Molecular Mechanisms of Grain Chalkiness Variation in Rice Panicles
Abstract
1. Introduction
2. Results
2.1. Chalkiness and Physicochemical Properties of Mature Grains
2.2. Micro-Scale Observations of the Endosperm Structure
2.3. Grain Filling Characteristics of Rice Gain from Different Positions in a Panicle
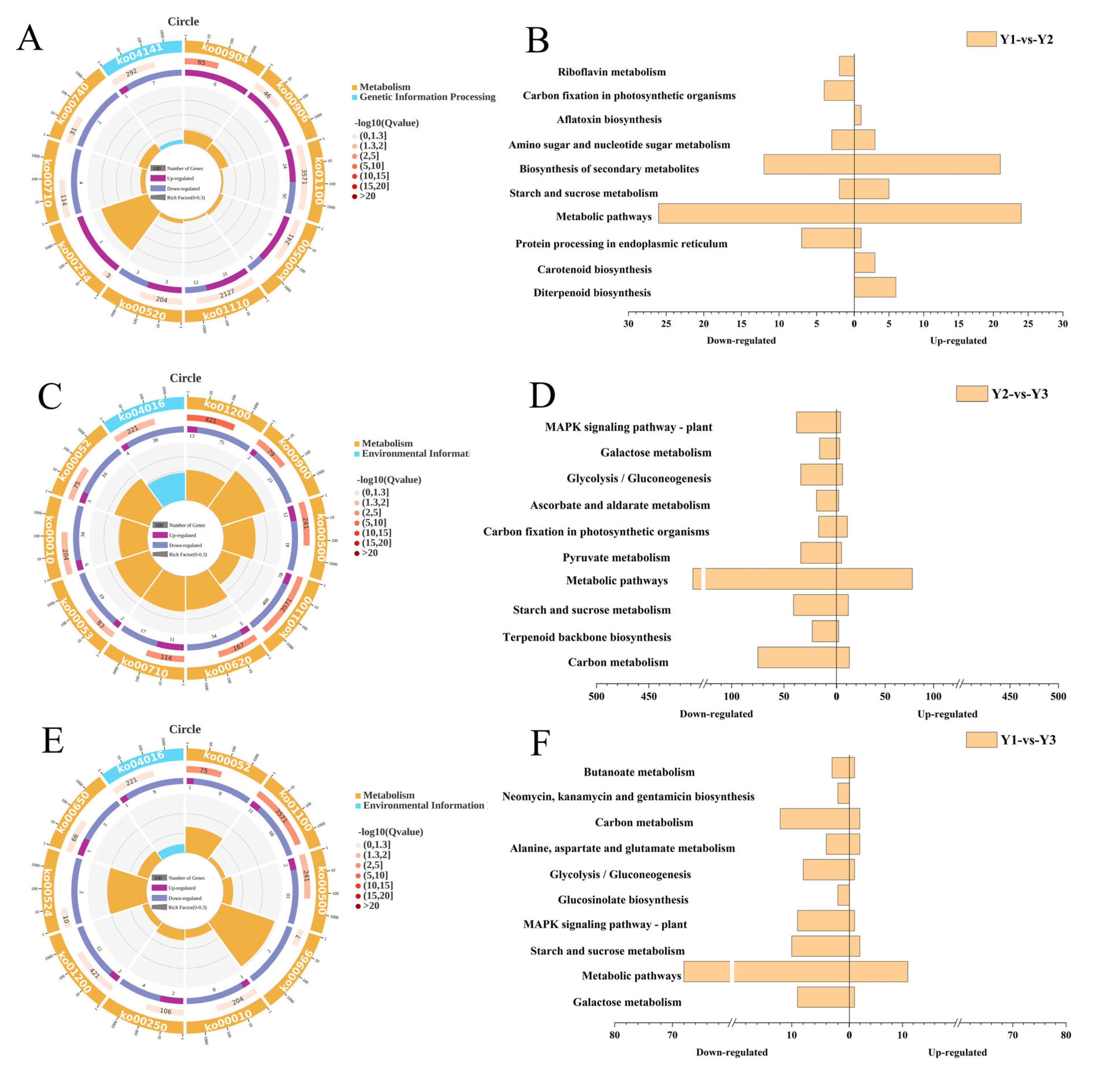
2.4. DEGs in Grains from Different Spikelet Positions in a Rice Panicle
2.5. Functional Classification of DEGs in Grains from Different Positions of the Panicle
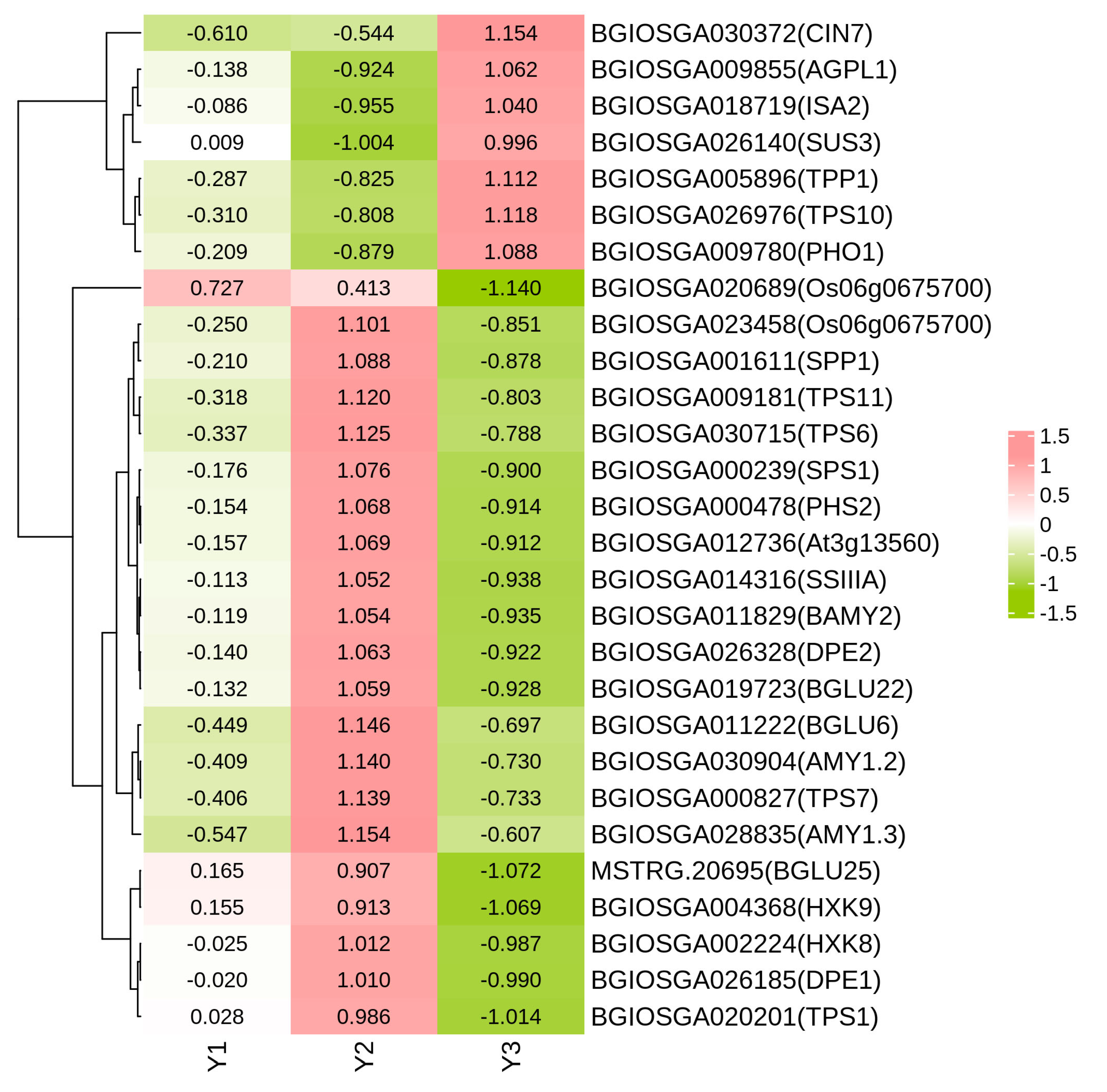
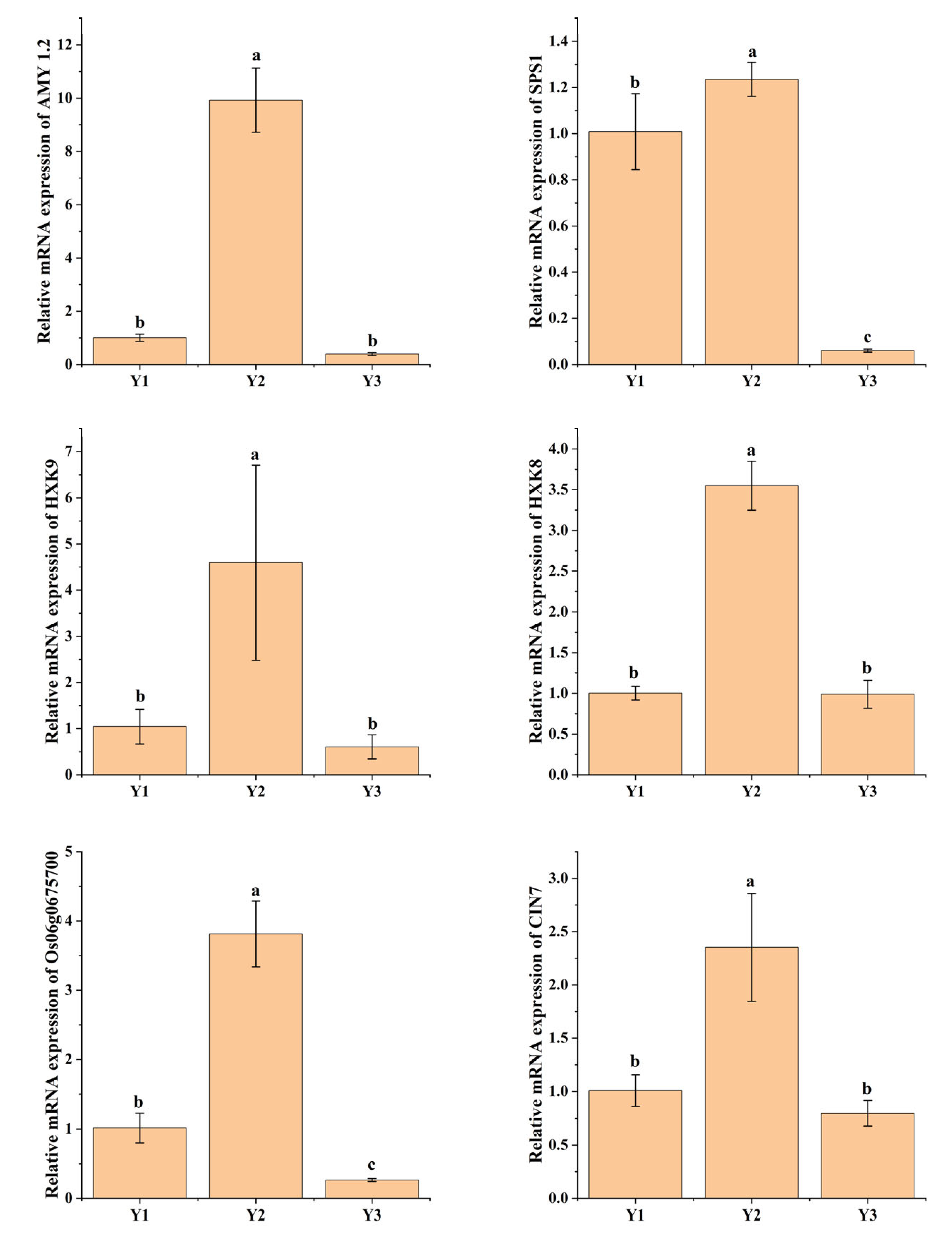
2.6. DEGs Related to Starch and Sucrose Metabolism
2.7. DEGs Related to Carbon Metabolism and Galactose Metabolism
2.8. DEGs Related to MAPK Signaling Pathways
3. Discussion
3.1. Positional Variation in Grain Chalkiness Within a Rice Panicle
3.2. Changes in Gene Expression and Its Relation to Grain Chalkiness Within a Rice Panicle
4. Materials and Methods
4.1. Plant Material and Growth Conditions
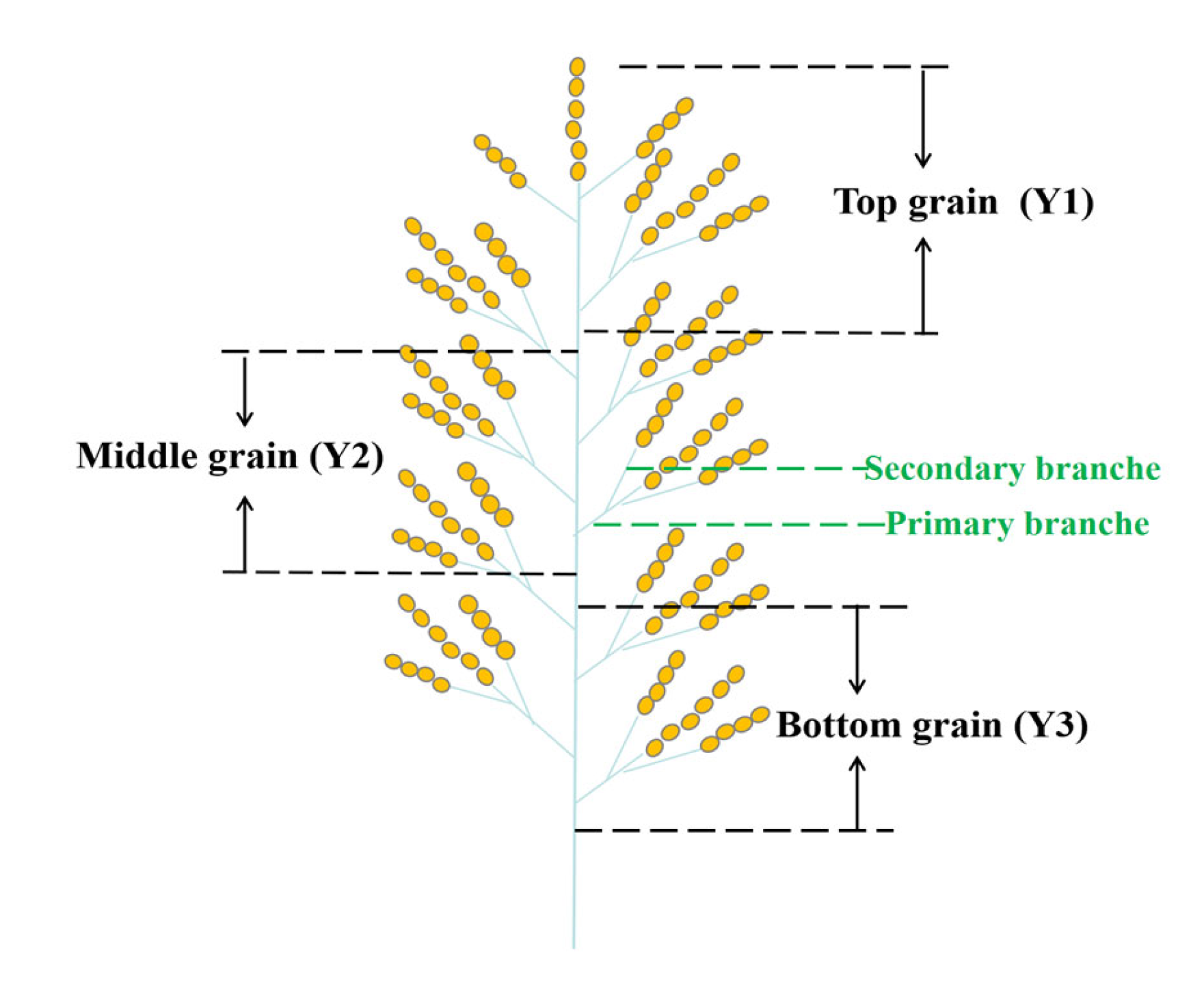
4.2. Sampling
4.3. Phenotype and Physicochemical Properties of the Matured Seeds
4.4. Gene Expression Analysis
4.5. DNA Database Search and Computer Analysis
4.6. qRT-PCR Analysis
4.7. Data Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Deng, F.; Li, Q.; Chen, H.; Zeng, Y.; Li, B.; Zhong, X.; Wang, L.; Ren, W. Relationship between chalkiness and the structural and thermal properties of rice starch after shading during grain-filling stage. Carbohydr. Polym. 2021, 252, 117212. [Google Scholar] [CrossRef] [PubMed]
- Tao, K.; Liu, X.; Yu, W.; Neoh, G.K.; Gilbert, R.G. Starch molecular structural differences between chalky and translucent parts of chalky rice grains. Food Chem. 2022, 394, 133471. [Google Scholar] [CrossRef] [PubMed]
- Xi, M.; Zhao, Y.; Lin, Z.; Zhang, X.; Ding, C.; Tang, S.; Liu, Z.; Wang, S.; Ding, Y. Comparison of physicochemical characteristics between white-belly and white-core rice grains. J. Cereal Sci. 2016, 69, 392–397. [Google Scholar] [CrossRef]
- Wang, C.; Guo, W.; Hu, P.; Wei, X.; Tang, S.; Jiao, G. Differences of physicochemical properties between chalky and translucent parts of rice grains. Rice Sci. 2022, 29, 577–588. [Google Scholar]
- Tang, S.; Zhang, H.; Liu, W.; Dou, Z.; Zhou, Q.; Chen, W.; Wang, S.; Ding, Y. Nitrogen fertilizer at heading stage effectively compensates for the deterioration of rice quality by affecting the starch-related properties under elevated temperatures. Food Chem. 2019, 277, 455–462. [Google Scholar] [CrossRef]
- Bai, S.; Hong, J.; Li, L.; Su, S.; Li, Z.; Wang, W.; Zhang, F.; Liang, W.; Zhang, D. Dissection of the genetic basis of rice panicle architecture using a genome-wide association study. Rice 2021, 14, 77. [Google Scholar] [CrossRef]
- Qiao, J.; Liu, Z.; Deng, S.; Ning, H.; Yang, X.; Lin, Z.; Li, G.; Wang, Q.; Wang, S.; Ding, Y. Occurrence of perfect and imperfect grains of six japonica rice cultivars as affected by nitrogen fertilization. Plant Soil 2011, 349, 191–202. [Google Scholar] [CrossRef]
- Dong, M.; Sang, S.; Wang, P.; Zhang, W.; Yang, J. Difference in chalky characters of the grains at different positions within a rice panicle. Crop J. 2006, 32, 103–111. (In Chinese) [Google Scholar]
- Mohapatra, P.K.; Patel, R.; Sahu, S.K. Time of Flowering Affects Grain Quality and Spikelet Partitioning Within the Rice Panicle. Aust. J. Plant Physiol. 1993, 20, 231–241. [Google Scholar] [CrossRef]
- Xi, M.; Wu, W.; Xu, Y.; Zhou, Y.; Chen, G.; Ji, Y.; Sun, X. Grain chalkiness traits is affected by the synthesis and dynamic accumulation of the storage protein in rice. J. Sci. Food Agric. 2021, 101, 6125–6133. [Google Scholar] [CrossRef]
- Zhang, X.; Lin, Z.; Liu, Z.; Li, G.; Wang, Q.; Wang, S.; Ding, Y. Analysis of variations in white-belly and white-core rice kernels within a panicle and the effect of panicle type. J. Integr. Agric. 2014, 13, 1672–1679. [Google Scholar] [CrossRef]
- Kido, M.; Yanatori, S. Studies on Positions in Panicle of Ventral white, Basal white, Milky white like white Core and Milky white kernels, and shapes of White opaque parts in these kernels. Jpn. J. Crop Sci. 2008, 37, 534–538. [Google Scholar] [CrossRef]
- Xi, M.; Wu, W.; Xu, Y.; Zhou, Y.; Chen, G.; Ji, Y.; Sun, X. iTRAQ-based quantitative proteomic analysis reveals the metabolic pathways of grain chalkiness in response to nitrogen topdressing in rice. Plant Physiol. Biochem. 2020, 154, 622–635. [Google Scholar] [CrossRef] [PubMed]
- Yamakawa, H.; Hakata, M. Atlas of rice grain filling-related metabolism under high temperature: Joint analysis of metabolome and transcriptome demonstrated inhibition of starch accumulation and induction of amino acid accumulation. Plant Cell Physiol. 2010, 51, 795–809. [Google Scholar] [CrossRef]
- Hakata, M.; Kuroda, M.; Miyashita, T.; Yamaguchi, T.; Kojima, M.; Sakakibara, H.; Mitsui, T.; Yamakawa, H. Suppression of α-amylase genes improves quality of rice grain ripened under high temperature. Plant Biotechnol. J. 2012, 10, 1110–1117. [Google Scholar] [CrossRef]
- Tang, S.; Chen, W.; Liu, W.; Zhou, Q.; Zhang, H.; Wang, S.; Ding, Y. Open-field warming regulates the morphological structure, protein synthesis of grain and affects the appearance quality of rice. J. Cereal Sci. 2018, 84, 20–29. [Google Scholar] [CrossRef]
- Yang, W.; Xu, P.; Zhang, J.; Zhang, S.; Li, Z.; Yang, K.; Chang, X.; Li, Y. OsbZIP60-mediated unfolded protein response regulates grain chalkiness in rice. J. Genet. Genom. 2022, 49, 414–426. [Google Scholar] [CrossRef]
- Kang, H.G.; Park, S.; Matsuoka, M.; An, G. White-core endosperm floury endosperm-4 in rice is generated by knockout mutations in the C4-type pyruvate orthophosphate dikinase gene (OsPPDKB). Plant J. 2005, 42, 901–911. [Google Scholar] [CrossRef]
- Lee, S.; Eom, J.; Hwang, S.; Shin, D.; An, G.; Okita, T.; Jeon, J. Plastidic phosphoglucomutase and ADP-glucose pyrophosphorylase mutants impair starch synthesis in rice pollen grains and cause male sterility. J. Exp. Bot. 2016, 67, 5557–5569. [Google Scholar] [CrossRef]
- Woo, M.O.; Ham, T.H.; Ji, H.S.; Choi, M.S.; Jiang, W.; Chu, S.H.; Piao, R.; Chin, J.H.; Kim, J.A.; Park, B.S. Inactivation of the UGPase1 gene causes genic male sterility and endosperm chalkiness in rice (Oryza sativa L.). Plant J. 2008, 54, 190–204. [Google Scholar] [CrossRef]
- Kim, Y.; Yeu, S.; Park, B.; Koh, H.; Song, J.; Seo, H. Protein disulfide isomerase-like protein 1-1 controls endosperm development through regulation of the amount and composition of seed proteins in rice. PLoS ONE 2012, 7, e44493. [Google Scholar] [CrossRef] [PubMed]
- Wakasa, Y.; Yasuda, H.; Oono, Y.; Kawakatsu, T.; Hirose, S.; Takahashi, H.; Hayashi, S.; Yang, L.; Takaiwa, F. Expression of ER quality control-related genes in response to changes in BiP1 levels in developing rice endosperm. Plant J. 2011, 65, 675–689. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Fan, C.; Xing, Y.; Yun, P.; Luo, L.; Yan, B.; Peng, B.; Xie, W.; Wang, G.; Li, X. Chalk5 encodes a vacuolar H+-translocating pyrophosphatase influencing grain chalkiness in rice. Nat. Genet. 2014, 46, 398–404. [Google Scholar] [CrossRef] [PubMed]
- Lin, Z.; Zhang, X.; Wang, Z.; Jiang, Y.; Liu, Z.; Alexander, D.; Li, G.; Wang, S.; Ding, Y. Metabolomic analysis of pathways related to rice grain chalkiness by a notched-belly mutant with high occurrence of white-belly grains. BMC Plant Biol. 2017, 17, 39. [Google Scholar] [CrossRef] [PubMed]
- Chandran, A.K.N.; Sandhu, J.; Irvin, L.; Paul, P.; Dhatt, B.K.; Hussain, W.; Gao, T.; Staswick, P.; Yu, H.; Morota, G. Rice Chalky Grain 5 regulates natural variation for grain quality under heat stress. Front. Plant Sci. 2022, 13, 1026472. [Google Scholar] [CrossRef]
- Wang, W.; Wang, W.; Pan, Y.; Tan, C.; Li, H.; Chen, Y.; Liu, X.; Wei, J.; Xu, N.; Han, Y. A new gain-of-function OsGS2/GRF4 allele generated by CRISPR/Cas9 genome editing increases rice grain size and yield. Crop J. 2022, 10, 1207–1212. [Google Scholar] [CrossRef]
- Lin, S.; Liu, Z.; Zhang, K.; Yang, W.; Zhan, P.; Tan, Q.; Gou, Y.; Ma, S.; Luan, X.; Huang, C.; et al. GL9 from Oryza glumaepatula controls grain size and chalkiness in rice. Crop J. 2023, 11, 198–207. [Google Scholar] [CrossRef]
- Zhao, D.; Li, Q.; Zhang, C.; Zhang, C.; Yang, Q.; Pan, L.; Ren, X.; Lu, J.; Gu, M.; Liu, Q. GS9 acts as a transcriptional activator to regulate rice grain shape and appearance quality. Nat. Commun. 2018, 9, 1240. [Google Scholar] [CrossRef]
- Su, Y.; Xiao, L. 3D visualization and volume-based quantification of rice chalkiness in vivo by using high resolution micro-CT. Rice 2020, 13, 69. [Google Scholar] [CrossRef]
- Wang, X.; Wang, K.; Yin, T.; Zhao, Y.; Liu, W.; Shen, Y.; Ding, Y.; Tang, S. Nitrogen Fertilizer Regulated Grain Storage Protein Synthesis and Reduced Chalkiness of Rice Under Actual Field Warming. Front. Plant Sci. 2021, 12, 715436. [Google Scholar] [CrossRef]
- You, C.; He, H.; Wu, L.; Wang, S.; Ding, Y.; Ma, C. iTRAQ-based proteome profile analysis of superior and inferior Spikelets at early grain filling stage in japonica Rice. BMC Plant Biol. 2017, 17, 100. [Google Scholar] [CrossRef] [PubMed]
- Kciuk, M.; Gielecińska, A.; Budzinska, A.; Mojzych, M.; Kontek, R. Metastasis and MAPK pathways. Int. J. Mol. Sci. 2022, 23, 3847. [Google Scholar] [CrossRef] [PubMed]
- Ronkina, N.; Gaestel, M. MAPK-activated protein kinases: Servant or partner? Annu. Rev. Biochem. 2022, 91, 505–540. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.; Ma, M.; Wang, Q.; Zhang, M.; Jing, G.; Li, C.; Ma, F. Arbuscular mycorrhizal fungi enhanced drought resistance in apple by regulating genes in the MAPK pathway. Plant Physiol. Biochem. 2020, 149, 245–255. [Google Scholar] [CrossRef]
- Kumar, K.; Raina, S.K.; Sultan, S.M. Arabidopsis MAPK signaling pathways and their cross talks in abiotic stress response. J. Plant Biochem. Biotechnol. 2020, 29, 700–714. [Google Scholar] [CrossRef]
- Pan, Y.; Gao, L.; Liang, Y.; Zhao, Y.; Deng, G. OrMKK3 Influences Morphology and Grain Size in Rice. J. Plant Biol. 2021, 66, 269–282. [Google Scholar] [CrossRef]
- Cheng, W.; Zhang, G.; Zhao, G.; Yao, H.; Xu, H. Variation in rice quality of different cultivars and grain positions as affected by water management. Field Crops Res. 2003, 80, 245–252. [Google Scholar] [CrossRef]
- Jeon, J.; Ryoo, N.; Hahn, T.; Walia, H.; Nakamura, Y. Starch biosynthesis in cereal endosperm. Plant Physiol. Biochem. 2010, 48, 383–392. [Google Scholar] [CrossRef]
- Lin, Z.; Zhang, X.; Yang, X.; Li, G.; Tang, S. Proteomic analysis of proteins related to rice grain chalkiness using iTRAQ and a novel comparison system based on a notched-belly mutant with white-belly. BMC Plant Biol. 2014, 14, 163–179. [Google Scholar] [CrossRef]
- Li, Q.; Deng, F.; Zeng, Y.; Li, B.; He, C.; Zhu, Y.; Zhou, X.; Zhang, Z.; Wang, L.; Tao, Y. Low light stress increases chalkiness by disturbing starch synthesis and grain filling of rice. Int. J. Mol. Sci. 2022, 23, 9153. [Google Scholar] [CrossRef]
- Fei, L.; Yang, S.; Ma, A.; Lunzhu, C.; Wang, M.; Wang, G.; Guo, S. Grain chalkiness is reduced by coordinating the biosynthesis of protein and starch in fragrant rice (Oryza sativa L.) grain under nitrogen fertilization. Field Crops Res. 2023, 302, 109098. [Google Scholar] [CrossRef]
- Liu, X.; Guo, T.; Wan, X.; Wang, H.; Zhu, M.; Li, A.; Su, N.; Shen, Y.; Mao, B.; Zhai, H. Transcriptome analysis of grain-filling caryopses reveals involvement of multiple regulatory pathways in chalky grain formation in rice. BMC Genom. 2010, 11, 730. [Google Scholar] [CrossRef] [PubMed]
- Nakata, M.; Fukamatsu, Y.; Miyashita, T.; Hakata, M.; Kimura, R.; Nakata, Y.; Kuroda, M.; Yamaguchi, T.; Yamakawa, H. High temperature-induced expression of rice α-amylases in developing endosperm produces chalky grains. Front. Plant Sci. 2017, 8, 2089. [Google Scholar] [CrossRef] [PubMed]
- Gann, P.; Esguerra, M.; Counce, P.; Srivastava, V. Genotype-dependent and heat-induced grain chalkiness in rice correlates with the expression patterns of starch biosynthesis genes. Plant-Environ. Interact. 2021, 2, 165–176. [Google Scholar] [CrossRef] [PubMed]
- Fujita, N.; Yoshida, M.; Kondo, T.; Saito, K.; Utsumi, Y.; Tokunaga, T.; Nishi, A.; Satoh, H.; Park, J.-H.; Jane, J.-L. Characterization of SSIIIa-deficient mutants of rice: The function of SSIIIa and pleiotropic effects by SSIIIa deficiency in the rice endosperm. Plant Physiol. 2007, 144, 2009–2023. [Google Scholar] [CrossRef]
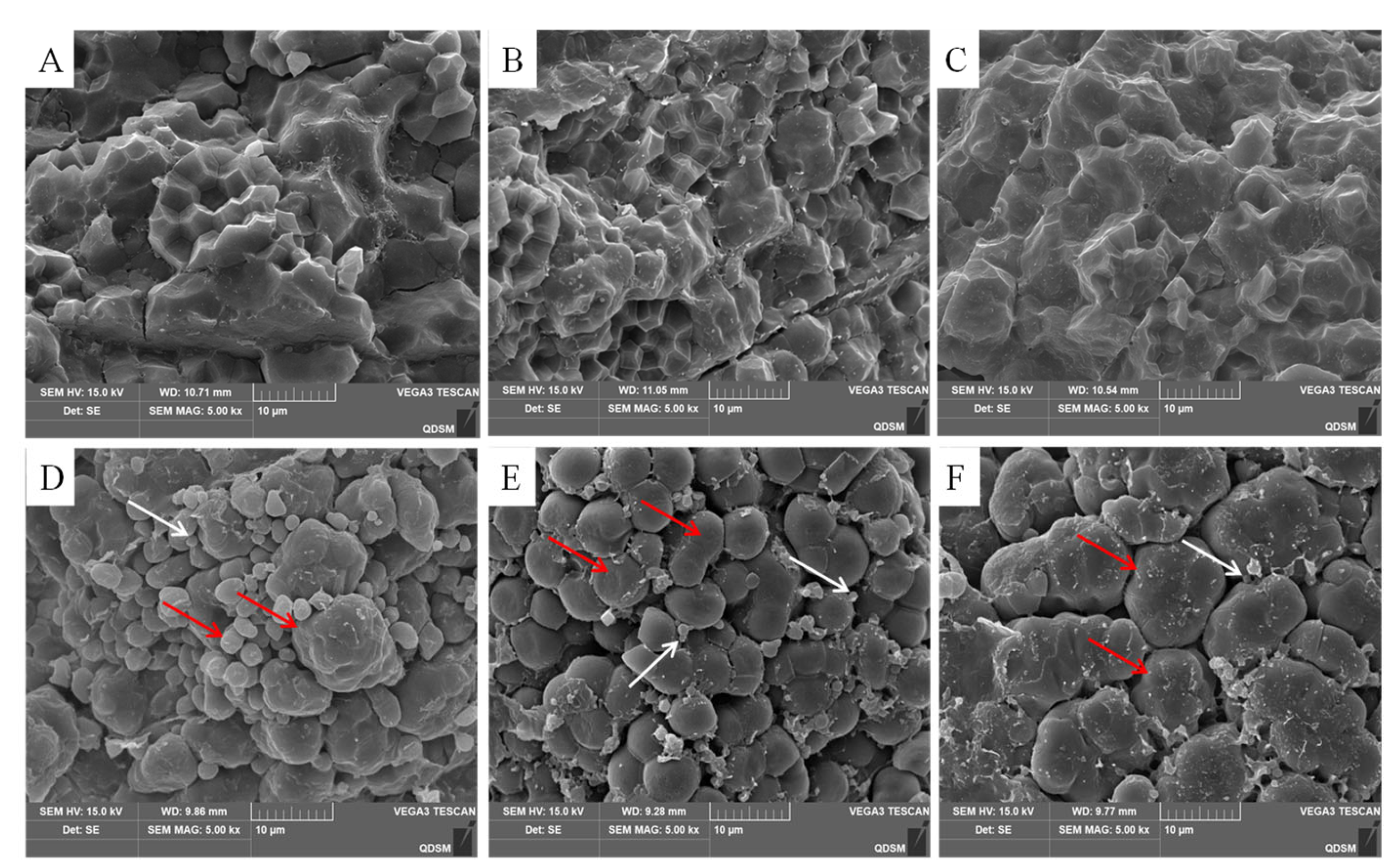

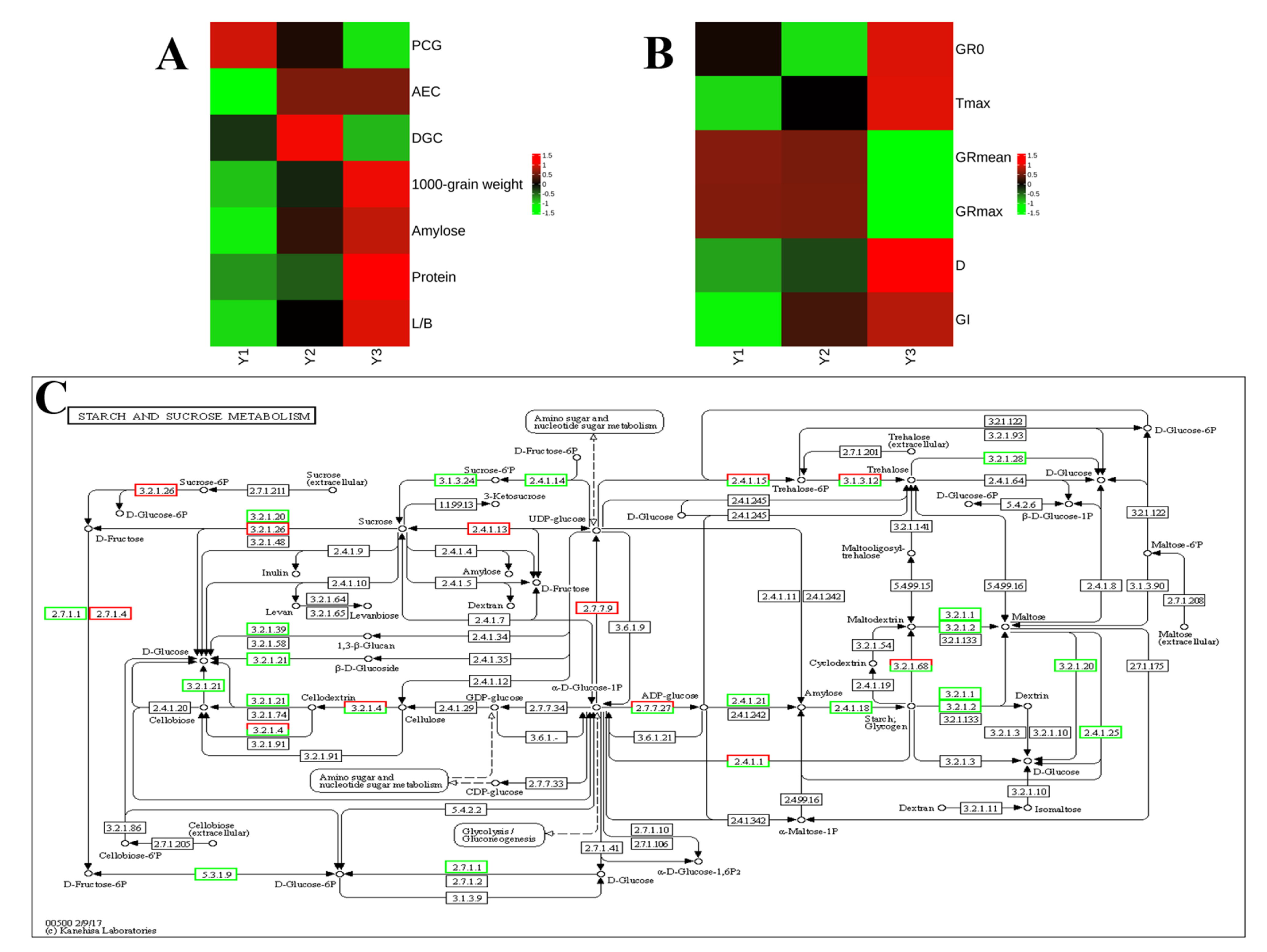
| Grain Position | PCG | AEC | DGC | 1000-Grain Weight(g) | Amylose (%) | Protein (%) | L/B |
|---|---|---|---|---|---|---|---|
| Y1 | 73.2 ± 2.68 a | 30.1 ± 1.60 b | 22.0 ± 0.71 b | 28.9 ± 0.02 c | 14.56 ± 0.112 b | 8.47 ± 0.140 b | 2.85 ± 0.012 c |
| Y2 | 62.5 ± 1.33 b | 41.7 ± 1.06 a | 26.1 ± 1.17 a | 29.2 ± 0.03 b | 15.39 ± 0.514 a | 8.53 ± 0.314 b | 2.92 ± 0.015 b |
| Y3 | 48.1 ± 1.92 c | 41.8 ± 0.81 a | 20.1 ± 1.18 b | 29.7 ± 0.04 a | 15.81 ± 0.042 a | 9.15 ± 0.406 a | 2.99 ± 0.010 a |
| Treatment | GR0 (mg.grain.d−1) | Tmax (d) | GRmean (mg.grain.d−1) | GRmax (mg.grain.d−1) | D (d) | GI (mg.grain−1) |
|---|---|---|---|---|---|---|
| Y1 | 3.22 ± 0.062 b | 3.53 ± 0.103 c | 1.48 ± 0.026 a | 2.96 ± 0.052 a | 14.86 ± 0.054 c | 22.00 ± 0.308 c |
| Y2 | 2.69 ± 0.479 c | 4.75 ± 0.018 b | 1.47 ± 0.025 a | 2.95 ± 0.050 a | 16.40 ± 0.361 b | 24.15 ± 0.123 b |
| Y3 | 3.68 ± 0.315 a | 6.00 ± 0.068 a | 1.14 ± 0.018 b | 2.28 ± 0.035 b | 21.96 ± 0.041 a | 24.94 ± 0.340 a |
| Enriched Pathway | ID | Y1-vs-Y2 | Y2-vs-Y3 | Y1-vs-Y3 |
|---|---|---|---|---|
| Metabolic pathways | ko01100 | + | + | + |
| Starch and sucrose metabolism | ko00500 | + | + | + |
| Carbon fixation in photosynthetic organisms | ko00710 | + | + | |
| Carbon metabolism | ko01200 | + | + | |
| Glycolysis/gluconeogenesis | ko00010 | + | + | |
| Galactose metabolism | ko00052 | + | + | |
| MAPK signaling pathway–plant | ko04016 | + | + | |
| Diterpenoid biosynthesis | ko00904 | + | ||
| Carotenoid biosynthesis | ko00906 | + | ||
| Protein processing in endoplasmic reticulum | ko04141 | + | ||
| Biosynthesis of secondary metabolites | ko01110 | + | ||
| Amino sugar and nucleotide sugar metabolism | ko00520 | + | ||
| Aflatoxin biosynthesis | ko00254 | + | ||
| Riboflavin metabolism | ko00740 | + | ||
| Terpenoid backbone biosynthesis | ko00900 | + | ||
| Pyruvate metabolism | ko00620 | + | ||
| Ascorbate and aldarate metabolism | ko00053 | + | ||
| Glucosinolate biosynthesis | ko00966 | + | ||
| Alanine, aspartate, and glutamate metabolism | ko00250 | + | ||
| Neomycin, kanamycin, and gentamicin biosynthesis | ko00524 | + | ||
| Butanoate metabolism | ko00650 | + |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Z.; Xi, M.; Xu, Y.; Sun, X.; Tu, D.; Zhou, Y.; Ji, Y.; Yang, L. Molecular Mechanisms of Grain Chalkiness Variation in Rice Panicles. Plants 2025, 14, 244. https://doi.org/10.3390/plants14020244
Li Z, Xi M, Xu Y, Sun X, Tu D, Zhou Y, Ji Y, Yang L. Molecular Mechanisms of Grain Chalkiness Variation in Rice Panicles. Plants. 2025; 14(2):244. https://doi.org/10.3390/plants14020244
Chicago/Turabian StyleLi, Zhong, Min Xi, Youzun Xu, Xueyuan Sun, Debao Tu, Yongjin Zhou, Yalan Ji, and Linsheng Yang. 2025. "Molecular Mechanisms of Grain Chalkiness Variation in Rice Panicles" Plants 14, no. 2: 244. https://doi.org/10.3390/plants14020244
APA StyleLi, Z., Xi, M., Xu, Y., Sun, X., Tu, D., Zhou, Y., Ji, Y., & Yang, L. (2025). Molecular Mechanisms of Grain Chalkiness Variation in Rice Panicles. Plants, 14(2), 244. https://doi.org/10.3390/plants14020244





