Genome-Wide Identification, Characterization, and Expression Analysis of BES1 Family Genes in ‘Tieguanyin’ Tea Under Abiotic Stress
Abstract
1. Introduction
2. Results
2.1. Identification and Characterization of CsBES1 Genes
2.2. The Motif, Domain, and Gene Structure of CsBES1 Family
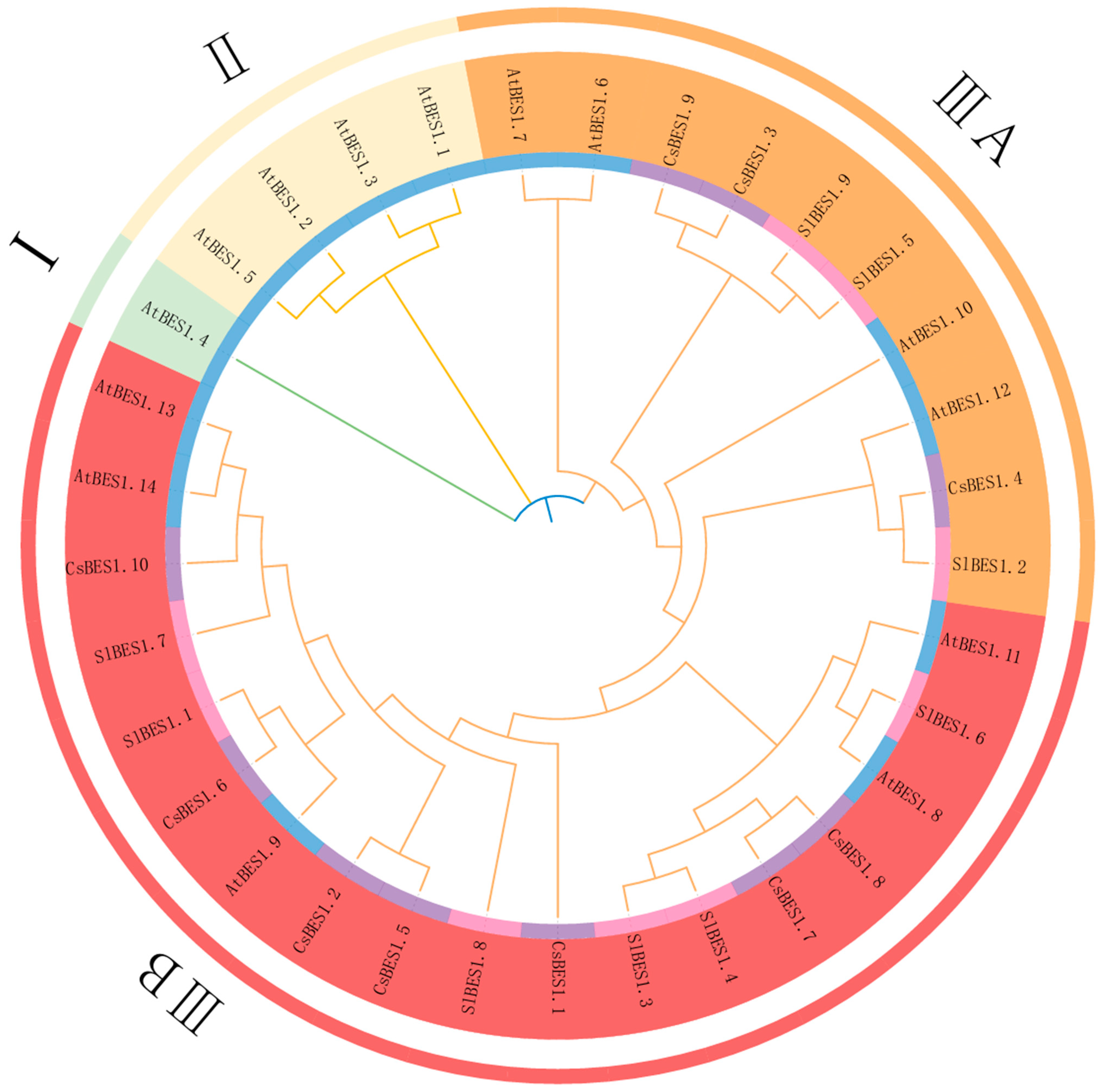
2.3. Phylogenetic Analysis of CsBES1s
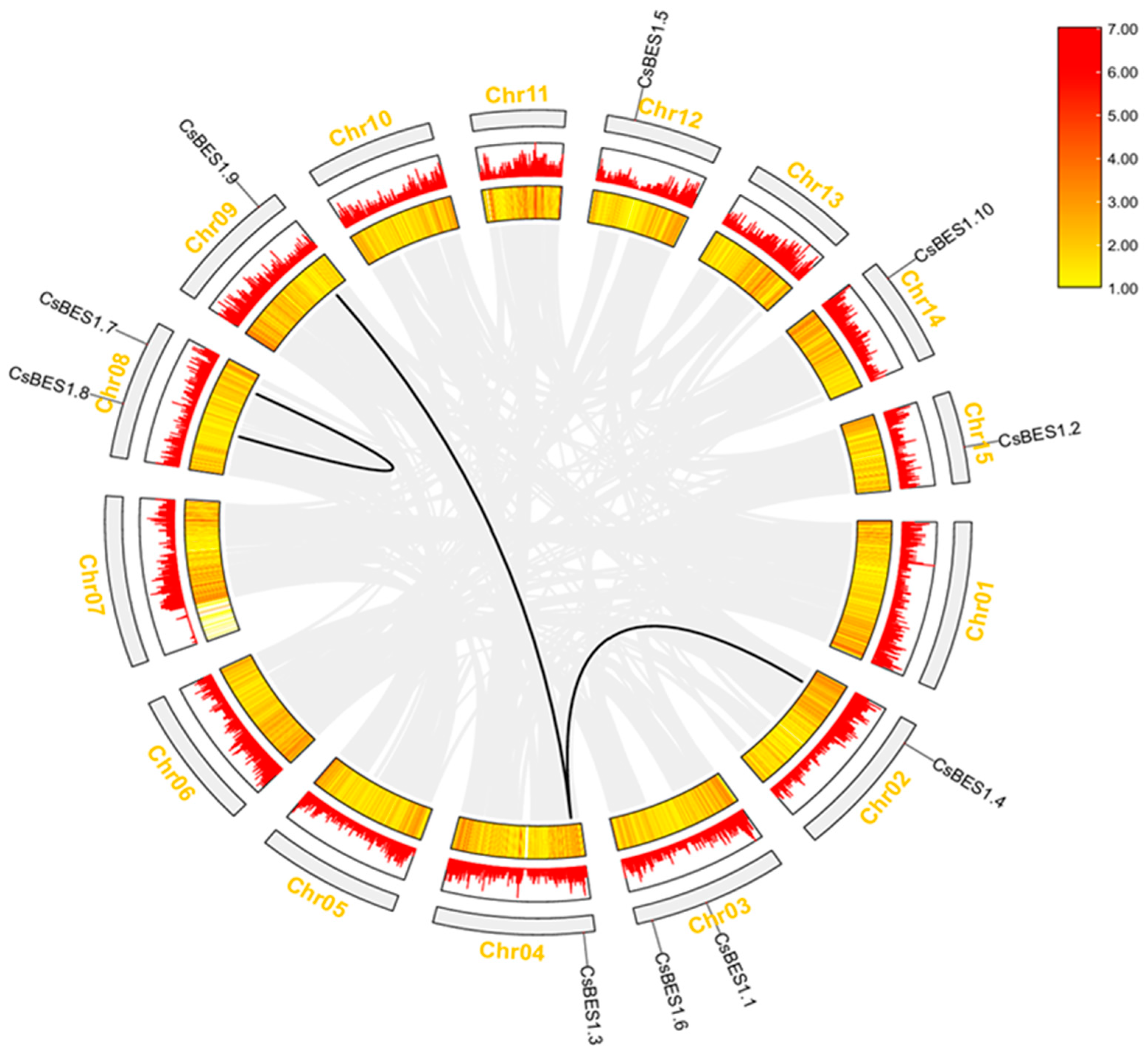
2.4. Chromosome Distribution of CsBES1s and Genomic Amplification in TGY
2.5. Cis-Regulatory Element Prediction of Promoters in 10 CsBES1 Genes
2.6. Gene Ontology (GO) and KEGG Analysis in CsBES1
2.7. Protein–Protein Interaction Network of CsBES1 Proteins
2.8. Expression Patterns of CsBES1 Genes in Response to Dark and Light Treatments
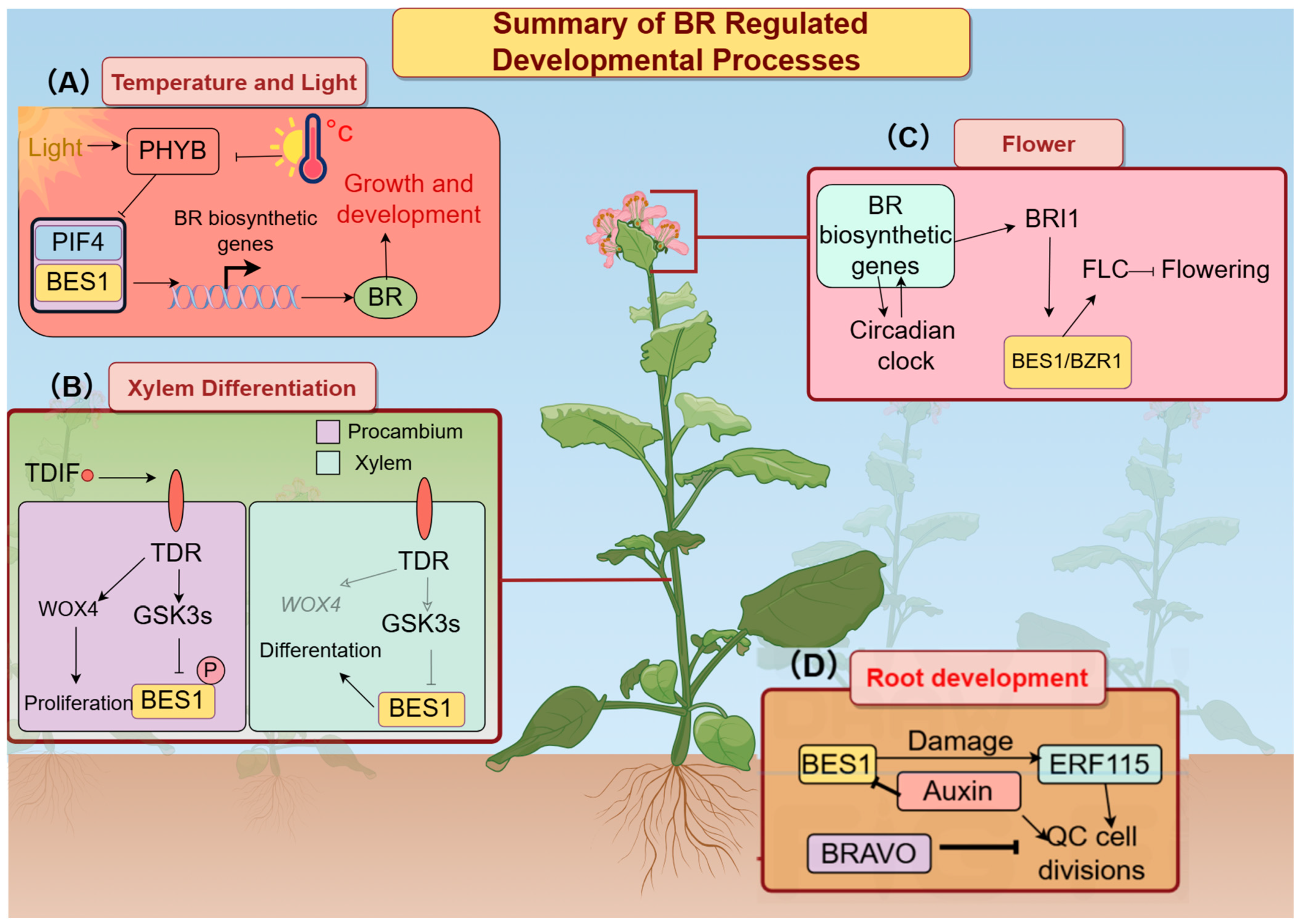
3. Discussion
4. Materials and Methods
4.1. Identification and Analysis of BES1 Gene Families in TGY
4.2. Physical and Chemical Properties of TGY
4.3. Phylogenetic Analysis of CsBES1s and BES1s from Other Species
4.4. Collinearity Analysis and Replication Event Analysis of BES1
4.5. GO and KEGG Analysis
4.6. Protein–Protein Interaction Network of CsBES1s
4.7. Abiotic Stress Therapy
4.8. RNA Extraction and Statistical Analysis
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Li, L.; Deng, X.W. It runs in the family: Regulation of brassinosteroid signaling by the BZR1-BES1 class of transcription factors. Trends Plant Sci. 2005, 10, 266–268. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.Y.; Bai, M.Y.; Oh, E.; Zhu, J.Y. Brassinosteroid signaling network and regulation of photomorphogenesis. Annu. Rev. Genet. 2012, 46, 701–724. [Google Scholar] [CrossRef] [PubMed]
- Anwar, A.; Liu, Y.; Dong, R.; Bai, L.; Yu, X.; Li, Y. The physiological and molecular mechanism of brassinosteroid in response to stress: A review. Biol. Res. 2018, 51, 46. [Google Scholar] [CrossRef] [PubMed]
- Feng, Z.; Shi, H.; Lv, M.; Ma, Y.; Li, J. Protein farnesylation negatively regulates brassinosteroid signaling via reducing BES1 stability in Arabidopsis thaliana. J. Integr. Plant Biol. 2021, 63, 1353–1366. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Xu, F.; Chen, Z.; Teng, Z.; Sun, K.; Li, X.; Yu, J.; Zhang, G.; Liang, Y.; Huang, X.; et al. Synergistic interplay of ABA and BR signal in regulating plant growth and adaptation. Nat. Plants 2021, 7, 1108–1118. [Google Scholar] [CrossRef]
- González-García, M.P.; Vilarrasa-Blasi, J.; Zhiponova, M.; Divol, F.; Mora-García, S.; Russinova, E.; Caño-Delgado, A.I. Brassinosteroids control meristem size by promoting cell cycle progression in Arabidopsis roots. Development 2011, 138, 849–859. [Google Scholar] [CrossRef]
- Planas-Riverola, A.; Gupta, A.; Betegón-Putze, I.; Bosch, N.; Ibañes, M.; Caño-Delgado, A.I. Brassinosteroid signaling in plant development and adaptation to stress. Development 2019, 146, dev151894. [Google Scholar] [CrossRef]
- Mitchell, J.W.; Mandava, N.; Worley, J.F.; Plimmer, J.R.; Smith, M.V. Brassins--a new family of plant hormones from rape pollen. Nature 1970, 225, 1065–1066. [Google Scholar] [CrossRef]
- Peres, A.; Soares, J.S.; Tavares, R.G.; Righetto, G.; Zullo, M.A.T.; Mandava, N.B.; Menossi, M. Brassinosteroids, the Sixth Class of Phytohormones: A Molecular View from the Discovery to Hormonal Interactions in Plant Development and Stress Adaptation. Int. J. Mol. Sci. 2019, 20, 331. [Google Scholar] [CrossRef]
- Nolan, T.M.; Vukašinović, N.; Hsu, C.W.; Zhang, J.; Vanhoutte, I.; Shahan, R.; Taylor, I.W.; Greenstreet, L.; Heitz, M.; Afanassiev, A.; et al. Brassinosteroid gene regulatory networks at cellular resolution in the Arabidopsis root. Science 2023, 379, eadf4721. [Google Scholar] [CrossRef]
- Kim, E.J.; Russinova, E. Brassinosteroid signalling. Curr. Biol. 2020, 30, R294–R298. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.; Tang, B.; Xie, Z.; Nolan, T.; Ye, H.; Song, G.Y.; Walley, J.; Yin, Y. GSK3-like kinase BIN2 phosphorylates RD26 to potentiate drought signaling in Arabidopsis. Plant J. 2019, 100, 923–937. [Google Scholar] [CrossRef] [PubMed]
- Ye, H.; Liu, S.; Tang, B.; Chen, J.; Xie, Z.; Nolan, T.M.; Jiang, H.; Guo, H.; Lin, H.-Y.; Li, L.; et al. RD26 mediates crosstalk between drought and brassinosteroid signalling pathways. Nat. Commun. 2017, 8, 14573. [Google Scholar] [CrossRef]
- Bulgakov, V.P.; Avramenko, T.V. Linking Brassinosteroid and ABA Signaling in the Context of Stress Acclimation. Int. J. Mol. Sci. 2020, 21, 5108. [Google Scholar] [CrossRef]
- Liu, Z.; Guo, J.T.; Li, T.; Xu, Y. Structure-based prediction of transcription factor binding sites using a protein-DNA docking approach. Proteins Struct. Funct. Genet. 2008, 72, 1114–1124. [Google Scholar] [CrossRef]
- Century, K.; Reuber, T.L.; Ratcliffe, O.J. Regulating the regulators: The future prospects for transcription-factor-based agricultural biotechnology products. Plant Physiol. 2008, 147, 20–29. [Google Scholar] [CrossRef]
- Chen, W.; Lv, M.; Wang, Y.; Wang, P.A.; Cui, Y.; Li, M.; Wang, R.; Gou, X.; Li, J. BES1 is activated by EMS1-TPD1-SERK1/2-mediated signaling to control tapetum development in Arabidopsis thaliana. Nat. Commun. 2019, 10, 4164. [Google Scholar] [CrossRef]
- Cui, X.Y.; Gao, Y.; Guo, J.; Yu, T.F.; Zheng, W.J.; Liu, Y.W.; Chen, J.; Xu, Z.S.; Ma, Y.Z. BES/BZR Transcription Factor TaBZR2 Positively Regulates Drought Responses by Activation of TaGST1. Plant Physiol. 2019, 180, 605–620. [Google Scholar] [CrossRef]
- Su, D.; Xiang, W.; Wen, L.; Lu, W.; Shi, Y.; Liu, Y.; Li, Z. Genome-wide identification, characterization and expression analysis of BES1 gene family in tomato. BMC Plant Biol. 2021, 21, 161. [Google Scholar] [CrossRef]
- Kono, A.; Yin, Y. Updates on BES1/BZR1 Regulatory Networks Coordinating Plant Growth and Stress Responses. Front. Plant Sci. 2020, 11, 617162. [Google Scholar] [CrossRef]
- Yin, Y.; Vafeados, D.; Tao, Y.; Yoshida, S.; Asami, T.; Chory, J. A new class of transcription factors mediates brassinosteroid-regulated gene expression in Arabidopsis. Cell 2005, 120, 249–259. [Google Scholar] [CrossRef] [PubMed]
- Guo, H.; Ye, H.; Li, L.; Yin, Y. A family of receptor-like kinases are regulated by BES1 and involved in plant growth in Arabidopsis thaliana. Plant Signal Behav. 2009, 4, 784–786. [Google Scholar] [CrossRef] [PubMed]
- Reinhold, H.; Soyk, S.; Simková, K.; Hostettler, C.; Marafino, J.; Mainiero, S.; Vaughan, C.K.; Monroe, J.D.; Zeeman, S.C. β-amylase-like proteins function as transcription factors in Arabidopsis, controlling shoot growth and development. Plant Cell 2011, 23, 1391–1403. [Google Scholar] [CrossRef]
- Lachowiec, J.; Mason, G.A.; Schultz, K.; Queitsch, C. Redundancy, Feedback, and Robustness in the Arabidopsis thaliana BZR/BEH Gene Family. Front. Genet. 2018, 9, 523. [Google Scholar] [CrossRef]
- Otani, Y.; Tomonaga, Y.; Tokushige, K.; Kamimura, M.; Sasaki, A.; Nakamura, Y.; Nakamura, T.; Matsuo, T.; Okamoto, S. Expression profiles of four BES1/BZR1 homologous genes encoding bHLH transcription factors in Arabidopsis. J. Pestic. Sci. 2020, 45, 95–104. [Google Scholar] [CrossRef]
- Yin, Y.; Wang, Z.Y.; Mora-Garcia, S.; Li, J.; Yoshida, S.; Asami, T.; Chory, J. BES1 accumulates in the nucleus in response to brassinosteroids to regulate gene expression and promote stem elongation. Cell 2002, 109, 181–191. [Google Scholar] [CrossRef]
- Mueller, A.K.; Labaied, M.; Kappe, S.H.; Matuschewski, K. Genetically modified Plasmodium parasites as a protective experimental malaria vaccine. Nature 2005, 433, 164–167. [Google Scholar] [CrossRef]
- Li, J.; Wen, J.; Lease, K.A.; Doke, J.T.; Tax, F.E.; Walker, J.C. BAK1, an Arabidopsis LRR receptor-like protein kinase, interacts with BRI1 and modulates brassinosteroid signaling. Cell 2002, 110, 213–222. [Google Scholar] [CrossRef]
- Kim, T.W.; Guan, S.; Burlingame, A.L.; Wang, Z.Y. The CDG1 kinase mediates brassinosteroid signal transduction from BRI1 receptor kinase to BSU1 phosphatase and GSK3-like kinase BIN2. Mol. Cell 2011, 43, 561–571. [Google Scholar] [CrossRef]
- He, J.-X.; Gendron, J.M.; Yang, Y.; Li, J.; Wang, Z.-Y. The GSK3-like kinase BIN2 phosphorylates and destabilizes BZR1, a positive regulator of the brassinosteroid signaling pathway in Arabidopsis. Proc. Natl. Acad. Sci. USA 2002, 99, 10185–10190. [Google Scholar] [CrossRef]
- Li, J.; Nam, K.H. Regulation of brassinosteroid signaling by a GSK3/SHAGGY-like kinase. Science 2002, 295, 1299–1301. [Google Scholar] [CrossRef] [PubMed]
- Toledo-Ortiz, G.; Huq, E.; Quail, P.H. The Arabidopsis basic/helix-loop-helix transcription factor family. Plant Cell 2003, 15, 1749–1770. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Wang, R.; Liu, M.; Yuan, W.; Zhao, Z.; Liu, X.; Peng, Y.; Yang, X.; Sun, Y.; Tang, W. Nucleocytoplasmic trafficking and turnover mechanisms of BRASSINAZOLE RESISTANT1 in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2021, 118, e2101838118. [Google Scholar] [CrossRef]
- Jiang, J.; Zhang, C.; Wang, X. A recently evolved isoform of the transcription factor BES1 promotes brassinosteroid signaling and development in Arabidopsis thaliana. Plant Cell 2015, 27, 361–374. [Google Scholar] [CrossRef]
- Saha, G.; Park, J.I.; Jung, H.J.; Ahmed, N.U.; Kayum, M.A.; Kang, J.G.; Nou, I.S. Molecular characterization of BZR transcription factor family and abiotic stress induced expression profiling in Brassica rapa. Plant Physiol. Biochem. 2015, 92, 92–104. [Google Scholar] [CrossRef]
- Setsungnern, A.; Muñoz, P.; Pérez-Llorca, M.; Müller, M.; Thiravetyan, P.; Munné-Bosch, S. A defect in BRI1-EMS-SUPPRESSOR 1 (bes1)-mediated brassinosteroid signaling increases photoinhibition and photo-oxidative stress during heat stress in Arabidopsis. Plant Sci. 2020, 296, 110470. [Google Scholar] [CrossRef]
- Yan, M.Y.; Xie, D.L.; Cao, J.J.; Xia, X.J.; Shi, K.; Zhou, Y.H.; Zhou, J.; Foyer, C.H.; Yu, J.Q. Brassinosteroid-mediated reactive oxygen species are essential for tapetum degradation and pollen fertility in tomato. Plant J. 2020, 102, 931–947. [Google Scholar] [CrossRef]
- Mecchia, M.A.; García-Hourquet, M.; Lozano-Elena, F.; Planas-Riverola, A.; Blasco-Escamez, D.; Marquès-Bueno, M.; Mora-García, S.; Caño-Delgado, A.I. The BES1/BZR1-family transcription factor MpBES1 regulates cell division and differentiation in Marchantia polymorpha. Curr. Biol. 2021, 31, 4860–4869.e4868. [Google Scholar] [CrossRef]
- Korwin Krukowski, P.; Visentin, I.; Russo, G.; Minerdi, D.; Bendahmane, A.; Schubert, A.; Cardinale, F. Transcriptome Analysis Points to BES1 as a Transducer of Strigolactone Effects on Drought Memory in Arabidopsis thaliana. Plant Cell Physiol. 2022, 63, 1873–1889. [Google Scholar] [CrossRef]
- Sun, Z.; Liu, X.; Zhu, W.; Lin, H.; Chen, X.; Li, Y.; Ye, W.; Yin, Z. Molecular Traits and Functional Exploration of BES1 Gene Family in Plants. Int. J. Mol. Sci. 2022, 23, 4242. [Google Scholar] [CrossRef]
- Yang, J.; Wu, Y.; Li, L.; Li, C. Comprehensive analysis of the BES1 gene family and its expression under abiotic stress and hormone treatment in Populus trichocarpa. Plant Physiol. Biochem. 2022, 173, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Nolan, T.M.; Ye, H.; Zhang, M.; Tong, H.; Xin, P.; Chu, J.; Chu, C.; Li, Z.; Yin, Y. Arabidopsis WRKY46, WRKY54, and WRKY70 Transcription Factors Are Involved in Brassinosteroid-Regulated Plant Growth and Drought Responses. Plant Cell 2017, 29, 1425–1439. [Google Scholar] [CrossRef] [PubMed]
- Franklin, K.A.; Lee, S.H.; Patel, D.; Kumar, S.V.; Spartz, A.K.; Gu, C.; Ye, S.; Yu, P.; Breen, G.; Cohen, J.D.; et al. PHYTOCHROME-INTERACTING FACTOR 4 (PIF4) regulates auxin biosynthesis at high temperature. Proc. Natl. Acad. Sci. USA 2011, 108, 20231–20235. [Google Scholar] [CrossRef] [PubMed]
- Song, X.; Ma, X.; Li, C.; Hu, J.; Yang, Q.; Wang, T.; Wang, L.; Wang, J.; Guo, D.; Ge, W.; et al. Comprehensive analyses of the BES1 gene family in Brassica napus and examination of their evolutionary pattern in representative species. BMC Genom. 2018, 19, 346. [Google Scholar] [CrossRef]
- Liu, Z.; Qanmber, G.; Lu, L.; Qin, W.; Liu, J.; Li, J.; Ma, S.; Yang, Z.; Yang, Z. Genome-wide analysis of BES1 genes in Gossypium revealed their evolutionary conserved roles in brassinosteroid signaling. Sci. China Life Sci. 2018, 61, 1566–1582. [Google Scholar] [CrossRef]
- Feng, W.; Zhang, H.; Cao, Y.; Liu, Y.; Zhao, Y.; Sun, F.; Yang, Q.; Zhang, X.; Zhang, Y.; Wang, Y.; et al. Maize ZmBES1/BZR1-1 transcription factor negatively regulates drought tolerance. Plant Physiol. Biochem. 2023, 205, 108188. [Google Scholar] [CrossRef]
- Gao, Y.; Hu, J.; Zhao, T.; Xu, X.; Jiang, J.; Li, J. Genome-wide Identification and Expression Pattern Analysis of BRI1-EMS–suppressor Transcription Factors in Tomato under Abiotic Stresses. J. Am. Soc. Hortic. Sci. 2018, 143, 84–90. [Google Scholar] [CrossRef]
- Song, X.; Li, Y.; Hou, X. Genome-wide analysis of the AP2/ERF transcription factor superfamily in Chinese cabbage (Brassica rapa ssp. pekinensis). BMC Genom. 2013, 14, 573. [Google Scholar] [CrossRef]
- Ma, S.; Ji, T.; Liang, M.; Li, S.; Tian, Y.; Gao, L. Genome-Wide Identification, Structural, and Gene Expression Analysis of BRI1-EMS-Suppressor 1 Transcription Factor Family in Cucumis sativus. Front. Genet. 2020, 11, 583996. [Google Scholar] [CrossRef]
- Xie, L.; Yang, C.; Wang, X. Brassinosteroids can regulate cellulose biosynthesis by controlling the expression of CESA genes in Arabidopsis. J. Exp. Bot. 2011, 62, 4495–4506. [Google Scholar] [CrossRef]
- Ye, Q.; Zhu, W.; Li, L.; Zhang, S.; Yin, Y.; Ma, H.; Wang, X. Brassinosteroids control male fertility by regulating the expression of key genes involved in Arabidopsis anther and pollen development. Proc. Natl. Acad. Sci. USA 2010, 107, 6100–6105. [Google Scholar] [CrossRef] [PubMed]
- Lozano-Durán, R.; Macho, A.P.; Boutrot, F.; Segonzac, C.; Somssich, I.E.; Zipfel, C. The transcriptional regulator BZR1 mediates trade-off between plant innate immunity and growth. Elife 2013, 2, e00983. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Liu, L.; Sun, S.; Li, Y.; Jia, L.; Ye, S.; Yu, Y.; Dossa, K.; Luan, Y. Leaf-transcriptome profiles of phoebe bournei provide insights into temporal drought stress responses. Front. Plant Sci. 2022, 13, 1010314. [Google Scholar] [CrossRef]
- Reygaert, W.C. The antimicrobial possibilities of green tea. Front. Microbiol. 2014, 5, 434. [Google Scholar] [CrossRef]
- Huang, Z.; Sanaeifar, A.; Tian, Y.; Liu, L.; Zhang, D.; Wang, H.; Ye, D.; Li, X. Improved generalization of spectral models associated with Vis-NIR spectroscopy for determining the moisture content of different tea leaves. J. Food Eng. 2021, 293, 110374. [Google Scholar] [CrossRef]
- Zeng, L.; Zhou, Y.; Fu, X.; Mei, X.; Cheng, S.; Gui, J.; Dong, F.; Tang, J.; Ma, S.; Yang, Z. Does oolong tea (Camellia sinensis) made from a combination of leaf and stem smell more aromatic than leaf-only tea? Contribution of the stem to oolong tea aroma. Food Chem. 2017, 237, 488–498. [Google Scholar] [CrossRef]
- Hara, Y. Elucidation of Physiological Functions of Tea Catechins and Their Practical Applications. J. Food Drug Anal. 2012, 20, 296–300. [Google Scholar] [CrossRef]
- Zhang, S.; Takano, J.; Murayama, N.; Tominaga, M.; Abe, T.; Park, I.; Seol, J.; Ishihara, A.; Tanaka, Y.; Yajima, K.; et al. Subacute Ingestion of Caffeine and Oolong Tea Increases Fat Oxidation without Affecting Energy Expenditure and Sleep Architecture: A Randomized, Placebo-Controlled, Double-Blinded Cross-Over Trial. Nutrients 2020, 12, 3671. [Google Scholar] [CrossRef]
- Wang, W.; Xin, H.; Wang, M.; Ma, Q.; Wang, L.; Kaleri, N.A.; Wang, Y.; Li, X. Transcriptomic Analysis Reveals the Molecular Mechanisms of Drought-Stress-Induced Decreases in Camellia sinensis Leaf Quality. Front. Plant Sci. 2016, 7, 385. [Google Scholar] [CrossRef]
- Yu, S.; Li, P.; Zhao, X.; Tan, M.; Ahmad, M.Z.; Xu, Y.; Tadege, M.; Zhao, J. CsTCPs regulate shoot tip development and catechin biosynthesis in tea plant (Camellia sinensis). Hortic. Res. 2021, 8, 104. [Google Scholar] [CrossRef]
- Zhang, Q.; Shi, Y.; Ma, L.; Yi, X.; Ruan, J. Metabolomic analysis using ultra-performance liquid chromatography-quadrupole-time of flight mass spectrometry (UPLC-Q-TOF MS) uncovers the effects of light intensity and temperature under shading treatments on the metabolites in tea. PLoS ONE 2014, 9, e112572. [Google Scholar] [CrossRef] [PubMed]
- Zheng, C.; Ma, J.Q.; Ma, C.L.; Shen, S.Y.; Liu, Y.F.; Chen, L. Regulation of Growth and Flavonoid Formation of Tea Plants (Camellia sinensis) by Blue and Green Light. J. Agric. Food Chem. 2019, 67, 2408–2419. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.F.; Huang, L.C.; Wei, K.; Yu, J.W.; Zhang, C.Q.; Liu, Q.Q. Light involved regulation of BZR1 stability and phosphorylation status to coordinate plant growth in Arabidopsis. Biosci. Rep. 2017, 37, BSR20170069. [Google Scholar] [CrossRef] [PubMed]
- Kim, B.; Jeong, Y.J.; Corvalán, C.; Fujioka, S.; Cho, S.; Park, T.; Choe, S. Darkness and gulliver2/phyB mutation decrease the abundance of phosphorylated BZR1 to activate brassinosteroid signaling in Arabidopsis. Plant J. 2014, 77, 737–747. [Google Scholar] [CrossRef]
- Roy, S.W.; Penny, D. A very high fraction of unique intron positions in the intron-rich diatom Thalassiosira pseudonana indicates widespread intron gain. Mol. Biol. Evol. 2007, 24, 1447–1457. [Google Scholar] [CrossRef]
- Kolodny, R. Searching protein space for ancient sub-domain segments. Curr. Opin. Struct. Biol. 2021, 68, 105–112. [Google Scholar] [CrossRef]
- Zhou, Q.; Zhao, M.; Xing, F.; Mao, G.; Wang, Y.; Dai, Y.; Niu, M.; Yuan, H. Identification and Expression Analysis of CAMTA Genes in Tea Plant Reveal Their Complex Regulatory Role in Stress Responses. Front. Plant Sci. 2022, 13, 910768. [Google Scholar] [CrossRef]
- Wang, Y.; Tang, H.; Debarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.H.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef]
- Panchy, N.; Lehti-Shiu, M.; Shiu, S.H. Evolution of Gene Duplication in Plants. Plant Physiol. 2016, 171, 2294–2316. [Google Scholar] [CrossRef]
- Holub, E.B. The arms race is ancient history in Arabidopsis, the wildflower. Nat. Rev. Genet. 2001, 2, 516–527. [Google Scholar] [CrossRef]
- Xiong, M.; Yu, J.; Wang, J.; Gao, Q.; Huang, L.; Chen, C.; Zhang, C.; Fan, X.; Zhao, D.; Liu, Q.-Q.; et al. Brassinosteroids regulate rice seed germination through the BZR1-RAmy3D transcriptional module. Plant Physiol. 2022, 189, 402–418. [Google Scholar] [CrossRef] [PubMed]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef] [PubMed]
- Gendron, J.M.; Wang, Z.Y. Multiple mechanisms modulate brassinosteroid signaling. Curr. Opin. Plant Biol. 2007, 10, 436–441. [Google Scholar] [CrossRef]
- Wang, X.; Niu, Y.; Zheng, Y. Multiple Functions of MYB Transcription Factors in Abiotic Stress Responses. Int. J. Mol. Sci. 2021, 22, 6125. [Google Scholar] [CrossRef]
- Kim, S.H.; Lee, S.H.; Park, T.K.; Tian, Y.; Yu, K.; Lee, B.H.; Bai, M.Y.; Cho, S.J.; Kim, T.W. Comparative analysis of BZR1/BES1 family transcription factors in Arabidopsis. Plant J. 2024, 117, 747–765. [Google Scholar] [CrossRef]
- Li, M.; Liu, C.; Hepworth, S.R.; Ma, C.; Li, H.; Li, J.; Wang, S.M.; Yin, H. SAUR15 interaction with BRI1 activates plasma membrane H+-ATPase to promote organ development of Arabidopsis. Plant Physiol. 2022, 189, 2454–2466. [Google Scholar] [CrossRef]
- Kojima, K.; Ichijo, H.; Naguro, I. Molecular functions of ASK family in diseases caused by stress-induced inflammation and apoptosis. J. Biochem. 2021, 169, 395–407. [Google Scholar] [CrossRef]
- Ohnishi, T.; Godza, B.; Watanabe, B.; Fujioka, S.; Hategan, L.; Ide, K.; Shibata, K.; Yokota, T.; Szekeres, M.; Mizutani, M. CYP90A1/CPD, a brassinosteroid biosynthetic cytochrome P450 of Arabidopsis, catalyzes C-3 oxidation. J. Biol. Chem. 2012, 287, 31551–31560. [Google Scholar] [CrossRef]
- Demuth, J.P.; Hahn, M.W. The life and death of gene families. Bioessays 2009, 31, 29–39. [Google Scholar] [CrossRef]
- Oudelaar, A.M.; Higgs, D.R. The relationship between genome structure and function. Nat. Rev. Genet. 2021, 22, 154–168. [Google Scholar] [CrossRef]
- Li, L.; Yu, X.; Thompson, A.; Guo, M.; Yoshida, S.; Asami, T.; Chory, J.; Yin, Y. Arabidopsis MYB30 is a direct target of BES1 and cooperates with BES1 to regulate brassinosteroid-induced gene expression. Plant J. 2009, 58, 275–286. [Google Scholar] [CrossRef] [PubMed]
- Saito, M.; Kondo, Y.; Fukuda, H. BES1 and BZR1 Redundantly Promote Phloem and Xylem Differentiation. Plant Cell Physiol. 2018, 59, 590–600. [Google Scholar] [CrossRef] [PubMed]
- Roy, S.W.; Gilbert, W. The evolution of spliceosomal introns: Patterns, puzzles and progress. Nat. Rev. Genet. 2006, 7, 211–221. [Google Scholar] [CrossRef] [PubMed]
- Song, X.M.; Huang, Z.N.; Duan, W.K.; Ren, J.; Liu, T.K.; Li, Y.; Hou, X.L. Genome-wide analysis of the bHLH transcription factor family in Chinese cabbage (Brassica rapa ssp. pekinensis). Mol. Genet. Genom. 2014, 289, 77–91. [Google Scholar] [CrossRef]
- Cao, X.; Khaliq, A.; Lu, S.; Xie, M.; Ma, Z.; Mao, J.; Chen, B. Genome-wide identification and characterization of the BES1 gene family in apple (Malus domestica). Plant Biol. 2020, 22, 723–733. [Google Scholar] [CrossRef]
- Cheng, M.; Yuan, H.; Wang, R.; Wang, W.; Zhang, L.; Fan, F.; Li, S. Identification and characterization of BES1 genes involved in grain size development of Oryza sativa L. Int. J. Biol. Macromol. 2023, 253, 127327. [Google Scholar] [CrossRef]
- Cao, X.; Ma, W.; Zeng, F.; Cheng, Y.; Ma, Z.; Mao, J.; Chen, B. Grape BES1 transcription factor gene VvBES1-3 confers salt tolerance in transgenic Arabidopsis. Gene 2023, 854, 147059. [Google Scholar] [CrossRef]
- Cartharius, K.; Frech, K.; Grote, K.; Klocke, B.; Haltmeier, M.; Klingenhoff, A.; Frisch, M.; Bayerlein, M.; Werner, T. MatInspector and beyond: Promoter analysis based on transcription factor binding sites. Bioinformatics 2005, 21, 2933–2942. [Google Scholar] [CrossRef]
- Yang, C.J.; Zhang, C.; Lu, Y.N.; Jin, J.Q.; Wang, X.L. The mechanisms of brassinosteroids’ action: From signal transduction to plant development. Mol. Plant 2011, 4, 588–600. [Google Scholar] [CrossRef]
- Ibañez, C.; Delker, C.; Martinez, C.; Bürstenbinder, K.; Janitza, P.; Lippmann, R.; Ludwig, W.; Sun, H.; James, G.V.; Klecker, M.; et al. Brassinosteroids Dominate Hormonal Regulation of Plant Thermomorphogenesis via BZR1. Curr. Biol. 2018, 28, 303–310.e303. [Google Scholar] [CrossRef]
- Yang, M.; Li, C.; Cai, Z.; Hu, Y.; Nolan, T.; Yu, F.; Yin, Y.; Xie, Q.; Tang, G.; Wang, X. SINAT E3 Ligases Control the Light-Mediated Stability of the Brassinosteroid-Activated Transcription Factor BES1 in Arabidopsis. Dev. Cell 2017, 41, 47–58.e44. [Google Scholar] [CrossRef] [PubMed]
- Oh, E.; Zhu, J.Y.; Wang, Z.Y. Interaction between BZR1 and PIF4 integrates brassinosteroid and environmental responses. Nat. Cell Biol. 2012, 14, 802–809. [Google Scholar] [CrossRef]
- Wang, W.; Lu, X.; Li, L.; Lian, H.; Mao, Z.; Xu, P.; Guo, T.; Xu, F.; Du, S.; Cao, X.; et al. Photoexcited CRYPTOCHROME1 Interacts with Dephosphorylated BES1 to Regulate Brassinosteroid Signaling and Photomorphogenesis in Arabidopsis. Plant Cell 2018, 30, 1989–2005. [Google Scholar] [CrossRef] [PubMed]
- Heijde, M.; Ulm, R. UV-B photoreceptor-mediated signalling in plants. Trends Plant Sci. 2012, 17, 230–237. [Google Scholar] [CrossRef] [PubMed]
- Jenkins, G.I. Structure and function of the UV-B photoreceptor UVR8. Curr. Opin. Struct. Biol. 2014, 29, 52–57. [Google Scholar] [CrossRef] [PubMed]
- Tilbrook, K.; Arongaus, A.B.; Binkert, M.; Heijde, M.; Yin, R.; Ulm, R. The UVR8 UV-B Photoreceptor: Perception, Signaling and Response. Arab. Book 2013, 11, e0164. [Google Scholar] [CrossRef]
- Rizzini, L.; Favory, J.J.; Cloix, C.; Faggionato, D.; O’Hara, A.; Kaiserli, E.; Baumeister, R.; Schäfer, E.; Nagy, F.; Jenkins, G.I.; et al. Perception of UV-B by the Arabidopsis UVR8 protein. Science 2011, 332, 103–106. [Google Scholar] [CrossRef]
- Huang, X.; Yang, P.; Ouyang, X.; Chen, L.; Deng, X.W. Photoactivated UVR8-COP1 module determines photomorphogenic UV-B signaling output in Arabidopsis. PLoS Genet. 2014, 10, e1004218. [Google Scholar] [CrossRef]
- Liang, T.; Mei, S.; Shi, C.; Yang, Y.; Peng, Y.; Ma, L.; Wang, F.; Li, X.; Huang, X.; Yin, Y.; et al. UVR8 Interacts with BES1 and BIM1 to Regulate Transcription and Photomorphogenesis in Arabidopsis. Dev. Cell 2018, 44, 512–523.e515. [Google Scholar] [CrossRef]
- Han, X.; Zhang, J.; Han, S.; Chong, S.L.; Meng, G.; Song, M.; Wang, Y.; Zhou, S.; Liu, C.; Lou, L.; et al. The chromosome-scale genome of Phoebe bournei reveals contrasting fates of terpene synthase (TPS)-a and TPS-b subfamilies. Plant Commun. 2022, 3, 100410. [Google Scholar] [CrossRef]
- Gasteiger, E.; Gattiker, A.; Hoogland, C.; Ivanyi, I.; Appel, R.D.; Bairoch, A. ExPASy: The proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 2003, 31, 3784–3788. [Google Scholar] [CrossRef]
- Chen, C.; Wu, Y.; Li, J.; Wang, X.; Zeng, Z.; Xu, J.; Liu, Y.; Feng, J.; Chen, H.; He, Y.; et al. TBtools-II: A “one for all, all for one“ bioinformatics platform for biological big-data mining. Molecular Plant 2023, 16, 1733–1742. [Google Scholar] [CrossRef]
- Cantalapiedra, C.P.; Hernández-Plaza, A.; Letunic, I.; Bork, P.; Huerta-Cepas, J. eggNOG-mapper v2: Functional Annotation, Orthology Assignments, and Domain Prediction at the Metagenomic Scale. Mol. Biol. Evol. 2021, 38, 5825–5829. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Furumichi, M.; Tanabe, M.; Sato, Y.; Morishima, K. KEGG: New perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017, 45, D353–D361. [Google Scholar] [CrossRef] [PubMed]
- Otasek, D.; Morris, J.H.; Bouças, J.; Pico, A.R.; Demchak, B. Cytoscape Automation: Empowering workflow-based network analysis. Genome Biol. 2019, 20, 185. [Google Scholar] [CrossRef] [PubMed]
- Winters, M.; Eskes, M.; Weir, A.; Moen, M.H.; Backx, F.J.; Bakker, E.W. Treatment of medial tibial stress syndrome: A systematic review. Sports Med. 2013, 43, 1315–1333. [Google Scholar] [CrossRef] [PubMed]
- Day, R.W.; Quinn, G.P. Comparisons of Treatments After an Analysis of Variance in Ecology. Ecol. Monogr. 1989, 59, 433–463. [Google Scholar] [CrossRef]







| Gene Name | Gene ID | Size/aa (Size in Amino Acids) | Molecular Weight/kDa (Molecular Weight in Kilodaltons) | Theoretical PI (Theoretical Isoelectric Point) | Grand Average of Hydropathicity | Instability Index | Subcellular Localization |
|---|---|---|---|---|---|---|---|
| CsBES1.1 | CsTGY03G0001903 | 495 | 55,357.08 | 10.02 | −0.765 | 72.8 | Nucleus |
| CsBES1.2 | CsTGY15G0001195 | 395 | 44,896.92 | 9.05 | −0.391 | 43.17 | Nucleus |
| CsBES1.3 | CsTGY04G0000351 | 377 | 40,575.01 | 8.92 | −0.654 | 81.82 | Nucleus |
| CsBES1.4 | CsTGY02G0000736 | 322 | 34,442.06 | 8.6 | −0.608 | 70.99 | Nucleus |
| CsBES1.5 | CsTGY12G0000463 | 315 | 35,172.59 | 8.13 | −0.604 | 37.02 | Chloroplast |
| CsBES1.6 | CsTGY03G0002935 | 699 | 78,007.57 | 5.37 | −0.454 | 37.34 | Nucleus |
| CsBES1.7 | CsTGY08G0002091 | 323 | 34,725.64 | 7.61 | −0.603 | 58.53 | nucleus |
| CsBES1.8 | CsTGY08G0000981 | 326 | 34,687.46 | 8.81 | −0.609 | 59.21 | nucleus |
| CsBES1.9 | CsTGY09G0002593 | 316 | 34,258.36 | 9.25 | −0.708 | 76.49 | nucleus |
| CsBES1.10 | CsTGY14G0000640 | 683 | 77,043.46 | 5.96 | −0.511 | 40.49 | nucleus |
| Gene Name | Forward Primer (5′-3′) | Reverse Primer (5′-3′) |
|---|---|---|
| CsBES1.2 | GGGTTAAGCAGATGGGTGGT | GTTCCGGTGAGGTCGGAATG |
| CsBES1.3 | GGAGGAGGGAGGAGGAAGCC | GTGCCATCGGGTTCGACTGT |
| CsBES1.4 | CAAACACTGCGACAACAACG | CGATAAGTGGTGCCATCGTC |
| CsBES1.6 | TGTGGAGGCAACGTTGGTGA | ATTCAGGGTTGCGCCTTCCC |
| CsBES1.7 | TGGACTCCTGGGCAAAGTGG | TCCCATGCCTTCACCAACCC |
| CsBES1.8 | GGGTCAGCTTCAGCAAGTCC | TGGCAGCGTAACGTGATGAG |
| CsBES1.9 | CGCCAACTATGGTCCCAACC | GCTTCACTGGGACGCTTTCA |
| CsBES1.10 | GTCAGGGCTGGTTCCGTGTC | ATCCTCGATCCCAGGCCGAA |
| GAPDH | TTGGCATCGTTGAGGGTCT | CAGTGGGAACACGGAAAGC |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Y.; Zhang, Y.; Yang, Z.; Li, Q.; Chen, W.; Wen, X.; Chen, H.; Cao, S. Genome-Wide Identification, Characterization, and Expression Analysis of BES1 Family Genes in ‘Tieguanyin’ Tea Under Abiotic Stress. Plants 2025, 14, 473. https://doi.org/10.3390/plants14030473
Zhang Y, Zhang Y, Yang Z, Li Q, Chen W, Wen X, Chen H, Cao S. Genome-Wide Identification, Characterization, and Expression Analysis of BES1 Family Genes in ‘Tieguanyin’ Tea Under Abiotic Stress. Plants. 2025; 14(3):473. https://doi.org/10.3390/plants14030473
Chicago/Turabian StyleZhang, Yanzi, Yanlin Zhang, Zhicheng Yang, Qingyan Li, Weixiang Chen, Xinyan Wen, Hao Chen, and Shijiang Cao. 2025. "Genome-Wide Identification, Characterization, and Expression Analysis of BES1 Family Genes in ‘Tieguanyin’ Tea Under Abiotic Stress" Plants 14, no. 3: 473. https://doi.org/10.3390/plants14030473
APA StyleZhang, Y., Zhang, Y., Yang, Z., Li, Q., Chen, W., Wen, X., Chen, H., & Cao, S. (2025). Genome-Wide Identification, Characterization, and Expression Analysis of BES1 Family Genes in ‘Tieguanyin’ Tea Under Abiotic Stress. Plants, 14(3), 473. https://doi.org/10.3390/plants14030473








