Genetic Diversity of the Collection of Far Eastern Actinidia spp. Revealed by RAD Sequencing Technology
Abstract
:1. Introduction
2. Results
2.1. Sequencing Data Analysis
2.2. Ploidy Level
2.3. Genetic Structure of the Far Eastern Actinidia Collection
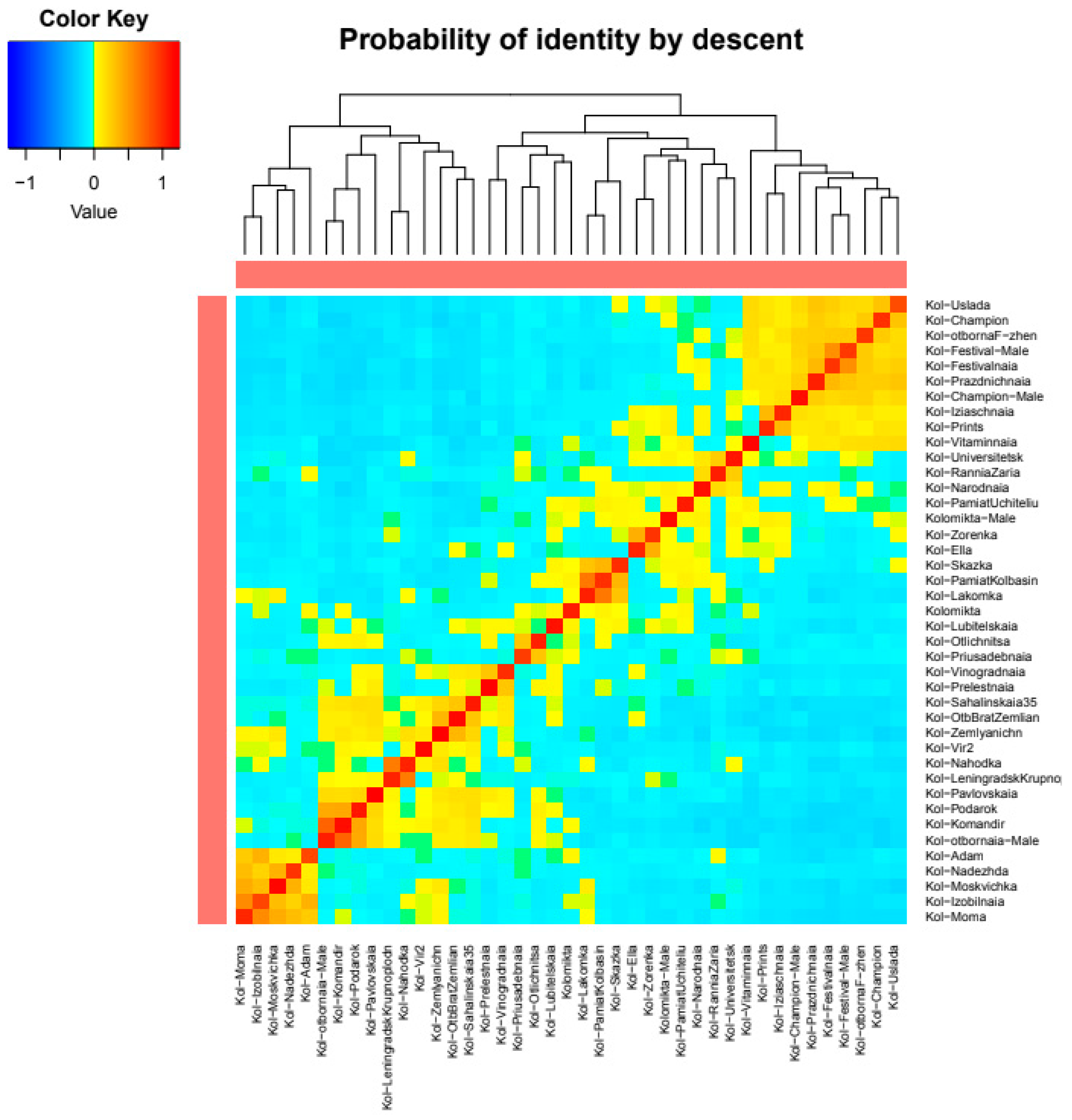
2.4. Intraspecific Genetic Structure of the Far Eastern Actinidia Collection
2.5. A. kolomikta Population with Non-Dropping Fruits
3. Discussion
4. Materials and Methods
4.1. Plant Material and DNA Extraction
4.2. Preparation of RAD Libraries and Sequencing
4.3. Data Analysis
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Huang, H. Natural distribution of genus actinidia. In Kiwifruit; Elsevier: Amsterdam, The Netherlands, 2016; pp. 169–190. ISBN 9780128030660. [Google Scholar]
- Huang, H.; Wang, Y.; Zhang, Z.; Jiang, Z.; Wang, S. Actinidia germplasm resources and kiwifruit industry in china. HortScience 2004, 39, 1165–1172. [Google Scholar] [CrossRef]
- Waswa, E.N.; Ding, S.-X.; Wambua, F.M.; Mkala, E.M.; Mutinda, E.S.; Odago, W.O.; Amenu, S.G.; Muthui, S.W.; Linda, E.L.; Katumo, D.M.; et al. The genus Actinidia Lindl. (Actinidiaceae): A comprehensive review on its ethnobotany, phytochemistry, and pharmacological properties. J. Ethnopharmacol. 2024, 319, 117222. [Google Scholar] [CrossRef] [PubMed]
- Pinto, D.; Delerue-Matos, C.; Rodrigues, F. Bioactivity, phytochemical profile and pro-healthy properties of Actinidia arguta: A review. Food Res. Int. 2020, 136, 109449. [Google Scholar] [CrossRef] [PubMed]
- Ferguson, A.R. 1904—The year that kiwifruit (Actinidia deliciosa) came to New Zealand. N. Z. J. Crop. Hortic. Sci. 2004, 32, 3–27. [Google Scholar] [CrossRef]
- Guroo, I.; Wani, S.A.; Wani, S.M.; Ahmad, M.; Mir, S.A.; Masoodi, F.A. A Review of Production and Processing of Kiwifruit. J. Food Process. Technol. 2017, 8, 1000699. [Google Scholar]
- Česonienė, L.; Januškevičė, V.; Saunoriūtė, S.; Liaudanskas, M.; Žvikas, V.; Krikštolaitis, R.; Viškelis, P.; Urbonavičienė, D.; Martusevičė, P.; Zych, M.; et al. Phenolic Compounds in Berries of Winter-Resistant Actinidia arguta Miq. and Actinidia kolomikta Maxim.: Evidence of Antioxidative Activity. Antioxidants 2024, 13, 372. [Google Scholar] [CrossRef]
- Baranowska-Wójcik, E.; Szwajgier, D. Characteristics and pro-health properties of mini kiwi (Actinidia arguta). Hortic. Environ. Biotechnol. 2019, 60, 217–225. [Google Scholar] [CrossRef]
- Sawicki, T.; Błaszczak, W.; Latocha, P. In vitro anticholinergic and antiglycaemic properties of frost-hardy Actinidia fruit extracts and their polyphenol profile, L-ascorbic acid content and antioxidant capacity. Food Res. Int. 2023, 173, 113324. [Google Scholar] [CrossRef] [PubMed]
- Sun, S.; Qi, X.; Wang, R.; Lin, M.; Fang, J. Evaluation of freezing tolerance in Actinidia germplasm based on relative electrolyte leakage. Hortic. Environ. Biotechnol. 2020, 61, 755–765. [Google Scholar] [CrossRef]
- Latocha, P. The Nutritional and Health Benefits of Kiwiberry (Actinidia arguta)—A Review. Plant Foods Hum. Nutr. 2017, 72, 325–334. [Google Scholar] [CrossRef]
- Latocha, P.; Krupa, T.; Wołosiak, R.; Worobiej, E.; Wilczak, J. Antioxidant activity and chemical difference in fruit of different Actinidia sp. Int. J. Food Sci. Nutr. 2010, 61, 381–394. [Google Scholar] [CrossRef] [PubMed]
- Kharkevich, S.S.; Kachura, N.N. Redkiye Vidy Rasteniy Sovetskogo Dal’nego Vostoka i ikh Okhrana; Nauka: Moscow, Russia, 1981; ISBN 9784739112502. (In Russian) [Google Scholar]
- Li, J.; Huang, H.; Sang, T. Molecular Phylogeny and Infrageneric Classification of Actinidia (Actinidiaceae). Syst. Bot. 2002, 27, 408–415. [Google Scholar]
- Chat, J.; Jáuregui, B.; Petit, R.J.; Nadot, S. Reticulate evolution in kiwifruit (Actinidia, Actinidiaceae) identified by comparing their maternal and paternal phylogenies. Am. J. Bot. 2004, 91, 736–747. [Google Scholar] [CrossRef]
- Ferguson, A.R.; Huang, H. Genetic resources of kiwifruit: Domestication and breeding. In Horticultural Reviews; Janick, J., Ed.; Wiley: Hoboken, NJ, USA, 2007; pp. 1–121. ISBN 9780470168011. [Google Scholar]
- Kolbasina, E.I.; Kozak, N.V.; Temirbekova, S.K. Genofond Aktinidii (Actinidia Lindl.) v Rossii; Kulikov, I.M., Ed.; IDK: Moscow, Russia, 2008; 74p, ISBN 978-5-85941-261-7. (In Russian) [Google Scholar]
- Molkanova, O.; Konovalova, L.; Kozak, N.; Malayeva, Y. Biologicheskiye osobennosti dal’nevostochnykh vidov roda Actinidia Lindl. Vestn. Udmurt. Univ. Series “Biology. Earth Sciences”. 2014. No. 1. Available online: https://cyberleninka.ru/article/n/biologicheskie-osobennosti-dalnevostochnyh-vidov-roda-actinidia-lindl (accessed on 21 November 2024). (In Russian).
- Ferguson, A.R. The need for characterisation and evaluation of germplasm: Kiwifruit as an example. Euphytica 2007, 154, 371–382. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhong, C.; Liu, Y.; Zhang, Q.; Sun, X.; Li, D. Agronomic trait variations and ploidy differentiation of kiwiberries in northwest china: Implication for breeding. Front. Plant Sci. 2017, 8, 711. [Google Scholar] [CrossRef] [PubMed]
- Yan, G.; Ferguson, A.R.; McNeilage, M.A.; Murray, B.G. Numerically unreduced (2n) gametes and sexual polyploidization in Actinidia. Euphytica 1997, 96, 267–272. [Google Scholar] [CrossRef]
- Malayeva, Y.; Ryzhova, N.; Molkanova, O.; Kochiyeva, Y. Molekulyarnyy RAPD-analiz genomnogo polimorfizma vidov roda Actinidia lindley, (sem. Actinidiaceae), proizrastayushchikh na territorii Rossii. Ekol. Genet. 2008, No. 3. Available online: https://cyberleninka.ru/article/n/molekulyarnyy-rapd-analiz-genomnogo-polimorfizma-vidov-roda-actinidia-lindley-sem-actinidiaceae-proizrastayuschih-na-territorii-rossii (accessed on 21 November 2024). (In Russian).
- Slobodova, N.; Sharko, F.; Gladysheva-Azgari, M.; Petrova, K.; Tsiupka, S.; Tsiupka, V.; Boulygina, E.; Rastorguev, S.; Tsygankova, S. Genetic Diversity of Common Olive (Olea europaea L.) Cultivars from Nikita Botanical Gardens Collection Revealed Using RAD-Seq Method. Genes 2023, 14, 1323. [Google Scholar] [CrossRef] [PubMed]
- Franchini, P.; Monné Parera, D.; Kautt, A.F.; Meyer, A. quaddRAD: A new high-multiplexing and PCR duplicate removal ddRAD protocol produces novel evolutionary insights in a nonradiating cichlid lineage. Mol. Ecol. 2017, 26, 2783–2795. [Google Scholar] [CrossRef]
- Melo, A.T.O.; Guthrie, R.S.; Hale, I. GBS-Based Deconvolution of the Surviving North American Collection of Cold-Hardy Kiwifruit (Actinidia spp.) Germplasm. PLoS ONE 2017, 12, e0170580. [Google Scholar] [CrossRef]
- Lang, P.L.M.; Weiß, C.L.; Kersten, S.; Latorre, S.M.; Nagel, S.; Nickel, B.; Meyer, M.; Burbano, H.A. Hybridization ddRAD-sequencing for population genomics of nonmodel plants using highly degraded historical specimen DNA. Mol. Ecol. Resour. 2020, 20, 1228–1247. [Google Scholar] [CrossRef]
- López de Heredia, U.; Mora-Márquez, F.; Goicoechea, P.G.; Guillardín-Calvo, L.; Simeone, M.C.; Soto, Á. ddRAD Sequencing-Based Identification of Genomic Boundaries and Permeability in Quercus ilex and Q. suber Hybrids. Front. Plant Sci. 2020, 11, 564414. [Google Scholar] [CrossRef] [PubMed]
- Yang, G.-Q.; Chen, Y.-M.; Wang, J.-P.; Guo, C.; Zhao, L.; Wang, X.-Y.; Guo, Y.; Li, L.; Li, D.-Z.; Guo, Z.-H. Development of a universal and simplified ddRAD library preparation approach for SNP discovery and genotyping in angiosperm plants. Plant Methods 2016, 12, 39. [Google Scholar] [CrossRef] [PubMed]
- Zheng, S.; Wang, Y.; Qu, D.; Sun, W.; Yu, Y.; Zhang, Y. Study on population structure of kiwifruit and GWAS for hairiness character. Gene 2022, 821, 146276. [Google Scholar] [CrossRef]
- Lin, M.; Fang, J.; Hu, C.; Qi, X.; Sun, S.; Chen, J.; Sun, L.; Zhong, Y. Genome-wide DNA polymorphisms in four Actinidia arguta genotypes based on whole-genome re-sequencing. PLoS ONE 2020, 15, e0219884. [Google Scholar] [CrossRef] [PubMed]
- Oh, S.; Lee, M.; Kim, K.; Han, H.; Won, K.; Kwack, Y.-B.; Shin, H.; Kim, D. Genetic diversity of kiwifruit (Actinidia spp.), including Korean native A. arguta, using single nucleotide polymorphisms derived from genotyping-by-sequencing. Hortic. Environ. Biotechnol. 2019, 60, 105–114. [Google Scholar] [CrossRef]
- Scaglione, D.; Fornasiero, A.; Pinto, C.; Cattonaro, F.; Spadotto, A.; Infante, R.; Meneses, C.; Messina, R.; Lain, O.; Cipriani, G.; et al. A RAD-based linkage map of kiwifruit (Actinidia chinensis Pl.) as a tool to improve the genome assembly and to scan the genomic region of the gender determinant for the marker-assisted breeding. Tree Genet. Genomes 2015, 11, 115. [Google Scholar] [CrossRef]
- Ding, F.; Zhang, L.; Wang, Y.; Wu, Y.; Wang, F.; Zhao, Y.; Zhang, Y. The complete chloroplast genome sequence of Actinidia arguta var. giraldii. Mitochondrial DNA Part B 2021, 6, 413–414. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Frankham, R.; Ballou, J.D.; Briscoe, D.A.; McInnes, K.H. Introduction to Conservation Genetics; Cambridge University Press: Cambridge, UK, 2002; ISBN 9780511808999. [Google Scholar]
- Khan, M.M.H.; Rafii, M.Y.; Ramlee, S.I.; Jusoh, M.; Al Mamun, M.; Halidu, J. DNA fingerprinting, fixation-index (Fst), and admixture mapping of selected Bambara groundnut (Vigna subterranea [L.] Verdc.) accessions using ISSR markers system. Sci. Rep. 2021, 11, 14527. [Google Scholar] [CrossRef]
- Lu, X.M.; Yu, X.F.; Li, G.Q.; Qu, M.H.; Wang, H.; Liu, C.; Man, Y.P.; Jiang, X.H.; Li, M.Z.; Wang, J.; et al. Genome assembly of autotetraploid Actinidia arguta highlights adaptive evolution and enables dissection of important economic traits. Plant Commun. 2024, 5, 100856. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Montefiori, M.; Comeskey, D.J.; Wohlers, M.; McGhie, T.K. Characterization and quantification of anthocyanins in red kiwifruit (Actinidia spp.). J. Agric. Food Chem. 2009, 57, 6856–6861. [Google Scholar] [CrossRef] [PubMed]
- Kolbasina, E.I. Aktinidii i Limonnik v Rossii; IDK: Moscow, Russia, 2000; 264p, ISBN 5-85941-021-2. (In Russian) [Google Scholar]
- Choi, H.R.; Baek, M.W.; Jeong, C.S.; Tilahun, S. Comparative Transcriptome Analysis of Softening and Ripening-Related Genes in Kiwifruit Cultivars Treated with Ethylene. Curr. Issues Mol. Biol. 2022, 44, 2593–2613. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Chen, Y.; Wang, L.; Li, D.; Liu, L.; Li, M. An ethylene response factor AcERF116 identified from A. catechu is involved in fruitlet abscission. Plant Sci. 2024, 344, 112091. [Google Scholar] [CrossRef] [PubMed]
- Lai, J.; Yu, B.; Cao, Z.; Chen, Y.; Wu, Q.; Huang, J.; Yang, C. Two homologous protein S-acyltransferases, PAT13 and PAT14, cooperatively regulate leaf senescence in Arabidopsis. J. Exp. Bot. 2015, 66, 6345–6353. [Google Scholar] [CrossRef]
- Evers, D.; Legay, S.; Lamoureux, D.; Hausman, J.F.; Hoffmann, L.; Renaut, J. Towards a synthetic view of potato cold and salt stress response by transcriptomic and proteomic analyses. Plant Mol. Biol. 2012, 78, 503–514. [Google Scholar] [CrossRef] [PubMed]
- Jiang, J.; Ren, X.; Li, L.; Hou, R.; Sun, W.; Jiao, C.; Yang, N.; Dong, Y. H2S Regulation of Metabolism in Cucumber in Response to Salt-Stress Through Transcriptome and Proteome Analysis. Front. Plant Sci. 2020, 11, 1283. [Google Scholar] [CrossRef]
- Famula, R.A.; Richards, J.H.; Famula, T.R.; Neale, D.B. Association genetics of carbon isotope discrimination and leaf morphology in a breeding population of Juglans regia L. Tree Genet. Genomes 2019, 15, 6. [Google Scholar] [CrossRef]
- Wang, L.; Lee, M.; Ye, B.; Yue, G.H. Genes, pathways and networks responding to drought stress in oil palm roots. Sci. Rep. 2020, 10, 21303. [Google Scholar] [CrossRef]
- ten Hove, C.A.; Bochdanovits, Z.; Jansweijer, V.M.A.; Koning, F.G.; Berke, L.; Sanchez-Perez, G.F.; Scheres, B.; Heidstra, R. Probing the roles of LRR RLK genes in Arabidopsis thaliana roots using a custom T-DNA insertion set. Plant Mol. Biol. 2011, 76, 69–83. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Yao, Z.; Cao, X.; Chen, M.; Chen, S.; Zhao, Q.; Zhao, S. A leucine-rich repeat receptor-like protein kinase enhances tomato resistance to Phelipanche aegyptiaca. Sci. Hortic. 2024, 337, 113353. [Google Scholar] [CrossRef]
- Li, H.; Chen, L.; Liu, R.; Cao, S.; Lu, Z. Comparative Proteomic Analysis of Floral Buds before and after Opening in Walnut (Juglans regia L.). Int. J. Mol. Sci. 2024, 25, 7878. [Google Scholar] [CrossRef]
- Hande, A.S.; Katageri, I.S.; Jadhav, M.P.; Adiger, S.; Gamanagatti, S.; Padmalatha, K.V.; Dhandapani, G.; Kanakachari, M.; Kumar, P.A.; Reddy, V.S. Transcript profiling of genes expressed during fibre development in diploid cotton (Gossypium arboreum L.). BMC Genom. 2017, 18, 675. [Google Scholar] [CrossRef] [PubMed]
- Iqbal, A.; Latif, A.; Galbraith, D.W.; Jabbar, B.; Ali, M.A.; Ahmed, M.; Gul, A.; Rao, A.Q.; Shahid, A.A.; Husnain, T. Structure-based prediction of protein–protein interactions between GhWlim5 Domain1 and GhACTIN-1 proteins: A practical evidence with improved fibre strength. J. Plant Biochem. Biotechnol. 2021, 30, 373–386. [Google Scholar] [CrossRef]
- Breuninger, H.; Lenhard, M. Expression of the central growth regulator BIG BROTHER is regulated by multiple cis-elements. BMC Plant Biol. 2012, 12, 41. [Google Scholar] [CrossRef] [PubMed]
- Zangir, N.M. Identifying New Interactors in the Cellulose Synthase Protein Complex Involved in Cellulose Biosynthesis in the Primary Plant Cell Wall. In Characterising the Cellulose Synthase Complexes of Cell Walls; Thesis; Wageningen University: Wageningen, The Netherlands, 2012; p. 91. [Google Scholar]
- Asakura, I.; Hoshino, Y. Distribution, ploidy levels, and fruit characteristics of three actinidia species native to hokkaido, Japan. Hortic. J. 2016, 85, 105–114. [Google Scholar] [CrossRef]
- Start, M.A.; Luby, J.; Filler, D.; Riera-Lizarazu, O.; Guthrie, R. Ploidy levels of cold-hardy actinidia accessions in the united states determined by flow cytometry. Acta Hortic. 2007, 753, 161–168. [Google Scholar] [CrossRef]
- Hijmans, R.J.; Gavrilenko, T.; Stephenson, S.; Bamberg, J.; Salas, A.; Spooner, D.M. Geographical and environmental range expansion through polyploidy in wild potatoes (Solanum section Petota). Glob. Ecol. Biogeogr. 2007, 16, 485–495. [Google Scholar] [CrossRef]
- Wendel, J.F.; Cronn, R.C. Polyploidy and the evolutionary history of cotton. In Advances in Agronomy; Elsevier: Amsterdam, The Netherlands, 2003; Volume 78, pp. 139–186. ISBN 9780120007967. [Google Scholar]
- Kataoka, I.; Mizugami, T.; Kim, J.G.; Beppu, K.; Fukuda, T.; Sugahara, S.; Tanaka, K.; Satoh, H.; Tozawa, K. Ploidy variation of hardy kiwifruit (Actinidia arguta) resources and geographic distribution in Japan. Sci. Hortic. 2010, 124, 409–414. [Google Scholar] [CrossRef]
- Piccolo, S.L.; Alfonzo, A.; Conigliaro, G.; Moschetti, G.; Barone, A. (PDF) A simple and rapid DNA extraction method from leaves of grapevine suitable for polymerase chain reaction analysis. Afr. J. Biotechnol. 2012, 11, 10305–10309. [Google Scholar]
- Gladysheva-Azgari, M.; Sharko, F.; Slobodova, N.; Petrova, K.; Boulygina, E.; Tsygankova, S.; Mitrofanova, I. Comparative Analysis Revealed Intrageneric and Intraspecific Genomic Variation in Chloroplast Genomes of Actinidia spp. (Actinidiaceae, Viridiplantae). Horticulturae 2023, 9, 1175. [Google Scholar] [CrossRef]
- Shalgimbayeva, G.; Volkov, A.; Slobodova, N.; Sharko, F.; Tsygankova, S.; Boulygina, E.; Nguyen, V.Q.; Pham, T.T.; Nguyen, D.T.; Assylbekova, S.Z.; et al. Genetic Investigation of Aral Wild Common Carp Populations (Cyprinus carpio) Using ddRAD Sequencing. Diversity 2021, 13, 295. [Google Scholar] [CrossRef]
- Rochette, N.C.; Rivera-Colón, A.G.; Catchen, J.M. Stacks 2: Analytical methods for paired-end sequencing improve RADseq-based population genomics. Mol. Ecol. 2019, 28, 4737–4754. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed]
- Knaus, B.J.; Grünwald, N.J. vcfr: A package to manipulate and visualize variant call format data in R. Mol. Ecol. Resour. 2017, 17, 44–53. [Google Scholar] [CrossRef] [PubMed]
- Kamvar, Z.N.; Brooks, J.C.; Grünwald, N.J. Novel R tools for analysis of genome-wide population genetic data with emphasis on clonality. Front. Genet. 2015, 6, 208. [Google Scholar] [CrossRef]
- Mijangos, J.L.; Gruber, B.; Berry, O.; Pacioni, C.; Georges, A. v2: An accessible genetic analysis platform for conservation, ecology and agriculture. Methods Ecol. Evol. 2022, 13, 2150–2158. [Google Scholar] [CrossRef]
- Jombart, T.; Ahmed, I. Adegenet 1.3-1: New tools for the analysis of genome-wide SNP data. Bioinformatics 2011, 27, 3070–3071. [Google Scholar] [CrossRef]
- Alexander, D.H.; Novembre, J.; Lange, K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009, 19, 1655–1664. [Google Scholar] [CrossRef]
- Cingolani, P.; Platts, A.; Wang, L.L.; Coon, M.; Nguyen, T.; Wang, L.; Land, S.J.; Lu, X.; Ruden, D.M. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff. Fly 2012, 6, 80–92. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
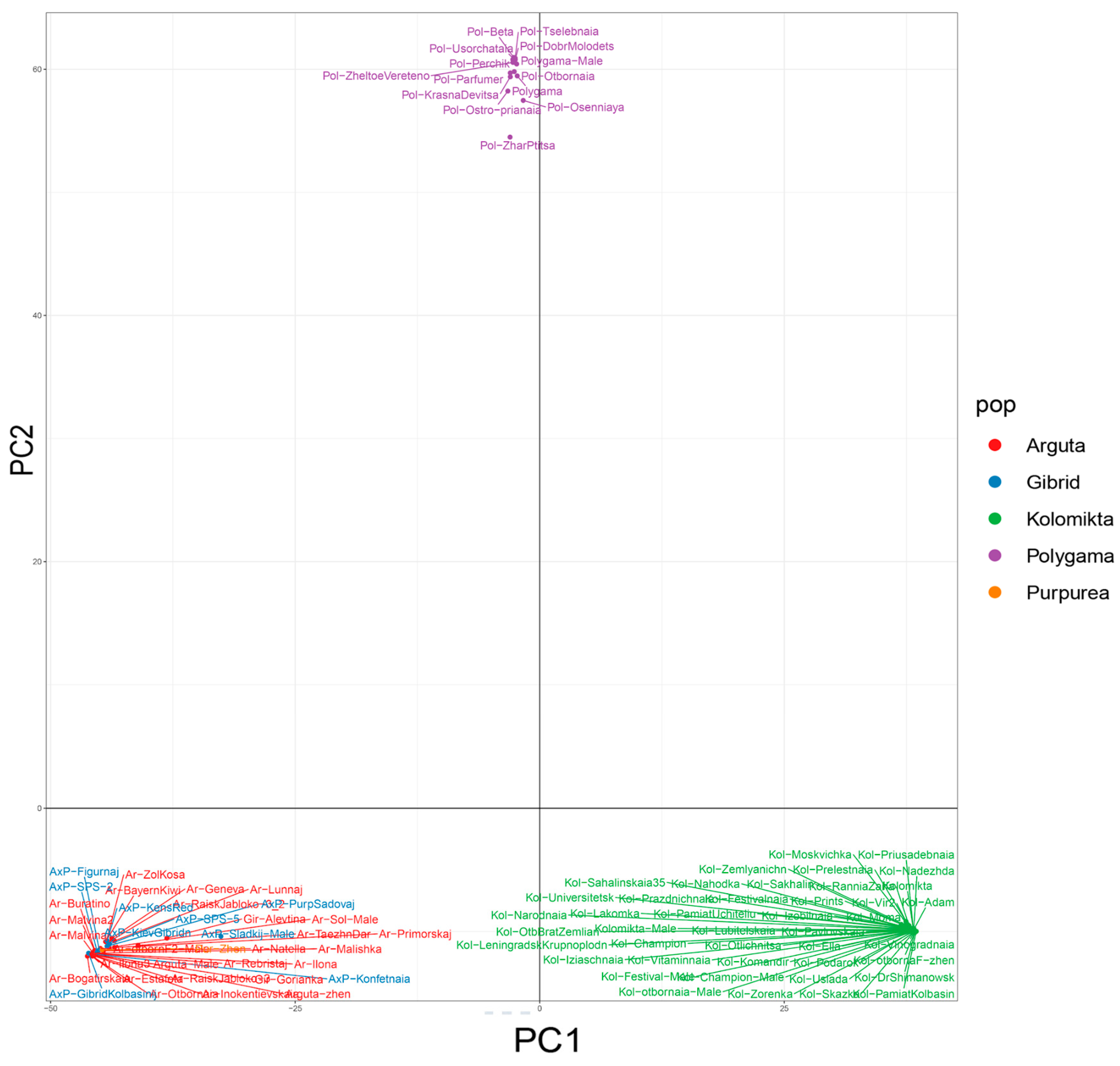


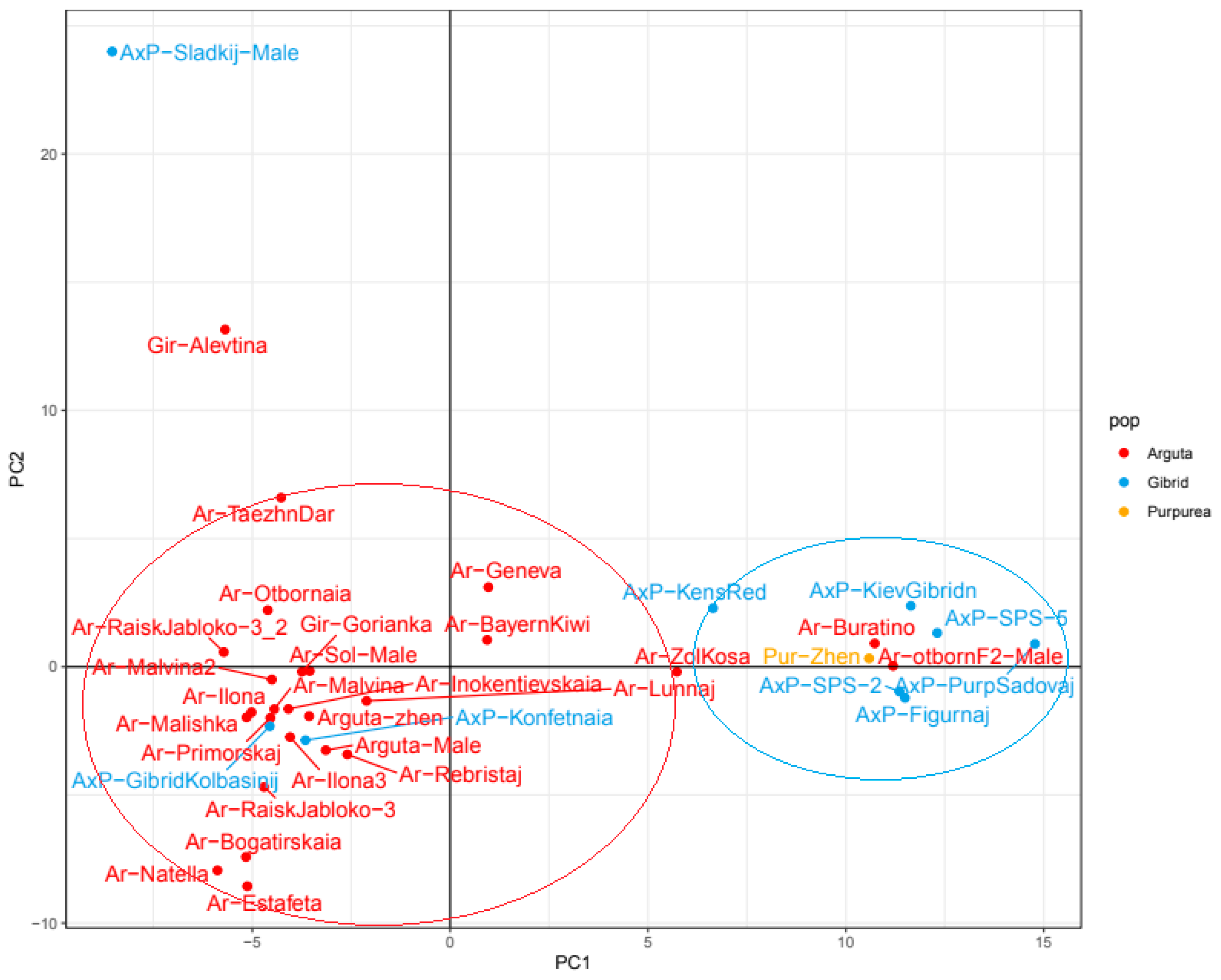
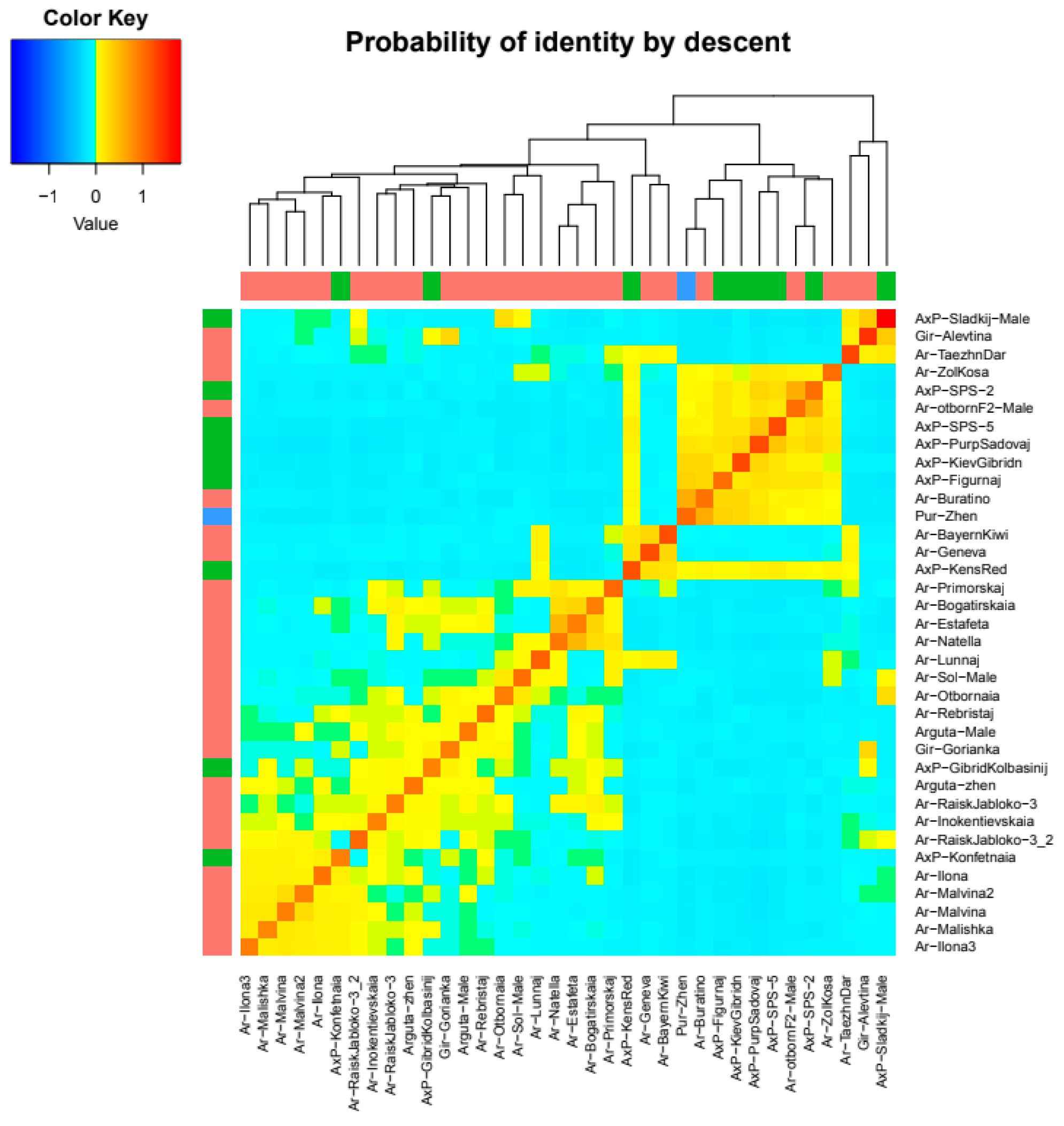

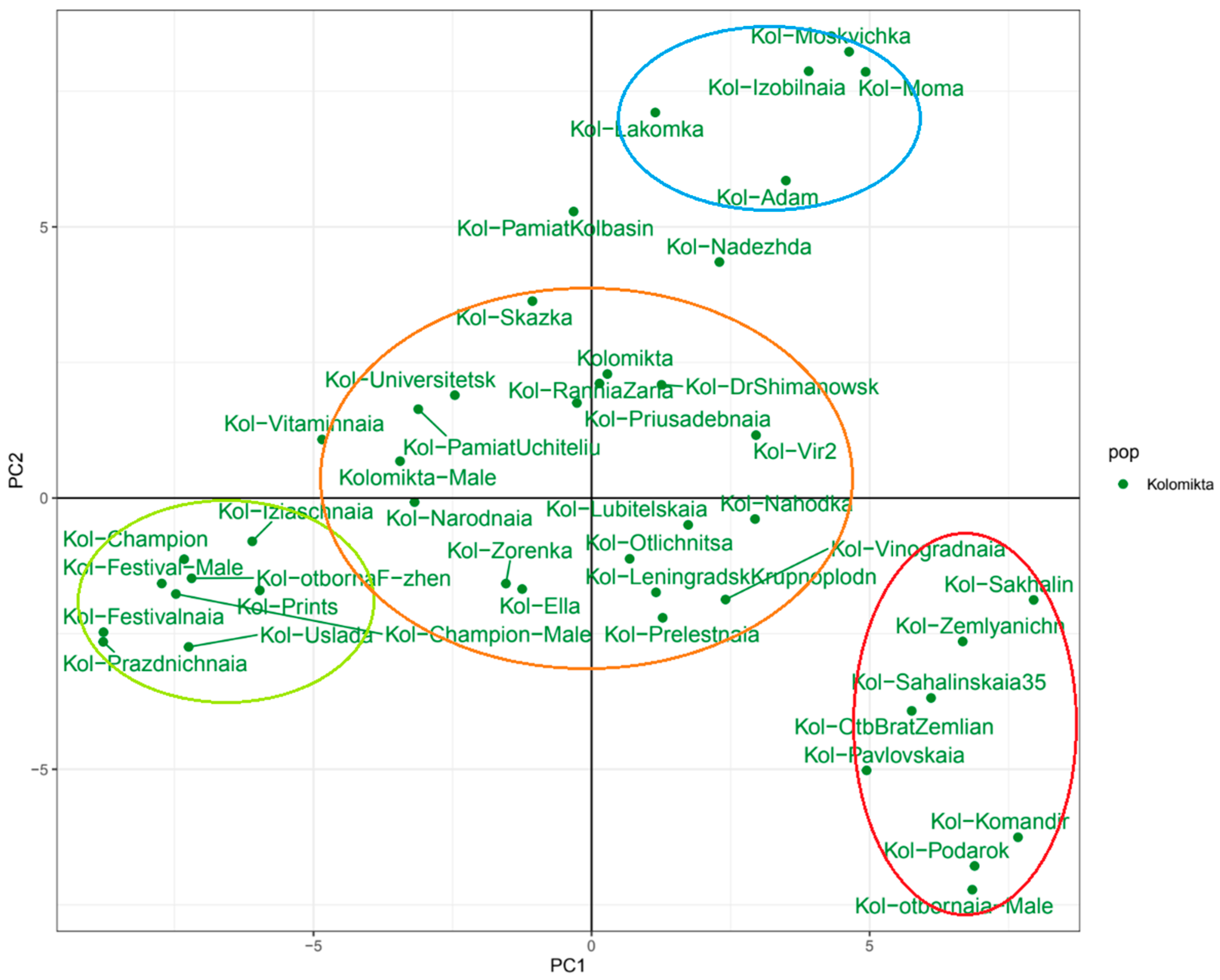
| A. arguta | A. kolomikta | A. polygama | A. purpurea | |
| A. arguta | 0 | |||
| A. kolomikta | 0.838 | 0 | ||
| A. polygama | 0.806 | 0.878 | 0 | |
| A. purpurea | 0.018 | 0.890 | 0.873 | 0 |
| Ho | Hs | FIS | |
|---|---|---|---|
| A. arguta | 0.0553 | 0.0631 | 0.1244 |
| A. kolomikta | 0.0266 | 0.0318 | 0.1642 |
| A. polygama | 0.0272 | 0.0311 | 0.1268 |
| Area of SNP Location | % SNP |
|---|---|
| intron_varian | 16.44% |
| intragenic_variant | 8.22% |
| missense_variant | 5.48% |
| 3_prime_UTR_variant | 1.37% |
| stop_gained | 1.37% |
| downstream_gene_variant | 19.18% |
| upstream_gene_variant | 19.18% |
| synonymous_variant | 28.77% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Slobodova, N.; Gladysheva-Azgari, M.; Sharko, F.; Petrova, K.; Boulygina, E.; Tsygankova, S.; Mitrofanova, I. Genetic Diversity of the Collection of Far Eastern Actinidia spp. Revealed by RAD Sequencing Technology. Plants 2025, 14, 7. https://doi.org/10.3390/plants14010007
Slobodova N, Gladysheva-Azgari M, Sharko F, Petrova K, Boulygina E, Tsygankova S, Mitrofanova I. Genetic Diversity of the Collection of Far Eastern Actinidia spp. Revealed by RAD Sequencing Technology. Plants. 2025; 14(1):7. https://doi.org/10.3390/plants14010007
Chicago/Turabian StyleSlobodova, Natalia, Maria Gladysheva-Azgari, Fedor Sharko, Kristina Petrova, Eugenia Boulygina, Svetlana Tsygankova, and Irina Mitrofanova. 2025. "Genetic Diversity of the Collection of Far Eastern Actinidia spp. Revealed by RAD Sequencing Technology" Plants 14, no. 1: 7. https://doi.org/10.3390/plants14010007
APA StyleSlobodova, N., Gladysheva-Azgari, M., Sharko, F., Petrova, K., Boulygina, E., Tsygankova, S., & Mitrofanova, I. (2025). Genetic Diversity of the Collection of Far Eastern Actinidia spp. Revealed by RAD Sequencing Technology. Plants, 14(1), 7. https://doi.org/10.3390/plants14010007








