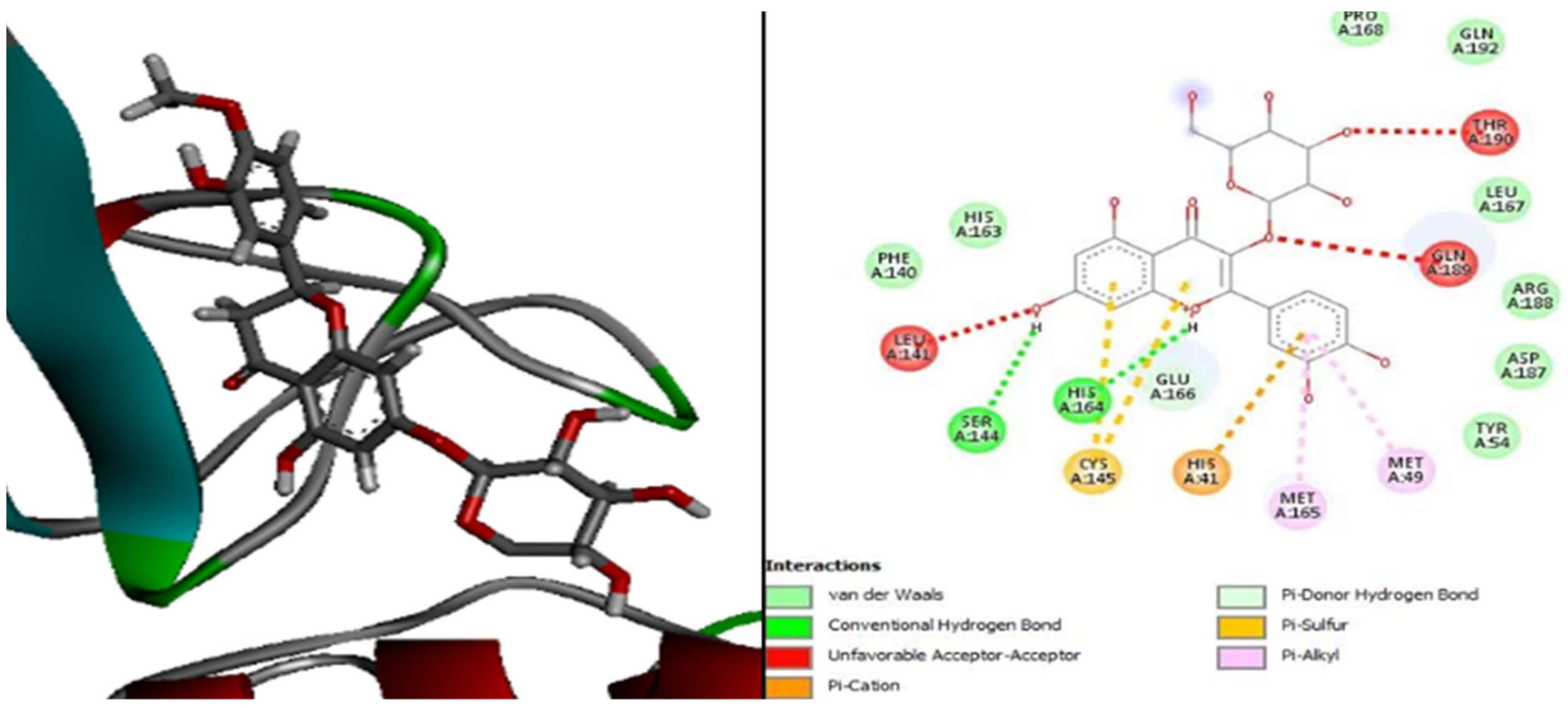

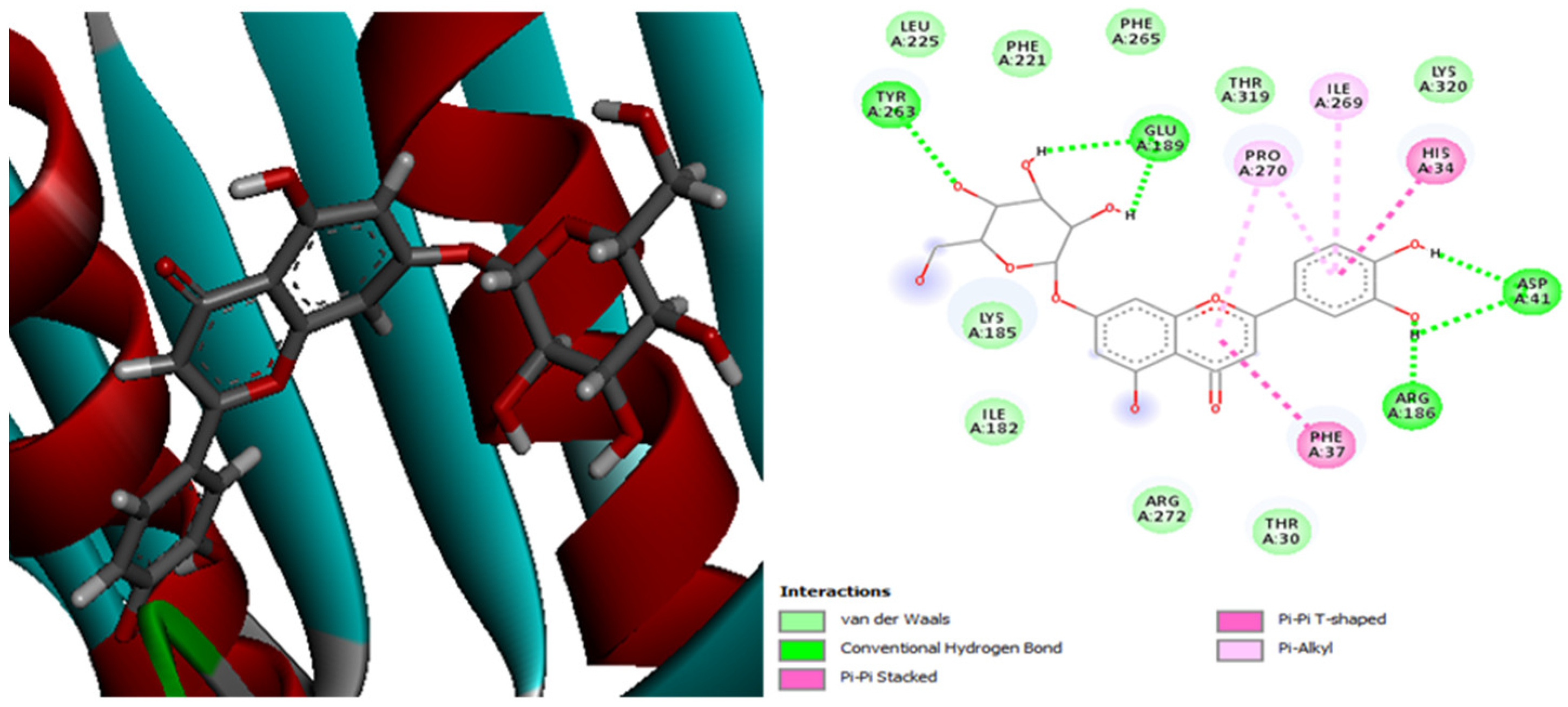

| Molecules | Score (kcal/mol) | Met A: 165 | His A: 41 | Met A: 49 | Leu A: 141 | His A: 164 | Gln A: 189 | His A: 163 | Glu A: 192 | Leu A: 167 | His A: 172 | Leu A: 27 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Hyperoside | −8.8 | + | + | + | + | + | - | - | - | - | - | - |
| Quercitrin | −8.8 | + | + | + | - | + | - | + | - | - | - | - |
| Astragalin | −8.6 | - | ++ | ++ | - | - | - | + | - | - | - | - |
| Rutin | −8.4 | + | +++ | + | - | - | - | - | - | - | - | - |
| Hesperidin | −8.2 | + | - | - | - | + | - | - | ++ | + | - | - |
| Apigetrin | −8 | + | - | - | + | - | + | - | - | - | - | - |
| Quercetin | −8 | + | + | + | + | + | + | - | - | - | - | - |
| Apigenin | −7.9 | + | + | + | + | + | - | + | - | - | - | - |
| Luteolin | −7.9 | + | + | + | + | + | - | - | - | - | - | - |
| Luteolin-7-glucoside | −7.9 | ++ | - | - | - | - | ++ | + | - | - | - | - |
| Remdesivir | −7.70 | ++ | +++ | + | - | - | + | - | - | - | - | - |
| Chlorogenic acid | −7.4 | + | - | - | + | - | - | - | - | - | - | - |
| Protocatechuic acid | −5.3 | - | - | - | + | - | - | - | - | - | - | - |
| Compounds | Score (kcal/mol) | Glu A: 189 | ArgA: 272 | Lys A: 185 | Arg A: 186 | Ile A: 182 | His A: 34 | Phe A: 37 | Thr A: 319 | Thr A: 30 | Phe A: 265 | Phe A: 221 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Luteolin-7-glucoside | −9.109 | ++ | - | - | + | - | + | + | - | - | - | - |
| Tigecycline | −9.027 | - | + | - | - | - | - | - | - | - | + | - |
| Rutin | −8.547 | +++ | + | - | - | - | - | - | - | - | - | - |
| Apigetrin | −8.483 | ++ | ++ | - | - | - | - | - | - | - | - | - |
| Hyperoside | −8.451 | + | - | - | - | - | - | - | - | - | - | - |
| Hesperidin | −8.443 | ++ | - | ++++ | - | - | - | - | - | - | - | - |
| Astragalin | −7.890 | + | + | + | - | - | - | - | - | + | - | - |
| Quercitrin | −7.765 | + | ++ | - | - | + | - | - | + | - | - | - |
| Luteolin | −7.656 | + | ++ | + | - | - | - | + | - | - | - | - |
| Quercetin | −7.632 | ++ | + | - | + | + | - | - | - | - | - | - |
| Apigenin | −7.614 | ++ | + | - | + | + | + | - | - | - | - | - |
| Chlorogenic Acid | −7.544 | + | + | + | - | - | - | - | - | - | - | - |
| Protocatechuic Acid | −7.030 | - | - | - | + | - | + | - | + | - | - | - |
| Compounds | Score (kcal/mol) | Arg A: 154 | Glu A: 25 | Asp A: 32 | Ile A: 61 | Arg A: 59 | Gly A: 81 | Glu A: 33 | Gly A: 83 | Ile A: 11 | Gly A: 58 | Ile A: 77 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Hesperidin | −7.941 | - | - | - | ++ | +++ | - | + | - | - | - | - |
| Luteolin-7-glucoside | −7.681 | ++ | + | - | - | - | - | - | - | + | - | - |
| Quercetin | −7.678 | ++ | - | - | - | - | - | - | - | - | - | - |
| Apigetrin | −7.645 | - | ++ | ++ | + | - | ++ | - | + | - | - | - |
| Quercitrin | −7.598 | - | - | + | ++ | + | - | + | - | - | - | - |
| Hyperoside | −7.473 | ++ | ++ | - | - | - | - | - | - | - | - | - |
| Apigenin | −7.471 | - | - | + | ++ | - | - | + | - | - | - | - |
| Chlorogenic acid | −6.798 | + | + | ++ | - | - | + | - | + | - | - | - |
| Penicillin | −6.704 | + | + | - | - | - | - | - | - | - | - | - |
| Luteolin | −6.660 | ++ | + | - | - | - | - | - | - | + | - | - |
| Astragalin | −6.587 | - | - | - | - | - | - | - | - | - | - | - |
| Rutin | −6.340 | + | + | + | - | - | - | - | - | - | - | - |
| Protocatechuic acid | −5.926 | - | - | - | - | - | - | - | - | - | - | - |
| No. | Analytes | RT a | Precursor ion (m/z) b | Fragmentations | Ionization mode | Equation | c R2 | U f | n-BuOH (µg/g) | EAF (µg/g) |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Protocatechuic acid | 7.00 | 153.4 | 109.0–108.0 | Neg | y = 590.460x + 120.26 | 0.9909 | 0.0215 | N.D | 71.62 |
| 2 | Chlorogenic acid | 8.03 | 353.3 | 191.2–85.0 | Neg | y = 697.935x + 87418.5 | 0.9910 | 0.0299 | 9.66 | 15.59 |
| 3 | Luteolin-7-glucoside | 13.20 | 447.0 | 285.1–284.1 | Neg | y = 215.412x + 36852.1 | 0.9939 | 0.0086 | 14.6 | 43.17 |
| 4 | Rutin | 13.67 | 609.1 | 300.1–301.1 | Neg | y = 469.333x + 30144.8 | 0.9902 | 0.0136 | 102.62 | 27.58 |
| 5 | Hesperidin | 13.68 | 611.1 | 303.0–449.3 | Poz | y = 2539.52x + 123981 | 0.9942 | 0.0162 | 42.02 | 7.83 |
| 6 | Hyperoside | 13.69 | 463.0 | 300.1–271.0 | Neg | y = 185.593x + 8126.67 | 0.9905 | 0.0126 | 399.91 | 3370.96 |
| 7 | Apigetrin | 14.54 | 431.0 | 268.1–269.1 | Neg | y = 1052.01x + 146897 | 0.9902 | 0.0132 | 23.19 | 192.56 |
| 8 | Quercitrin | 14.98 | 447.0 | 300.0–301.1 | Neg | y = 175.298x + 33626.6 | 0.9918 | 0.0133 | 20.44 | 2300.33 |
| 9 | Astragalin | 15.13 | 447.0 | 284.1–227.1 | Neg | y = 329.506x + 44598.6 | 0.9900 | 0.0153 | 147.22 | 3391.36 |
| 10 | Quercetin | 17.10 | 301.2 | 151.1–179.1 | Neg | y = 1826.89x − 146948 | 0.9962 | 0.0573 | N.D | 24.75 |
| 11 | Luteolin | 17.78 | 285.2 | 133.1–151.0 | Neg | y = 3166.03x + 495252 | 0.9901 | 0.0188 | N.D | 2.41 |
| 12 | Apigenin | 19.20 | 269.2 | 117.0–151.1 | Neg | y = 3115.89x + 483037 | 0.9910 | 0.0181 | N.D | 6.64 |
| 13 | Pseudohypericin | 26.34 | 519.0 | 487.1–475.1 | Neg | y = 2548.96x + 468900 | 0.9908 | 0.0172 | N.D | N.D. |
| 14 | Hyperforin | 28.97 | 535.3 | 383.3–315.2 | Neg | y = 44260.6x + 203394 | 0.9901 | 0.0418 | N.D | N.D. |
| 15 | Hypericin | 30.18 | 503.0 | 405.1–433.1 | Neg | y = 7676.03x + 605593 | 0.9925 | 0.0189 | N.D | N.D. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Benouchenne, D.; Bellil, I.; Bendjedid, S.; Ramos, A.; Nieto, G.; Akkal, S.; Khelifi, D. The First Records of the In Silico Antiviral and Antibacterial Actions of Molecules Detected in Extracts of Algerian Fir (Abies numidica De Lannoy) Using LC-MS/MS Analysis. Plants 2024, 13, 1246. https://doi.org/10.3390/plants13091246
Benouchenne D, Bellil I, Bendjedid S, Ramos A, Nieto G, Akkal S, Khelifi D. The First Records of the In Silico Antiviral and Antibacterial Actions of Molecules Detected in Extracts of Algerian Fir (Abies numidica De Lannoy) Using LC-MS/MS Analysis. Plants. 2024; 13(9):1246. https://doi.org/10.3390/plants13091246
Chicago/Turabian StyleBenouchenne, Djamila, Ines Bellil, Samira Bendjedid, Ana Ramos, Gema Nieto, Salah Akkal, and Douadi Khelifi. 2024. "The First Records of the In Silico Antiviral and Antibacterial Actions of Molecules Detected in Extracts of Algerian Fir (Abies numidica De Lannoy) Using LC-MS/MS Analysis" Plants 13, no. 9: 1246. https://doi.org/10.3390/plants13091246
APA StyleBenouchenne, D., Bellil, I., Bendjedid, S., Ramos, A., Nieto, G., Akkal, S., & Khelifi, D. (2024). The First Records of the In Silico Antiviral and Antibacterial Actions of Molecules Detected in Extracts of Algerian Fir (Abies numidica De Lannoy) Using LC-MS/MS Analysis. Plants, 13(9), 1246. https://doi.org/10.3390/plants13091246






