Targeted Regulation of AhGRF3b by ahy-miR396 Modulates Leaf Growth and Cold Tolerance in Peanut
Abstract
1. Introduction
2. Results
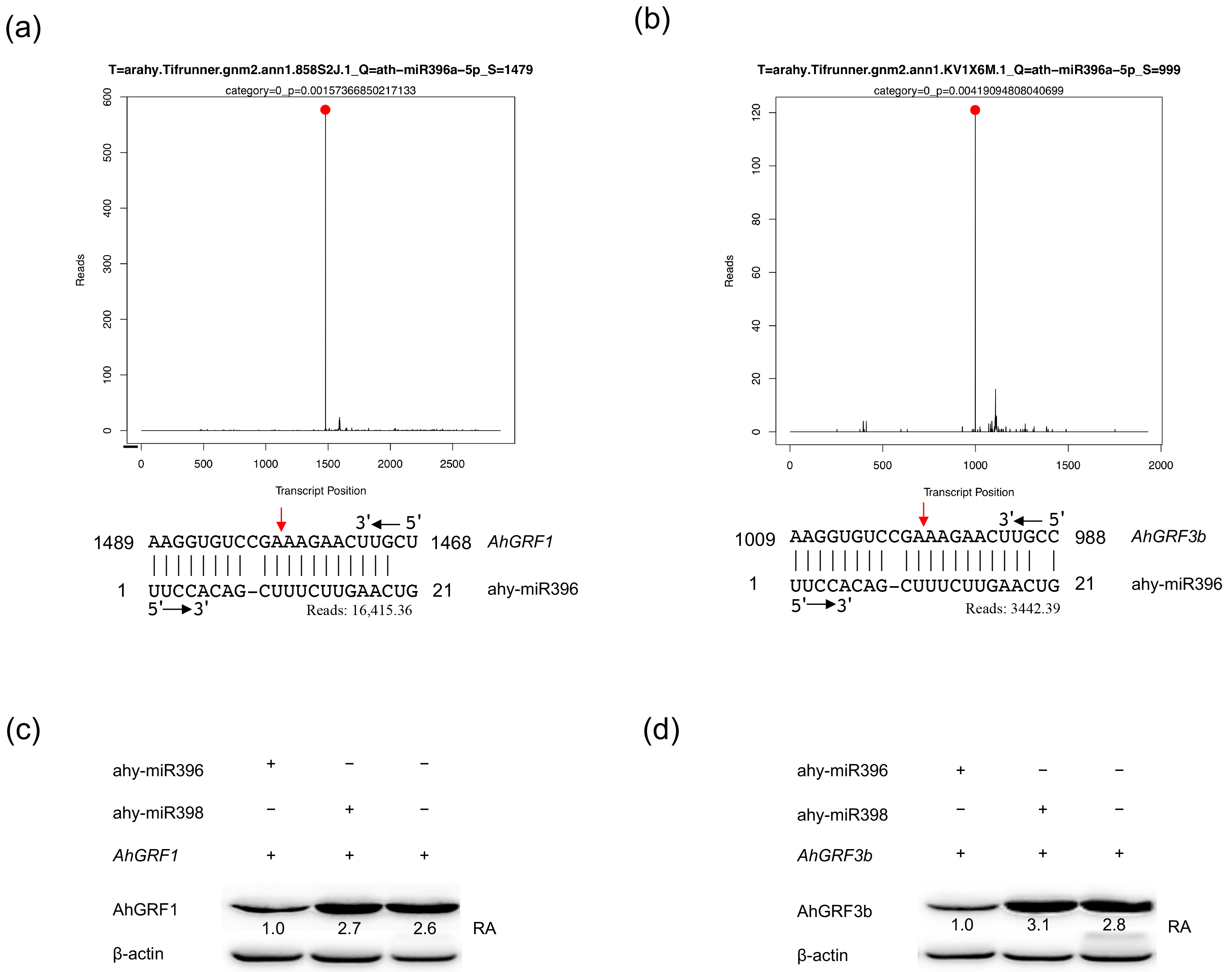
2.1. The AhGRF3b Gene Targeted by ahy-miR396
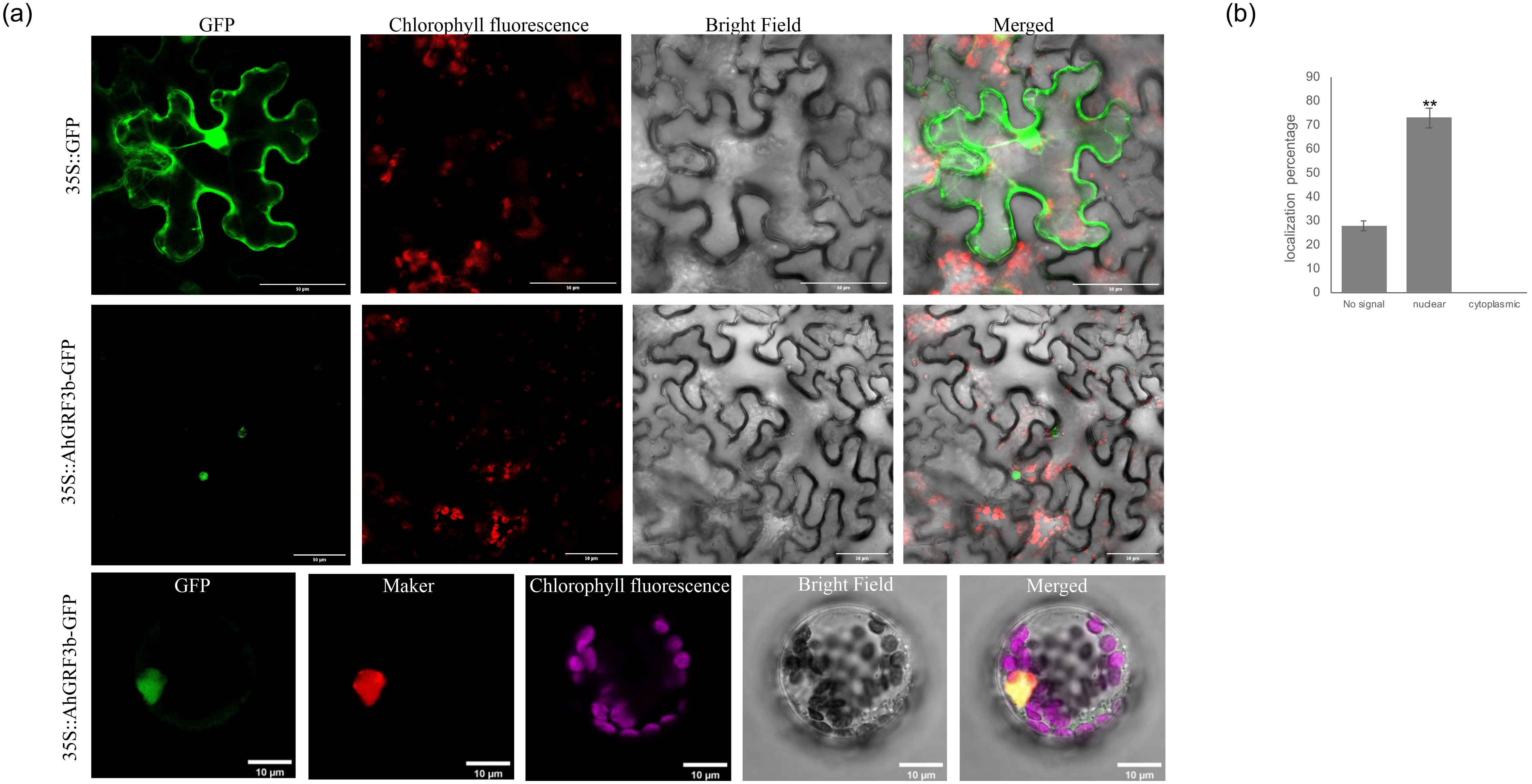
2.2. Subcellular Localization of AhGRF3b
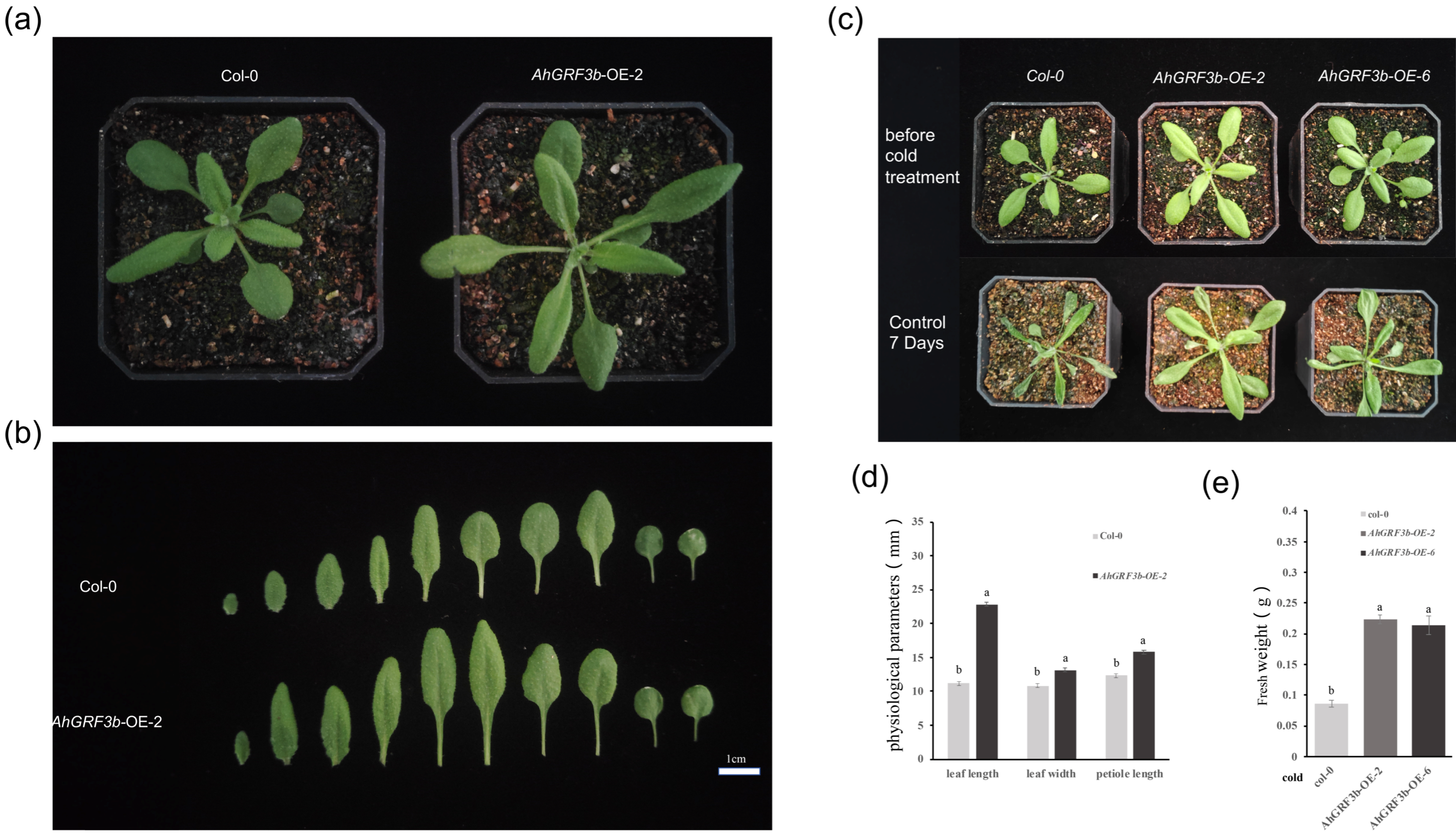
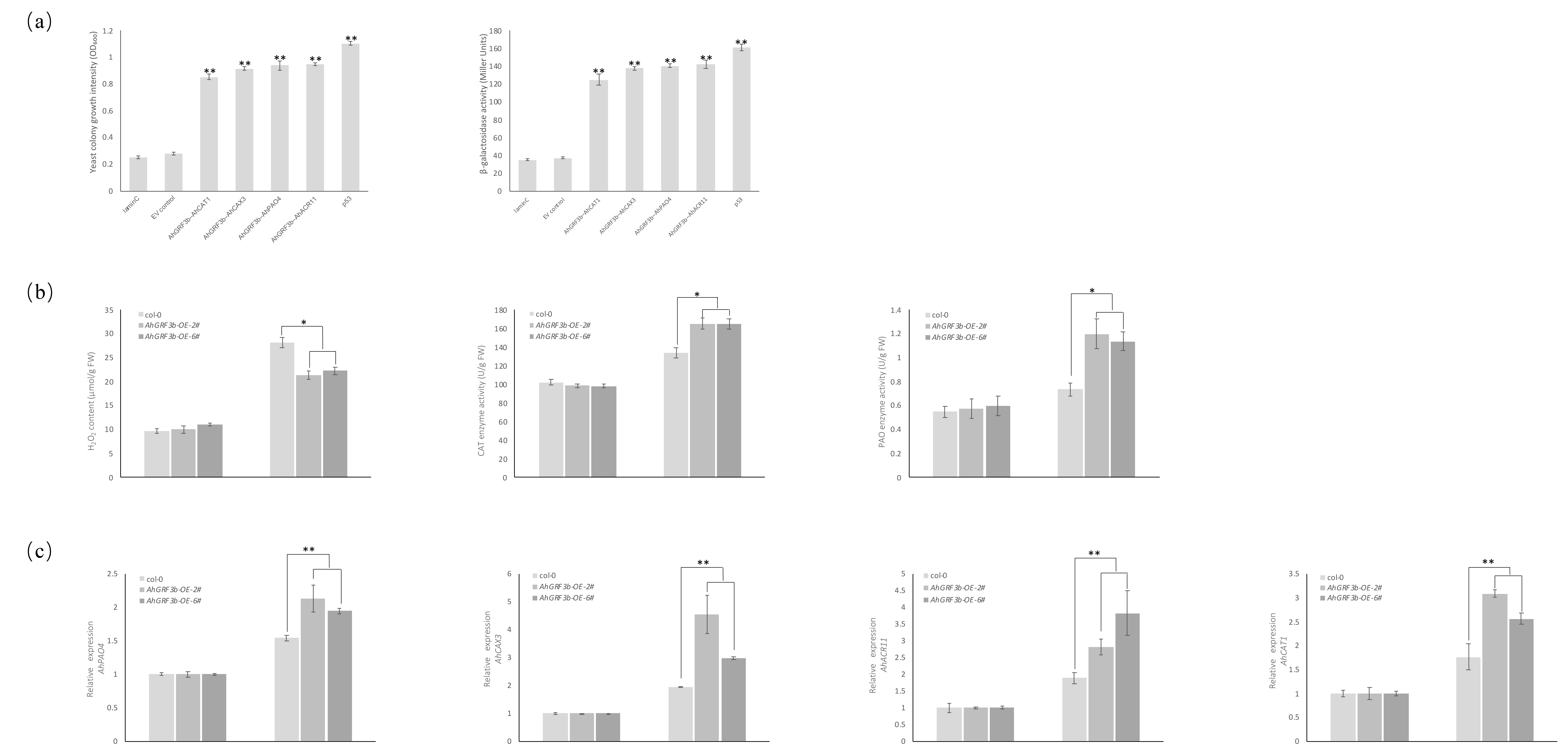
2.3. Physiological Parameter Measurement and Cold Tolerance Analysis of AhGRF3b-Overexpressing Plants
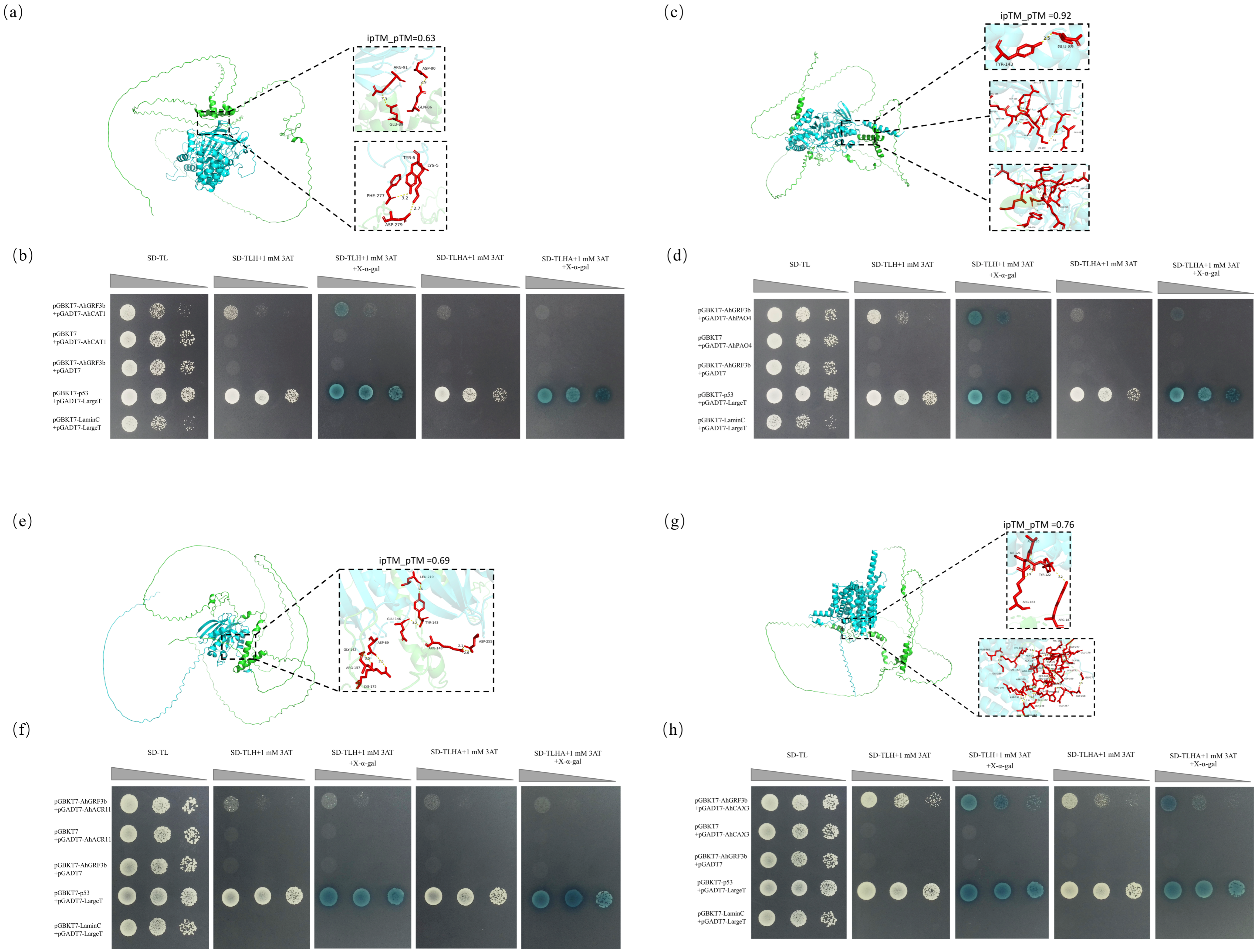
2.4. Interactions Between AhGRF3b and the Proteins AhCAT1, AhACR11, AhCAX3, and AhPAO4
3. Discussion
4. Materials and Methods
4.1. Degradome Library Construction and Sequencing
4.2. Western Blot
4.3. Cloning and Genetic Transformation of the AhGRF3b Gene
4.4. Subcellular Localization
4.5. Physiological Parameters and Cold Tolerance Assessment of Transgenic Plants
4.6. Yeast Two-Hybrid (Y2H) Assay
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| GRFs | Growth-regulating factors |
| CAT | Catalase |
| PAO | Probable Polyamine Oxidase |
| ACR | ACT Domain Repeat protein |
| CAX | Vacuolar cation/proton exchanger |
References
- Wu, X.; Zhang, M.; Zheng, Z.; Sun, Z.; Qi, F.; Liu, H.; Wang, J.; Wang, M.; Zhao, R.; Wu, Y.; et al. Fine-mapping of a candidate gene for web blotch resistance in Arachis hypogaea L. J. Integr. Agric. 2024, 23, 1494–1506. [Google Scholar] [CrossRef]
- Jeon, J.; Kim, J. Cold stress signaling networks in Arabidopsis. J. Plant Biol. 2013, 56, 69–76. [Google Scholar] [CrossRef]
- Jin, H. Endogenous small RNAs and antibacterial immunity in plants. FEBS Lett. 2008, 582, 2679–2684. [Google Scholar] [CrossRef]
- Omidbakhshfard, M.A.; Proost, S.; Fujikura, U.; Mueller-Roeber, B. Growth-regulating factors (GRFs): A small transcription factor family with important functions in plant biology. Mol. Plant 2015, 8, 998–1010. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, R.E.; Mecchia, M.A.; Debernardi, J.M.; Schommer, C.; Weigel, D.; Palatnik, J.F. Control of cell proliferation in Arabidopsis thaliana by microRNA miR396. Development 2010, 137, 103–112. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Luan, Y.; Zhai, J. Sp-miR396a-5p acts as a stress-responsive gene regulator by conferring tolerance to abiotic stresses and susceptibility to Phytophthora nicotianae infection in transgenic tobacco. Plant Cell Rep. 2015, 34, 2013–2025. [Google Scholar] [PubMed]
- Van der Knaap, E.; Kim, J.H.; Kende, H. A novel gibberellin-induced gene from rice and its potential regulatory role in stem growth. Plant Physiol. 2000, 122, 695–704. [Google Scholar] [CrossRef]
- Kim, J.H.; Kende, H. A transcriptional coactivator, AtGIF1, is involved in regulating leaf growth and morphology in Arabidopsis. Proc. Natl. Acad. Sci. USA 2004, 101, 13374–13379. [Google Scholar] [CrossRef]
- Huang, J.; Chen, G.Z.; Ahmad, S.; Hao, Y.; Chen, J.L.; Zhou, Y.Z.; Lan, S.R.; Liu, Z.J.; Peng, D.H. Genome-wide identification and characterization of the GRF gene family in Melastoma dodecandrum. Int. J. Mol. Sci. 2023, 24, 1261. [Google Scholar] [CrossRef]
- Lazzara, F.E.; Rodriguez, R.E.; Palatnik, J.F. Molecular mechanisms regulating growth-regulating factors activity in plant growth, development, and environmental responses. J. Exp. Bot. 2024, 75, 4360–4372. [Google Scholar] [CrossRef]
- Juneja, S.; Saini, R.; Mukit, A.; Kumar, S. Drought priming modulates ABF, GRFs, related microRNAs and induces metabolic adjustment during heat stress in chickpea. Plant Physiol. Biochem. 2023, 203, 108007. [Google Scholar] [CrossRef]
- Hu, Q.; Jiang, B.; Wang, L.; Song, Y.; Tang, X.; Zhao, Y.; Fan, X.; Gu, Y.; Zheng, Q.; Cheng, J.; et al. Genome-wide analysis of growth-regulating factor genes in grape (Vitis vinifera L.): Identification, characterization and their responsive expression to osmotic stress. Plant Cell Rep. 2023, 42, 107–121. [Google Scholar] [CrossRef]
- Wang, P.; Xiao, Y.; Yan, M.; Yan, Y.; Lei, X.; Di, P.; Wang, Y. Whole-genome identification and expression profiling of growth-regulating factor (GRF) and GRF-interacting factor (GIF) gene families in Panax ginseng. BMC Genom. 2023, 24, 334. [Google Scholar] [CrossRef]
- Guo, M.; Wu, F.; Ye, K.; Jiang, C.; An, Y.; Chen, N.; Huang, L.; Lu, M.; Zhang, J. Analysis of the 14–3-3/GRF gene family reveals the role of PagGRF12a in leaf development in poplar. Plant Sci. 2025, 359, 112661. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.J.; Wang, W.L.; Zhuang, J. Developmental processes and responses to hormonal stimuli in tea plant (Camellia sinensis) leaves are controlled by GRF and GIF gene families. Funct. Integr. Genom. 2017, 17, 503–512. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Xie, Q.; Yan, J.; Chen, J.; Chen, Q. Genome-wide identification and characterization of the abiotic-stress-responsive GRF gene family in diploid woodland strawberry (Fragaria vesca). Plants 2021, 10, 1916. [Google Scholar] [CrossRef] [PubMed]
- Khisti, M.; Avuthu, T.; Yogendra, K.; Kumar Valluri, V.; Kudapa, H.; Reddy, P.S.; Tyagi, W. Genome-wide identification and expression profiling of growth-regulating factor (GRF) and GRF-interacting factor (GIF) gene families in chickpea and pigeonpea. Sci. Rep. 2024, 14, 17178. [Google Scholar] [CrossRef]
- Wang, P.; Wang, Z.; Cao, H.; He, J.; Qin, C.; He, L.; Liu, B.; Wang, J.; Kong, L.; Ren, W.; et al. Genome-wide identification and expression pattern analysis of the GRF transcription factor family in Astragalus mongholicus. Mol. Biol. Rep. 2024, 51, 618. [Google Scholar] [CrossRef]
- Wei, C.; Yan, J.; Xu, P.; Wu, X.; Yi, Y.; Yue, X.; Chen, C.; Yan, L.; Yin, M. Genome-wide analysis of the potato GRF gene family and their expression profiles in response to hormone and Ralstonia solanacearum infection. Genes Genom. 2024, 46, 1423–1436. [Google Scholar] [CrossRef]
- Kiyak, A. GRF gene family in apricot (Prunus armeniaca L.): Genome-wide identification, characterization, and expression patterns during different developmental stages. Genet. Resour. Crop Evol. 2025, 72, 5831–5851. [Google Scholar] [CrossRef]
- Cao, J.F.; Huang, J.Q.; Liu, X.; Huang, C.C.; Zheng, Z.S.; Zhang, X.F.; Shangguan, X.X.; Wang, L.J.; Zhang, Y.G.; Wendel, J.F.; et al. Genome-wide characterization of the GRF family and their roles in response to salt stress in Gossypium. BMC Genom. 2020, 21, 575. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Wang, X.; Wang, J.; Niu, J.; Du, R.; Ji, G.; Zhu, L.; Zhang, J.; Lv, P.; Cao, J. Systematical characterization of GRF gene family in sorghum, and their potential functions in aphid resistance. Gene 2022, 836, 146669. [Google Scholar] [CrossRef]
- Zan, T.; Zhang, L.; Xie, T.; Li, L. Genome-wide identification and analysis of the growth-regulating factor (GRF) gene family and GRF-interacting factor family in Triticum aestivum L. Biochem. Genet. 2020, 58, 705–724. [Google Scholar] [CrossRef]
- Khatun, K.; Robin, A.H.K.; Park, J.I.; Nath, U.K.; Kim, C.K.; Lim, K.B.; Nou, I.S.; Chung, M.Y. Molecular characterization and expression profiling of tomato GRF transcription factor family genes in response to abiotic stresses and phytohormones. Int. J. Mol. Sci. 2017, 18, 1056. [Google Scholar] [CrossRef]
- Fu, M.K.; He, Y.N.; Yang, X.Y.; Tang, X.; Wang, M.; Dai, W.S. Genome-wide identification of the GRF family in sweet orange (Citrus sinensis) and functional analysis of CsGRF04 in response to multiple abiotic stresses. BMC Genom. 2024, 25, 37. [Google Scholar] [CrossRef]
- Deng, H.; Wen, Z.; Hou, Q.; Yu, R.; Cai, X.; Liu, K.; Qiao, G. Genome-wide identification and analysis of the growth-regulating factor (GRF) family in sweet cherry. Genet. Resour. Crop Evol. 2024, 71, 3881–3899. [Google Scholar] [CrossRef]
- Tang, Y.; Cheng, W.; Li, S.; Li, Y.; Wang, X.; Xie, J.; He, Y.; Wang, Y.; Niu, Y.; Bao, X.; et al. Genome-wide identification and expression analysis of the growth-regulating factor (GRF) family in Jatropha curcas. PLoS ONE 2021, 16, e0254711. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Ren, C.; Xue, Y.; Tian, Y.; Zhang, H.; Li, N.; Sheng, C.; Jiang, H.; Bai, D. Small RNA and degradome deep sequencing reveals the roles of microRNAs in peanut (Arachis hypogaea L.) cold response. Front. Plant Sci. 2022, 13, 920195. [Google Scholar] [CrossRef]
- Fonini, L.S.; Lazzarotto, F.; Barros, P.M.; Cabreira-Cagliari, C.; Martins, M.A.B.; Saibo, N.J.M.; Turchetto-Zolet, A.C.; Margis-Pinheiro, M. Molecular evolution and diversification of the GRF transcription factor family. Genet. Mol. Biol. 2020, 43, e20200080. [Google Scholar] [CrossRef]
- Cheng, Z.; Wen, S.; Wu, Y.; Shang, L.; Wu, L.; Lyu, D.; Yu, H.; Wang, J.; Jian, H. Comparative evolution and expression analysis of GRF transcription factor genes in seven plant species. Plants 2023, 12, 2790. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Tian, Y.; Wu, K.; Ye, Y.; Yu, J.; Zhang, J.; Liu, Q.; Hu, M.; Li, H.; Tong, Y.; et al. Modulating plant growth–metabolism coordination for sustainable agriculture. Nature 2018, 560, 595–600. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wu, Z.; Jia, M.; Li, J.; Song, T.; Jin, H.; Sun, J.; Qiu, C.; Lu, X.; Yuan, Y.; et al. The identification and characterization of the PeGRF gene family in Populus euphratica Oliv. Heteromorphic leaves provide a theoretical basis for the functional study of PeGRF9. Int. J. Mol. Sci. 2025, 26, 66. [Google Scholar] [CrossRef]
- Zhu, R.; Cao, B.; Sun, M.; Wu, J.; Li, J. Genome-wide identification and evolution of the GRF gene family and functional characterization of PbGRF18 in pear. Int. J. Mol. Sci. 2023, 24, 14690. [Google Scholar] [CrossRef]
- Lantzouni, O.; Alkofer, A.; Falter-Braun, P.; Schwechheimer, C. Growth-regulating factors interact with DELLAs and regulate growth in cold stress. Plant Cell 2020, 32, 1018–1034. [Google Scholar] [CrossRef]
- Cai, X.; Zhang, L.; Xiao, L.; Wen, Z.; Hou, Q.; Yang, K. Genome-wide identification of GRF gene family and their contribution to abiotic stress response in pitaya (Hylocereus polyrhizus). Int. J. Biol. Macromol. 2022, 223, 618–635. [Google Scholar] [CrossRef]
- Du, X.Q.; Sun, S.S.; Zhou, T.; Zhang, L.; Feng, Y.N.; Zhang, K.L.; Hua, Y.P. Genome-wide identification of the CAT genes and molecular characterization of their transcriptional responses to various nutrient stresses in allotetraploid rapeseed. Int. J. Mol. Sci. 2024, 25, 12658. [Google Scholar] [CrossRef] [PubMed]
- Hammes, U.Z.; Nielsen, E.; Honaas, L.A.; Taylor, C.G.; Schachtman, D.P. AtCAT6, a sink-tissue-localized transporter for essential amino acids in Arabidopsis. Plant J. 2006, 48, 414–427. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.; Wen, Y.; Deng, H.; Yu, R.; Tang, J.; Fu, Q.; Shao, Y.; Qiao, G. Overexpression of PavGRF5 in sweet cherry negatively regulates cold stress tolerance in transgenic Arabidopsis. Sci. Hortic. 2025, 350, 114328. [Google Scholar] [CrossRef]
- Cheng, N.H.; Pittman, J.K.; Shigaki, T.; Lachmansingh, J.; LeClere, S.; Lahner, B.; Salt, D.E.; Hirschi, K.D. Functional association of Arabidopsis CAX1 and CAX3 is required for normal growth and ion homeostasis. Plant Physiol. 2005, 138, 2048–2060. [Google Scholar] [CrossRef]
- Benkő, P.; Kaszler, N.; Gémes, K.; Fehér, A. Subfunctionalization of parental polyamine oxidase (PAO) genes in the allopolyploid tobacco Nicotiana tabacum (L.). Genes 2023, 14, 2025. [Google Scholar] [CrossRef]
- Sung, T.Y.; Chung, T.Y.; Hsu, C.P.; Hsieh, M.H. The ACR11 encodes a novel type of chloroplastic ACT domain repeat protein that is coordinately expressed with GLN2 in Arabidopsis. BMC Plant Biol. 2011, 11, 118. [Google Scholar] [CrossRef] [PubMed]
- Addo-Quaye, C.; Eshoo, T.W.; Bartel, D.P.; Axtell, M.J. Endogenous siRNA and miRNA targets identified by sequencing of the Arabidopsis degradome. Curr. Biol. 2008, 18, 758–762. [Google Scholar] [CrossRef]
- Zhang, L.; He, Y.; Chen, X.; Cao, Y.; Hou, Y.; Li, J.; Gao, C.; Wei, C. Characterization of B- and C-class MADS-box genes in medicinal plant Epimedium sagittatum. Med. Plant Biol. 2023, 2, 1–12. [Google Scholar]
- Folkers, U.; Kirik, V.; Schöbinger, U.; Falk, S.; Krishnakumar, S.; Pollock, M.A.; Oppenheimer, D.G.; Day, I.; Reddy, A.R.; Jürgens, G.; et al. The cell morphogenesis gene ANGUSTIFOLIA encodes a CtBP/BARS-like protein and is involved in the control of the microtubule cytoskeleton. EMBO J. 2002, 21, 1280–1288. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Zeng, X.; Zhao, Y.; Chen, R. AlphaFold2 and its applications in the fields of biology and medicine. Signal Transduct. Target. Ther. 2023, 8, 115. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, X.; Liu, Q.; Liu, X.; Lin, H.; Zhang, X.; Zhang, R.; Chen, Z.; Zhang, X.; Tian, Y.; Xue, Y.; et al. Targeted Regulation of AhGRF3b by ahy-miR396 Modulates Leaf Growth and Cold Tolerance in Peanut. Plants 2025, 14, 3203. https://doi.org/10.3390/plants14203203
Zhang X, Liu Q, Liu X, Lin H, Zhang X, Zhang R, Chen Z, Zhang X, Tian Y, Xue Y, et al. Targeted Regulation of AhGRF3b by ahy-miR396 Modulates Leaf Growth and Cold Tolerance in Peanut. Plants. 2025; 14(20):3203. https://doi.org/10.3390/plants14203203
Chicago/Turabian StyleZhang, Xin, Qimei Liu, Xinyu Liu, Haoyu Lin, Xiaoyu Zhang, Rui Zhang, Zhenbo Chen, Xiaoji Zhang, Yuexia Tian, Yunyun Xue, and et al. 2025. "Targeted Regulation of AhGRF3b by ahy-miR396 Modulates Leaf Growth and Cold Tolerance in Peanut" Plants 14, no. 20: 3203. https://doi.org/10.3390/plants14203203
APA StyleZhang, X., Liu, Q., Liu, X., Lin, H., Zhang, X., Zhang, R., Chen, Z., Zhang, X., Tian, Y., Xue, Y., Zhang, H., Li, N., Nie, P., & Bai, D. (2025). Targeted Regulation of AhGRF3b by ahy-miR396 Modulates Leaf Growth and Cold Tolerance in Peanut. Plants, 14(20), 3203. https://doi.org/10.3390/plants14203203



