Genome-Wide Identification, Phylogenetic Classification, and Expression Profiling of the HSF Gene Family in Rosa hybrida Under Heat and Drought Stress
Abstract
1. Introduction
2. Results
2.1. Identification of HSF Genes in Rosa hybrida
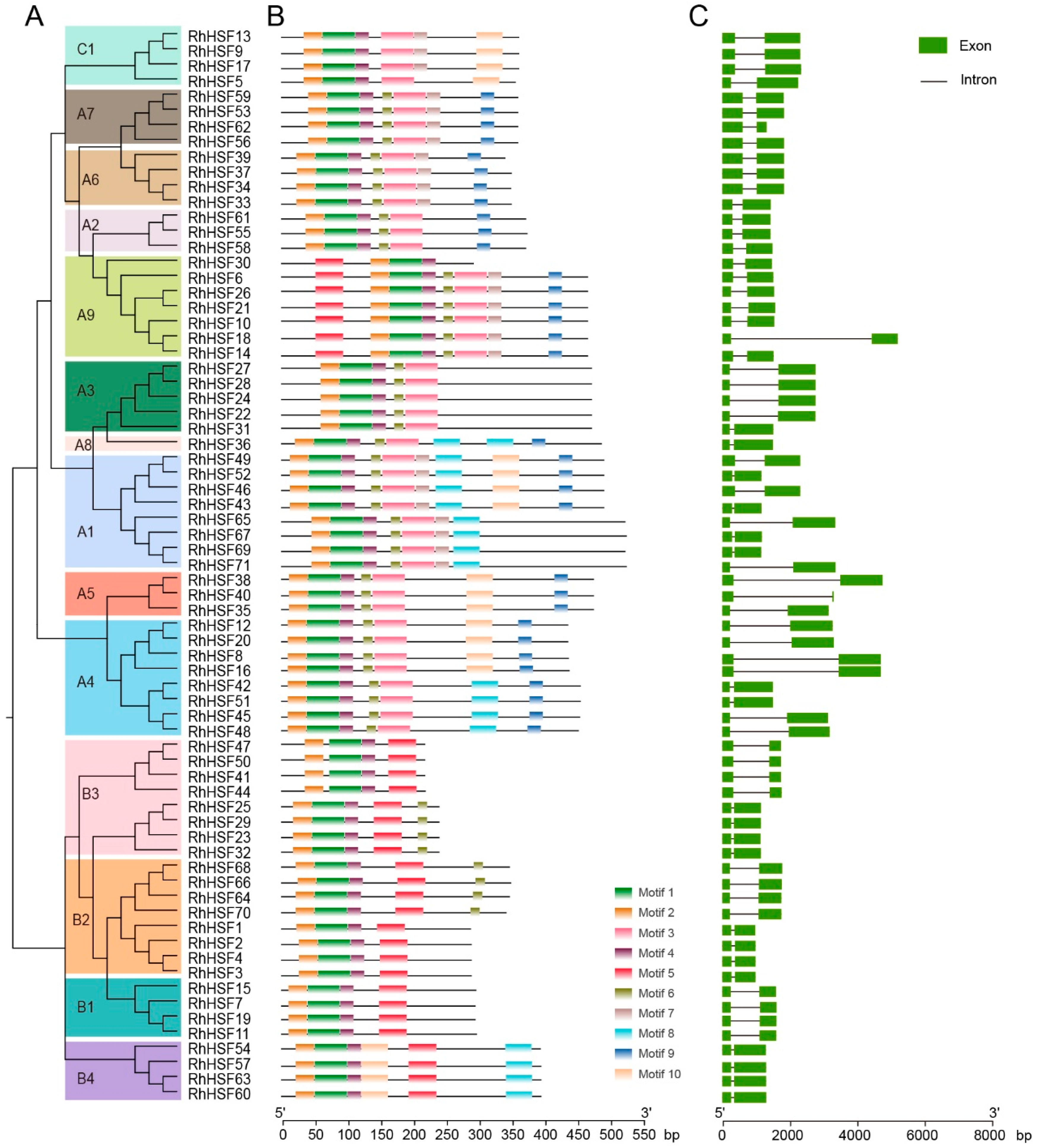
2.2. Phylogenetic Relationships and Classification of RhHSFs
2.3. Conserved Motif Composition and Gene Structure of RhHSF Family
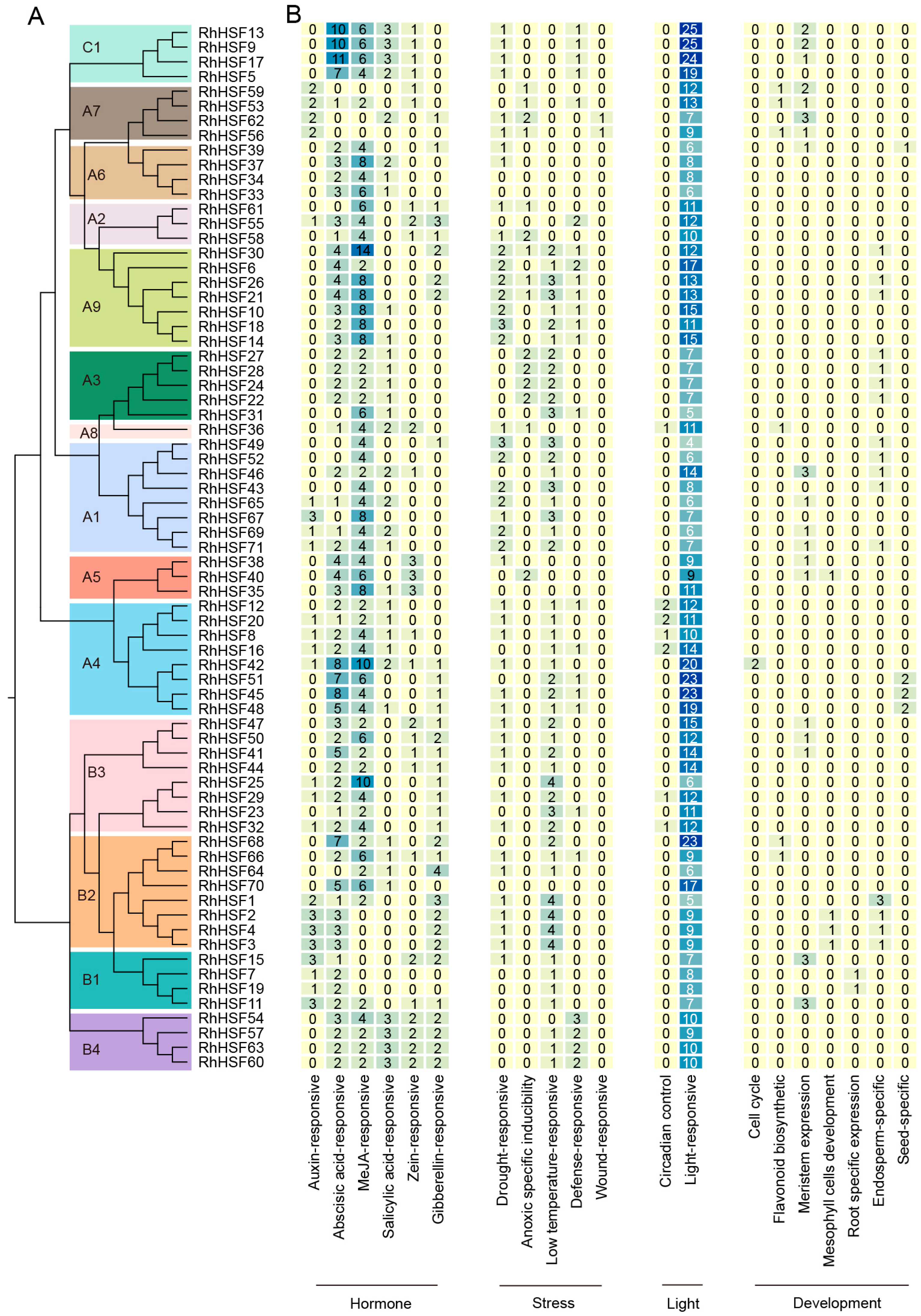
2.4. Integrated Analysis of Cis-Element Distribution and Enrichment Patterns
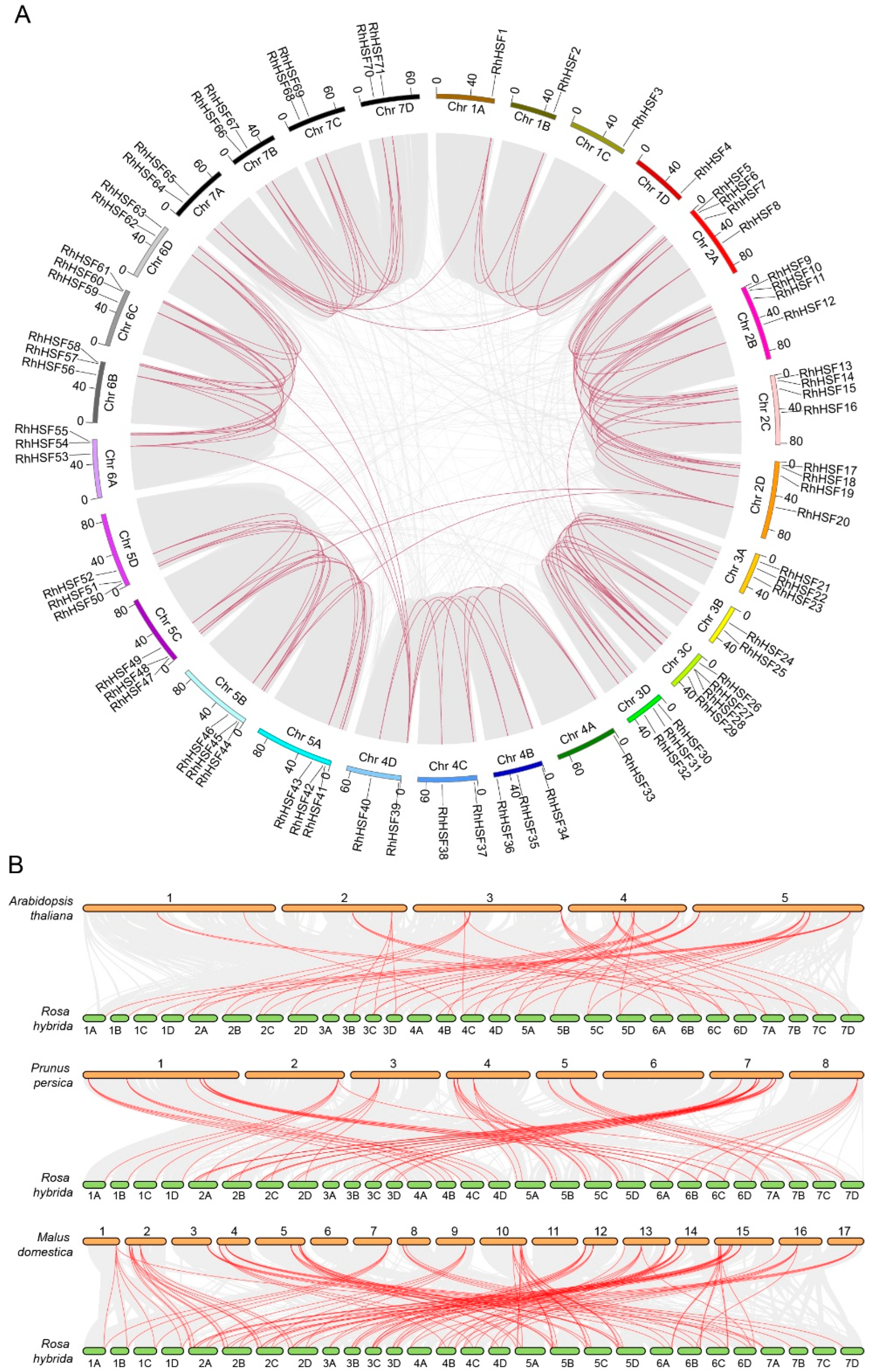
2.5. Chromosomal Distribution and Synteny Analysis of RhHSFs
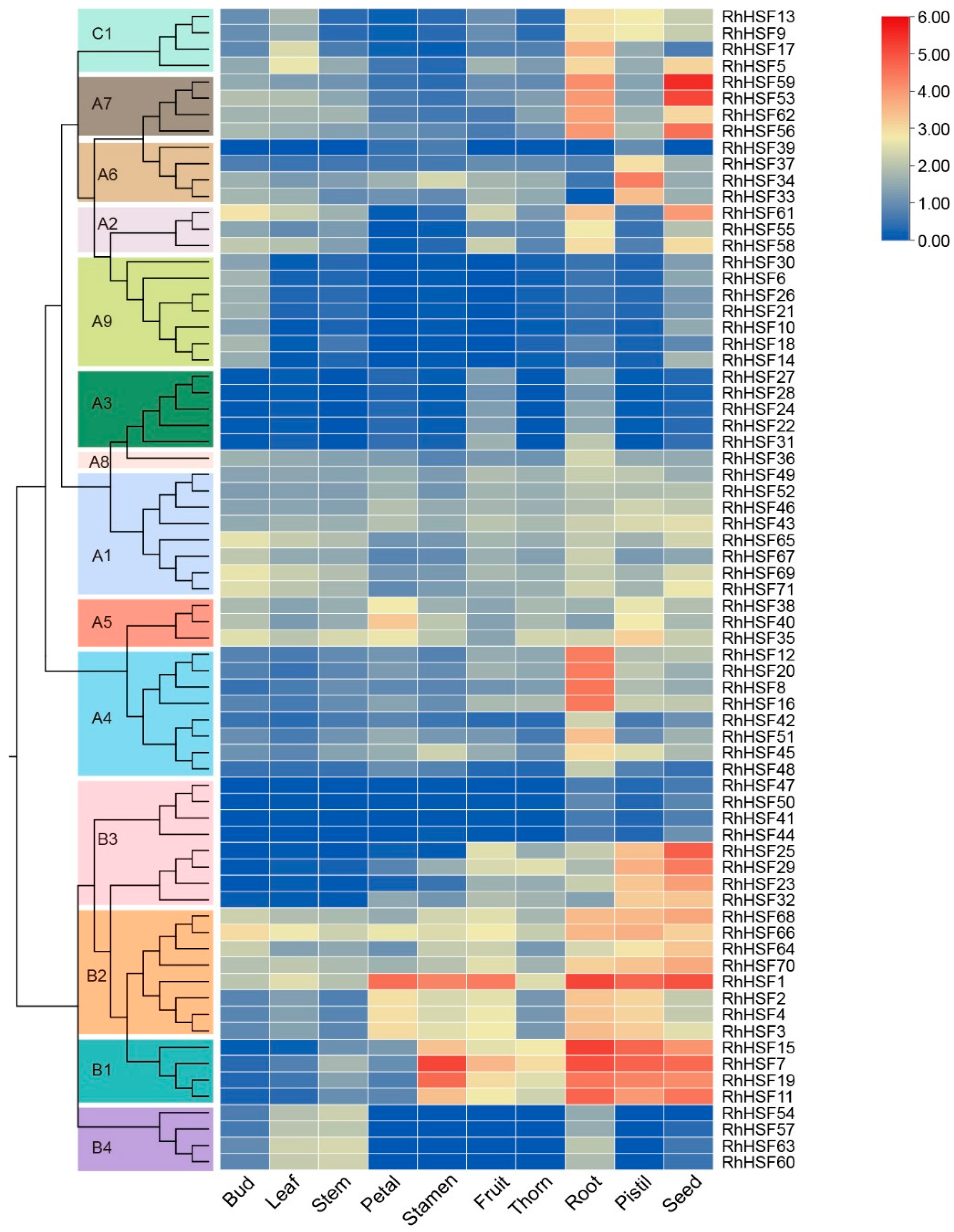
2.6. Tissue-Specific Expression Profiles of RhHSFs
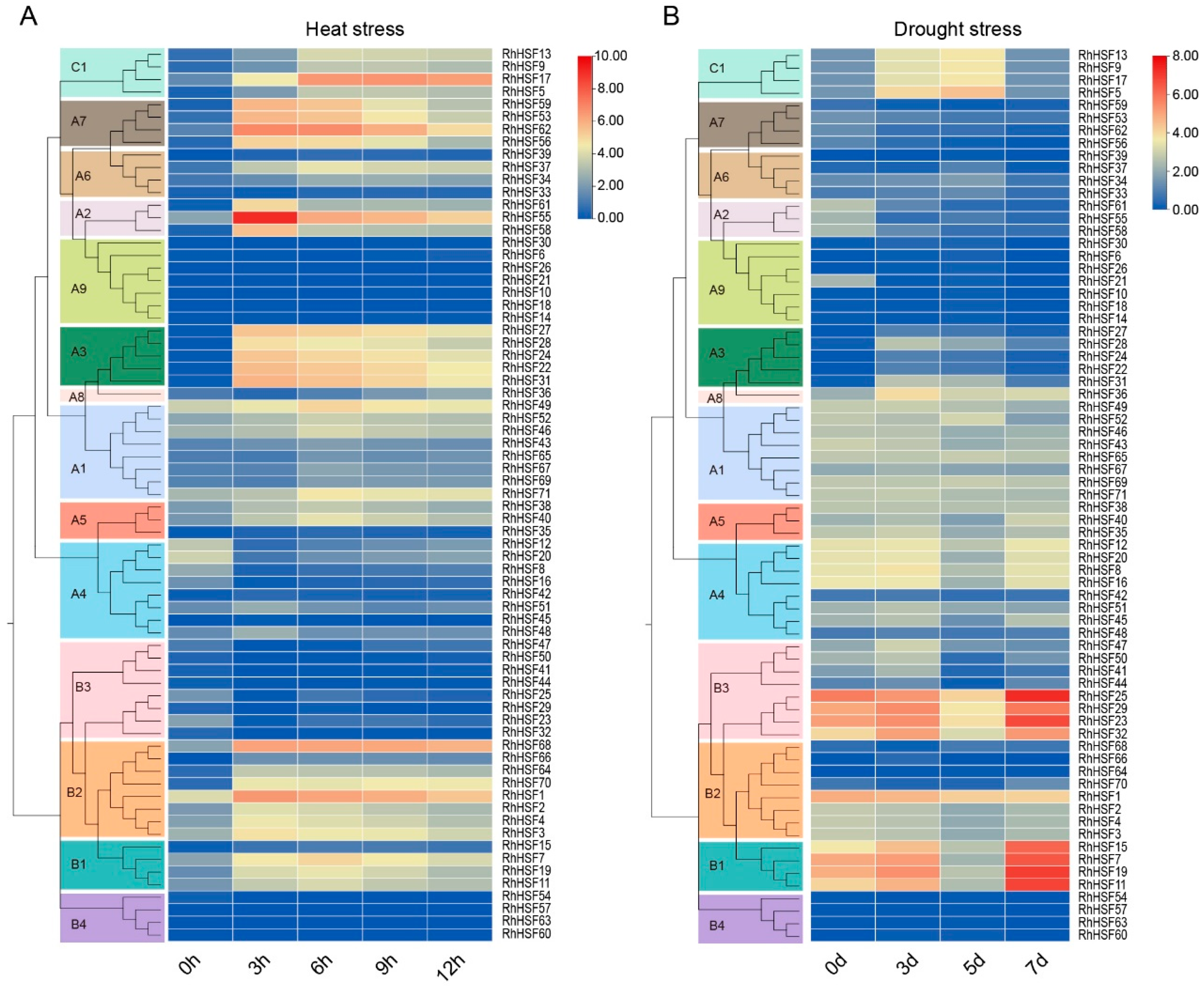
2.7. Dynamics of RhHSF Gene Expression Under Heat Stress
2.8. Expression Profiles of RhHSFs Under Drought Stress
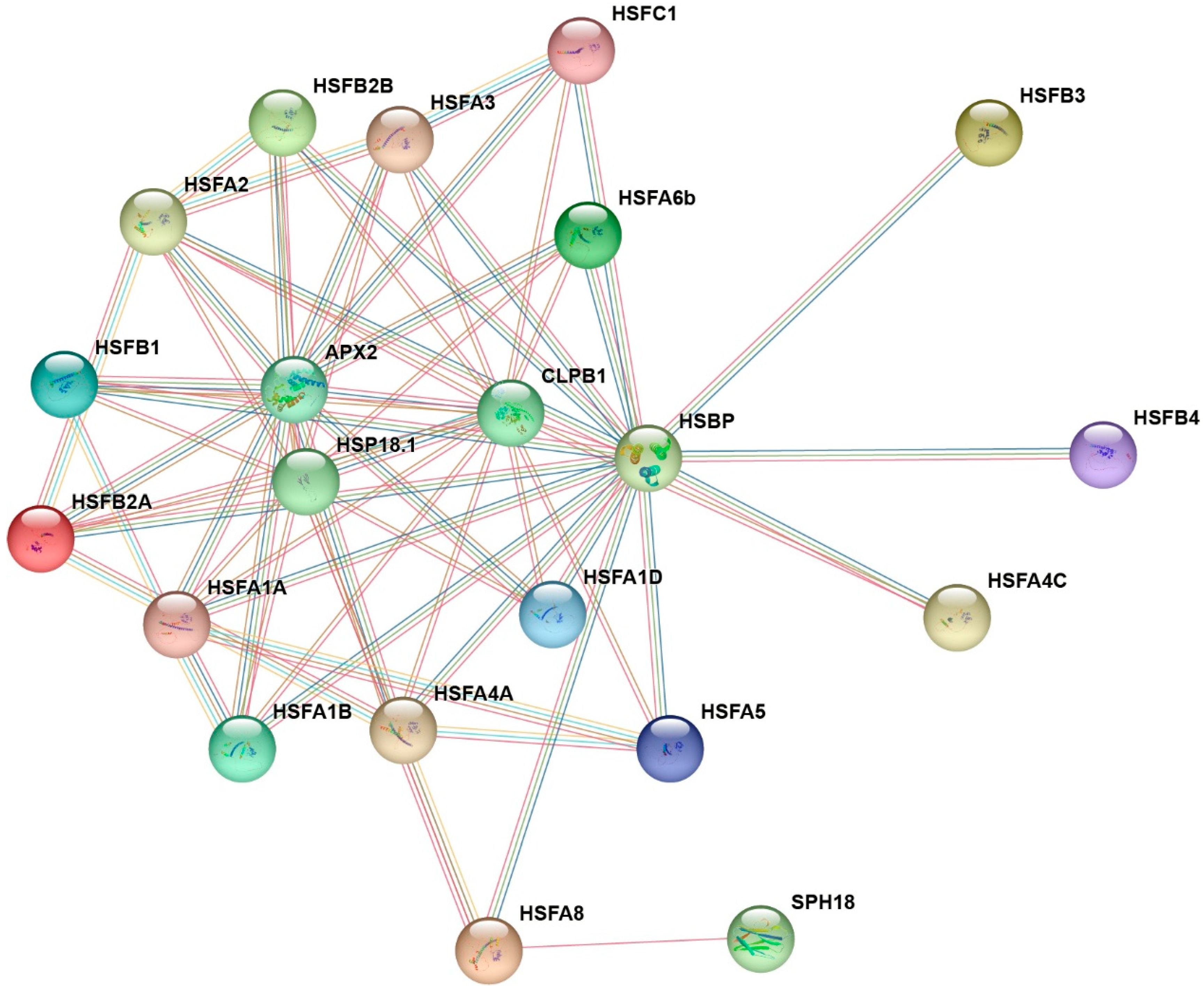
2.9. Protein–Protein Interaction Network Analysis of RhHSFs
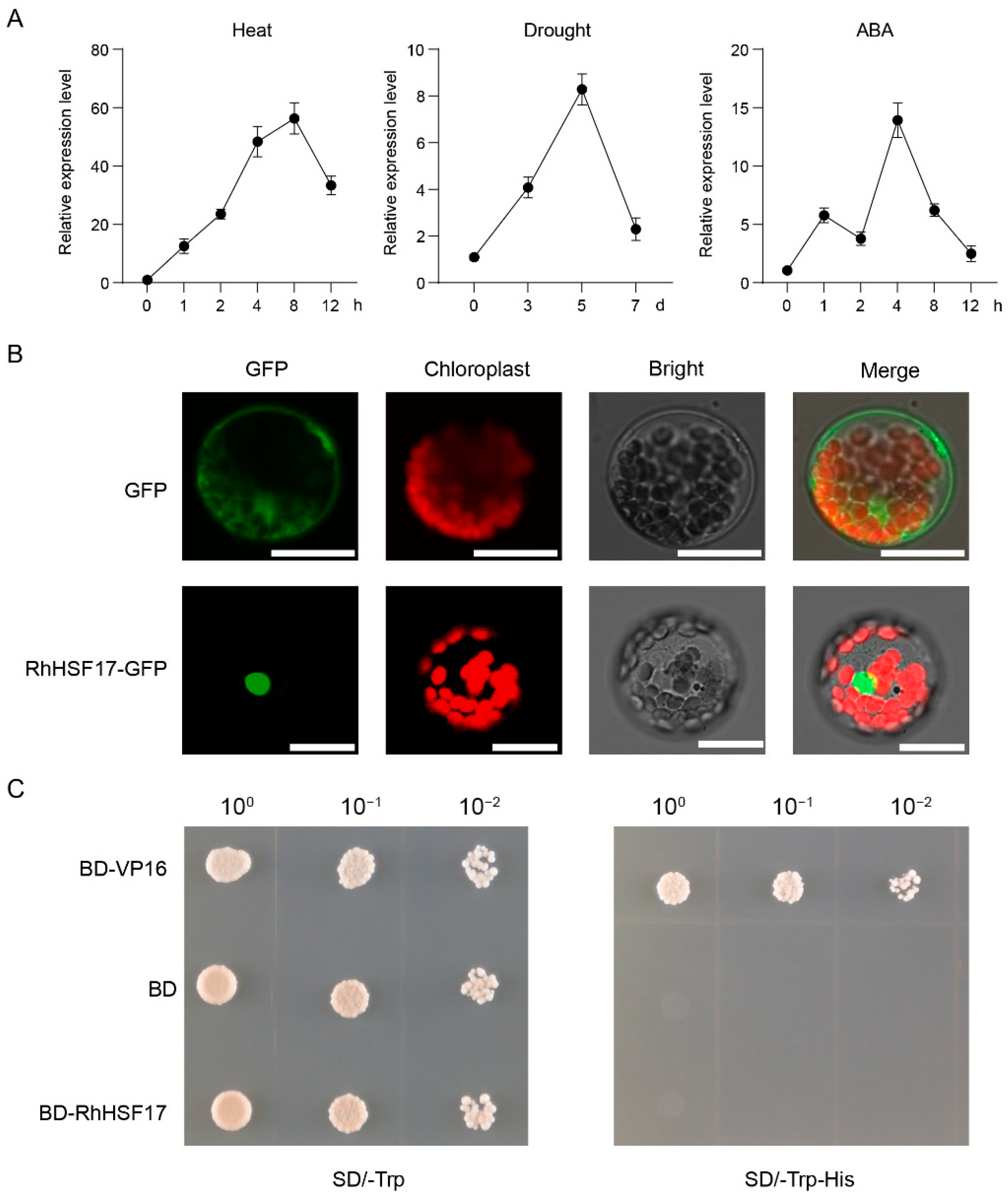
2.10. The Functional Analysis of RhHSF17
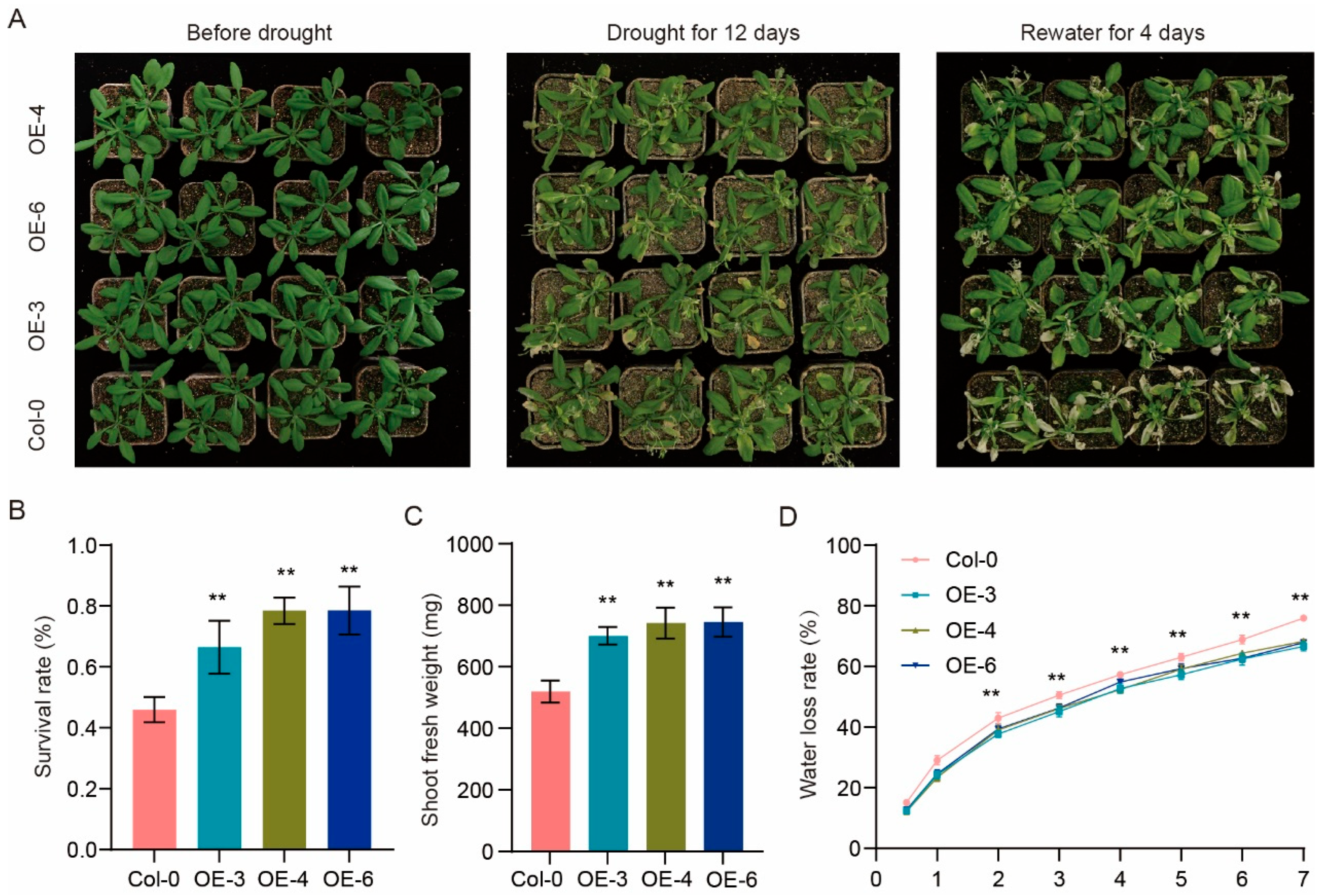
2.11. RhHSF17 Overexpression Enhances Drought Tolerance in Arabidopsis
3. Discussion
4. Materials and Methods
4.1. Genome-Wide Identification of HSF Genes in Rosa hybrida
4.2. Phylogenetic Analysis and Classification of RhHSF Genes
4.3. Conserved Motif and Gene Structure Analysis
4.4. Cis-Acting Element Prediction in Promoters
4.5. Chromosomal Mapping and Synteny Analysis
4.6. RNA-Seq Data Processing and Expression Profiling
4.7. Protein–Protein Interaction Network Construction
4.8. Subcellular Localization of RhHSF17
4.9. Transcription Activation Assay
4.10. Heat Stress and ABA Treatments
4.11. Quantitative Real-Time PCR (qRT-PCR) Analysis of RhHSF17
4.12. Generation of Transgenic Arabidopsis Overexpressing RhHSF17
4.13. Drought Treatment at the Seedling Stage in Arabidopsis Transgenic Lines
4.14. Primer Information
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Chaudhry, S.; Sidhu, G.P.S. Climate change regulated abiotic stress mechanisms in plants: A comprehensive review. Plant Cell Rep. 2022, 41, 1–31. [Google Scholar] [CrossRef]
- Zandalinas, S.I.; Mittler, R. Plant responses to multifactorial stress combination. New Phytol. 2022, 234, 1161–1167. [Google Scholar] [CrossRef] [PubMed]
- Murmu, K.; Sarkar, A.; Sarma, S.S.; Jana, K.; Murmu, S. Mitigating the impact of drought and heat stress on crop productivity and environmental sustainability. In Drought and Heat Stress in Agriculture: Implications, Mitigation and Policy Approaches; Springer: Singapore, 2025; pp. 155–173. [Google Scholar]
- Kopecká, R.; Kameniarová, M.; Černý, M.; Brzobohatý, B.; Novák, J. Abiotic stress in crop production. Int. J. Mol. Sci. 2023, 24, 6603. [Google Scholar] [CrossRef]
- Hofmann, T.A.; Atkinson, W.; Fan, M.; Simkin, A.J.; Jindal, P.; Lawson, T. Impact of climate-driven changes in temperature on stomatal anatomy and physiology. Philos. Trans. B 2025, 380, 20240244. [Google Scholar] [CrossRef]
- Haghpanah, M.; Hashemipetroudi, S.; Arzani, A.; Araniti, F. Drought tolerance in plants: Physiological and molecular responses. Plants 2024, 13, 2962. [Google Scholar] [CrossRef]
- Chaves, M.M.; Costa, J.M.; Zarrouk, O.; Pinheiro, C.; Lopes, C.M.; Pereira, J.S. Controlling stomatal aperture in semi-arid regions—The dilemma of saving water or being cool? Plant Sci. 2016, 251, 54–64. [Google Scholar] [CrossRef]
- Vihervaara, A.; Duarte, F.M.; Lis, J.T. Molecular mechanisms driving transcriptional stress responses. Nat. Rev. Genet. 2018, 19, 385–397. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Xu, J. The heat shock transcription factors regulate response mechanisms to abiotic stresses in plants. Crop Des. 2025, 4, 100109. [Google Scholar] [CrossRef]
- Scharf, K.; Berberich, T.; Ebersberger, I.; Nover, L. The plant heat stress transcription factor (hsf) family: Structure, function and evolution. Biochim. Biophys. Acta-Gene Regul. Mech. 2012, 1819, 104–119. [Google Scholar] [CrossRef]
- Åkerfelt, M.; Morimoto, R.I.; Sistonen, L. Heat shock factors: Integrators of cell stress, development and lifespan. Nat. Rev. Mol. Cell Biol. 2010, 11, 545–555. [Google Scholar] [CrossRef] [PubMed]
- Guo, M.; Liu, J.; Ma, X.; Luo, D.; Gong, Z.; Lu, M. The plant heat stress transcription factors (HSFs): Structure, regulation, and function in response to abiotic stresses. Front. Plant Sci. 2016, 7, 114. [Google Scholar] [CrossRef]
- Kotak, S.; Port, M.; Ganguli, A.; Bicker, F.; Von Koskull Döring, P. Characterization of c-terminal domains of arabidopsis heat stress transcription factors (hsfs) and identification of a new signature combination of plant class a hsfs with AHA and NES motifs essential for activator function and intracellular localization. Plant. J. 2004, 39, 98–112. [Google Scholar] [CrossRef] [PubMed]
- Kmiecik, S.W.; Mayer, M.P. Molecular mechanisms of heat shock factor 1 regulation. Trends Biochem. Sci. 2022, 47, 218–234. [Google Scholar] [CrossRef] [PubMed]
- Andrási, N.; Pettkó-Szandtner, A.; Szabados, L. Diversity of plant heat shock factors: Regulation, interactions, and functions. J. Exp. Bot. 2020, 72, 1558–1575. [Google Scholar] [CrossRef] [PubMed]
- Guertin, M.J.; Lis, J.T. Chromatin landscape dictates HSF binding to target DNA elements. PLoS Genet. 2010, 6, e1001114. [Google Scholar] [CrossRef]
- Yurina, N.P. Heat shock proteins in plant protection from oxidative stress. Mol. Biol. 2023, 57, 951–964. [Google Scholar] [CrossRef]
- Huang, L.; Zhou, M.; Ding, Y.; Zhu, C. Gene networks involved in plant heat stress response and tolerance. Int. J. Mol. Sci. 2022, 23, 11970. [Google Scholar] [CrossRef]
- Yoshida, T.; Ohama, N.; Nakajima, J.; Kidokoro, S.; Mizoi, J.; Nakashima, K.; Maruyama, K.; Kim, J.; Seki, M.; Todaka, D. Arabidopsis HsfA1 transcription factors function as the main positive regulators in heat shock-responsive gene expression. Mol. Genet. Genom. 2011, 286, 321–332. [Google Scholar] [CrossRef]
- LIU, H.C.; LIAO, H.T.; CHARNG, Y.Y. The role of class a1 heat shock factors (HSFA1s) in response to heat and other stresses in Arabidopsis. Plant Cell Environ. 2011, 34, 738–751. [Google Scholar] [CrossRef]
- Zhang, L.; Li, Y.; Xing, D.; Gao, C. Characterization of mitochondrial dynamics and subcellular localization of ROS reveal that HsfA2 alleviates oxidative damage caused by heat stress in arabidopsis. J. Exp. Bot. 2009, 60, 2073–2091. [Google Scholar] [CrossRef]
- Mishra, S.K.; Chaudhary, C.; Baliyan, S.; Poonia, A.K.; Sirohi, P.; Kanwar, M.; Gazal, S.; Kumari, A.; Sircar, D.; Germain, H.; et al. Heat-stress-responsive HvHSFA2e gene regulates the heat and drought tolerance in barley through modulation of phytohormone and secondary metabolic pathways. Plant Cell Rep. 2024, 43, 172. [Google Scholar] [CrossRef]
- Kang, Y.; Sun, P.; Yang, Y.; Li, M.; Wang, H.; Sun, X.; Jin, W. Genome-wide analysis of the hsf gene family in rosa chinensis and RcHsf17 function in thermotolerance. Int. J. Mol. Sci. 2024, 26, 287. [Google Scholar] [CrossRef] [PubMed]
- Bi, H.; Zhao, Y.; Li, H.; Liu, W. Wheat heat shock factor TaHsfA6f increases ABA levels and enhances tolerance to multiple abiotic stresses in transgenic plants. Int. J. Mol. Sci. 2020, 21, 3121. [Google Scholar] [CrossRef] [PubMed]
- Kumar, R.R.; Goswami, S.; Singh, K.; Dubey, K.; Rai, G.K.; Singh, B.; Singh, S.; Grover, M.; Mishra, D.; Kumar, S. Characterization of novel heat-responsive transcription factor (TaHSFA6e) gene involved in regulation of heat shock proteins (HSPs)—A key member of heat stress-tolerance network of wheat. J. Biotechnol. 2018, 279, 1–12. [Google Scholar] [CrossRef]
- Darras, A.I. Implementation of sustainable practices to ornamental plant cultivation worldwide: A critical review. Agronomy 2020, 10, 1570. [Google Scholar] [CrossRef]
- Wang, H.; Xu, W.; Zhang, X.; Wang, L.; Jia, S.; Zhao, S.; Li, W.; Lu, R.; Ren, A.; Zhang, S. Transcriptomics and metabolomics analyses of Rosa hybrida to identify heat stress response genes and metabolite pathways. BMC Plant Biol. 2024, 24, 874. [Google Scholar] [CrossRef]
- Wang, H.; Shen, Y.; Wang, K.; He, S.; Kim, W.; Shang, W.; Wang, Z.; Shi, L. Effect of exogenous calcium on the heat tolerance in rosa hybrida ‘carolla’. Horticulturae 2022, 8, 980. [Google Scholar] [CrossRef]
- Zhang, Z.; Yang, T.; Liu, Y.; Wu, S.; Sun, H.; Wu, J.; Li, Y.; Zheng, Y.; Ren, H.; Yang, Y. Haplotype-resolved genome assembly and resequencing provide insights into the origin and breeding of modern rose. Nat. Plants 2024, 10, 1659–1671. [Google Scholar] [CrossRef]
- Bian, X.H.; Li, W.; Niu, C.F.; Wei, W.; Hu, Y.; Han, J.Q.; Lu, X.; Tao, J.J.; Jin, M.; Qin, H. A class b heat shock factor selected for during soybean domestication contributes to salt tolerance by promoting flavonoid biosynthesis. New Phytol. 2020, 225, 268–283. [Google Scholar] [CrossRef]
- Hao, X.; He, S. Genome-wide identification, classification and expression analysis of the heat shock transcription factor family in garlic (Allium sativum L.). BMC Plant Biol. 2024, 24, 421. [Google Scholar] [CrossRef]
- Yang, Y.; Yin, J.; Zhu, L.; Xu, L.; Wu, W.; Lu, Y.; Chen, J.; Shi, J.; Hao, Z. Genome-wide analysis of the liriodendron chinense hsf gene family under abiotic stress and characterization of the LcHsfA2a gene. Int. J. Mol. Sci. 2024, 25, 2733. [Google Scholar] [CrossRef]
- Li, S.; Sun, Y.; Hu, Z.; Dong, F.; Zhu, J.; Cao, M.; Wang, C. Cloning and expression analysis of RhHsf24 gene in rose (Rosa hybrida). Sci. Rep. 2025, 15, 8182. [Google Scholar] [CrossRef]
- Wang, H.; Li, M.; Yang, Y.; Sun, P.; Zhou, S.; Kang, Y.; Xu, Y.; Yuan, X.; Feng, Z.; Jin, W. Physiological and molecular responses of different rose (rosa hybrida l.) Cultivars to elevated ozone levels. Plant Direct 2023, 7, e513. [Google Scholar] [CrossRef]
- Jiao, Y.; Wickett, N.J.; Ayyampalayam, S.; Chanderbali, A.S.; Landherr, L.; Ralph, P.E.; Tomsho, L.P.; Hu, Y.; Liang, H.; Soltis, P.S. Ancestral polyploidy in seed plants and angiosperms. Nature 2011, 473, 97–100. [Google Scholar] [CrossRef]
- Li, G.; Shi, X.; Lin, Q.; Lv, M.; Chen, J.; Wen, Y.; Feng, Z.; Azam, S.M.; Cheng, Y.; Wang, S. Genome-wide identification and expression analysis of heat shock transcription factors in camellia sinensis under abiotic stress. Plants 2025, 14, 697. [Google Scholar] [CrossRef]
- Liao, Y.; Liu, Z.; Gichira, A.W.; Yang, M.; Mbichi, R.W.; Meng, L.; Wan, T. Deep evaluation of the evolutionary history of the heat shock factor (HSF) gene family and its expansion pattern in seed plants. PeerJ 2022, 10, e13603. [Google Scholar] [CrossRef]
- Finkelstein, R. Abscisic acid synthesis and response. Arab. Book/Am. Soc. Plant Biol. 2013, 11, e0166. [Google Scholar] [CrossRef] [PubMed]
- Cutler, S.R.; Rodriguez, P.L.; Finkelstein, R.R.; Abrams, S.R. Abscisic acid: Emergence of a core signaling network. Annu. Rev. Plant Biol. 2010, 61, 651–679. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Zhang, H.; Zhao, H.; Gao, T.; Song, A.; Jiang, J.; Chen, F.; Chen, S. Chrysanthemum cm HSFA 4 gene positively regulates salt stress tolerance in transgenic chrysanthemum. Plant Biotechnol. J. 2018, 16, 1311–1321. [Google Scholar] [CrossRef] [PubMed]
- Kotak, S.; Vierling, E.; Baumlein, H.; Koskull-Döring, P.V. A novel transcriptional cascade regulating expression of heat stress proteins during seed development of Arabidopsis. The Plant Cell 2007, 19, 182–195. [Google Scholar] [CrossRef]
- Qi, C.; Dong, D.; Li, Y.; Wang, X.; Guo, L.; Liu, L.; Dong, X.; Li, X.; Yuan, X.; Ren, S. Heat shock-induced cold acclimation in cucumber through CsHSFA1d-activated JA biosynthesis and signaling. Plant. J. 2022, 111, 85–102. [Google Scholar] [CrossRef]
- Dickinson, P.J.; Kumar, M.; Martinho, C.; Yoo, S.J.; Lan, H.; Artavanis, G.; Charoensawan, V.; Schöttler, M.A.; Bock, R.; Jaeger, K.E.; et al. Chloroplast signaling gates thermotolerance in Arabidopsis. Cell Rep. 2018, 22, 1657–1665. [Google Scholar] [CrossRef]
- Liu, A.; Zou, J.; Liu, C.; Zhou, X.; Zhang, X.; Luo, G.; Chen, X. Over-expression of OsHsfA7 enhanced salt and drought tolerance in transgenic rice. BMB Rep. 2013, 46, 31. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Yang, L.; Fang, Y.; Wang, G.; Lyu, S.; Deng, S. A genome-wide-level insight into the HSF gene family of rhodomyrtus tomentosa and the functional divergence of RtHSFA2a and RtHSFA2b in thermal adaptation. Plant Physiol. Biochem. 2025, 220, 109460. [Google Scholar] [CrossRef]
- Guo, M.; Lu, J.; Zhai, Y.; Chai, W.; Gong, Z.; Lu, M. Genome-wide analysis, expression profile of heat shock factor gene family (CaHsfs) and characterisation of CaHsfA2 in pepper (Capsicum annuum L.). BMC Plant Biol. 2015, 15, 151. [Google Scholar] [CrossRef] [PubMed]
- Giorno, F.; Guerriero, G.; Baric, S.; Mariani, C. Heat shock transcriptional factors in malus domestica: Identification, classification and expression analysis. BMC Genom. 2012, 13, 639. [Google Scholar] [CrossRef] [PubMed]
- Mittal, D.; Chakrabarti, S.; Sarkar, A.; Singh, A.; Grover, A. Heat shock factor gene family in rice: Genomic organization and transcript expression profiling in response to high temperature, low temperature and oxidative stresses. Plant Physiol. Biochem. 2009, 47, 785–795. [Google Scholar] [CrossRef]
- Fragkostefanakis, S.; Schleiff, E.; Scharf, K. Back to the basics: The molecular blueprint of plant heat stress transcription factors. Biol. Chem. 2025, 406, 173–187. [Google Scholar] [CrossRef]
- Ramegowda, V.; Senthil, A.; Senthil-Kumar, M. Stress combinations and their interactions in crop plants. Plant Physiol. Rep. 2024, 29, 1–5. [Google Scholar] [CrossRef]
- Wei, J.T.; Zheng, L.; Ma, X.J.; Yu, T.F.; Gao, X.; Hou, Z.H.; Liu, Y.W.; Cao, X.Y.; Chen, J.; Zhou, Y.B. An ABF5b-HsfA2h/HsfC2a-NCED2b/POD4/HSP26 module integrates multiple signaling pathway to modulate heat stress tolerance in wheat. Plant Biotechnol. J. 2025. [Google Scholar] [CrossRef]
- Hu, X.; Chen, D.; Lynne Mclntyre, C.; Fernanda Dreccer, M.; Zhang, Z.; Drenth, J.; Kalaipandian, S.; Chang, H.; Xue, G. Heat shock factor c2a serves as a proactive mechanism for heat protection in developing grains in wheat via an ABA-mediated regulatory pathway. Plant Cell Environ. 2018, 41, 79–98. [Google Scholar] [CrossRef] [PubMed]
- Jiao, S.; Guo, C.; Yao, W.; Zhang, N.; Zhang, J.; Xu, W. An amur grape VaHsfC1 is involved in multiple abiotic stresses. Sci. Hortic. 2022, 295, 110785. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA x: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, B.; Gao, S.; Lercher, M.J.; Hu, S.; Chen, W. Evolview v3: A webserver for visualization, annotation, and management of phylogenetic trees. Nucleic Acids Res. 2019, 47, W270–W275. [Google Scholar] [CrossRef]
- Wang, Y.; Tang, H.; DeBarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.; Jin, H.; Marler, B.; Guo, H. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef]
- Pertea, M.; Kim, D.; Pertea, G.M.; Leek, J.T.; Salzberg, S.L. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and ballgown. Nat. Protoc. 2016, 11, 1650–1667. [Google Scholar] [CrossRef]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef]
- Chen, C.; Wu, Y.; Li, J.; Wang, X.; Zeng, Z.; Xu, J.; Liu, Y.; Feng, J.; Chen, H.; He, Y. TBtools-II: A “one for all, all for one” bioinformatics platform for biological big-data mining. Mol. Plant. 2023, 16, 1733–1742. [Google Scholar] [CrossRef]
- Yoo, S.; Cho, Y.; Sheen, J. Arabidopsis mesophyll protoplasts: A versatile cell system for transient gene expression analysis. Nat. Protoc. 2007, 2, 1565–1572. [Google Scholar] [CrossRef]
- Liu, L.; Guo, Y.; Wu, Z.; Ren, H.; Jiang, Y.; Ma, N.; Gao, J.; Sun, X. RhAGL24 regulating auxin-related gene RhARF18 affects stamen petaloidy in rose. Horticulturae 2022, 8, 407. [Google Scholar] [CrossRef]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef] [PubMed]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for agrobacterium-mediated transformation of Arabidopsis thaliana. Plant. J. 1998, 16, 735–743. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhu, J.; Fan, S.; Li, R.; Dong, F.; Liu, Y.; Wang, C. Genome-Wide Identification, Phylogenetic Classification, and Expression Profiling of the HSF Gene Family in Rosa hybrida Under Heat and Drought Stress. Plants 2025, 14, 3167. https://doi.org/10.3390/plants14203167
Zhu J, Fan S, Li R, Dong F, Liu Y, Wang C. Genome-Wide Identification, Phylogenetic Classification, and Expression Profiling of the HSF Gene Family in Rosa hybrida Under Heat and Drought Stress. Plants. 2025; 14(20):3167. https://doi.org/10.3390/plants14203167
Chicago/Turabian StyleZhu, Jiao, Shikai Fan, Rongchong Li, Fei Dong, Yiyang Liu, and Chengpeng Wang. 2025. "Genome-Wide Identification, Phylogenetic Classification, and Expression Profiling of the HSF Gene Family in Rosa hybrida Under Heat and Drought Stress" Plants 14, no. 20: 3167. https://doi.org/10.3390/plants14203167
APA StyleZhu, J., Fan, S., Li, R., Dong, F., Liu, Y., & Wang, C. (2025). Genome-Wide Identification, Phylogenetic Classification, and Expression Profiling of the HSF Gene Family in Rosa hybrida Under Heat and Drought Stress. Plants, 14(20), 3167. https://doi.org/10.3390/plants14203167





