Infiltration-RNAseq Reveals Enhanced Defense Responses in Nicothiana benthamiana Leaves Overexpressing the Banana Gene MaWRKY45
Abstract
1. Introduction
2. Results
2.1. Analysis of N. benthamiana De Novo Reference Transcriptome Under Banana MaWRKY45 Overexpression
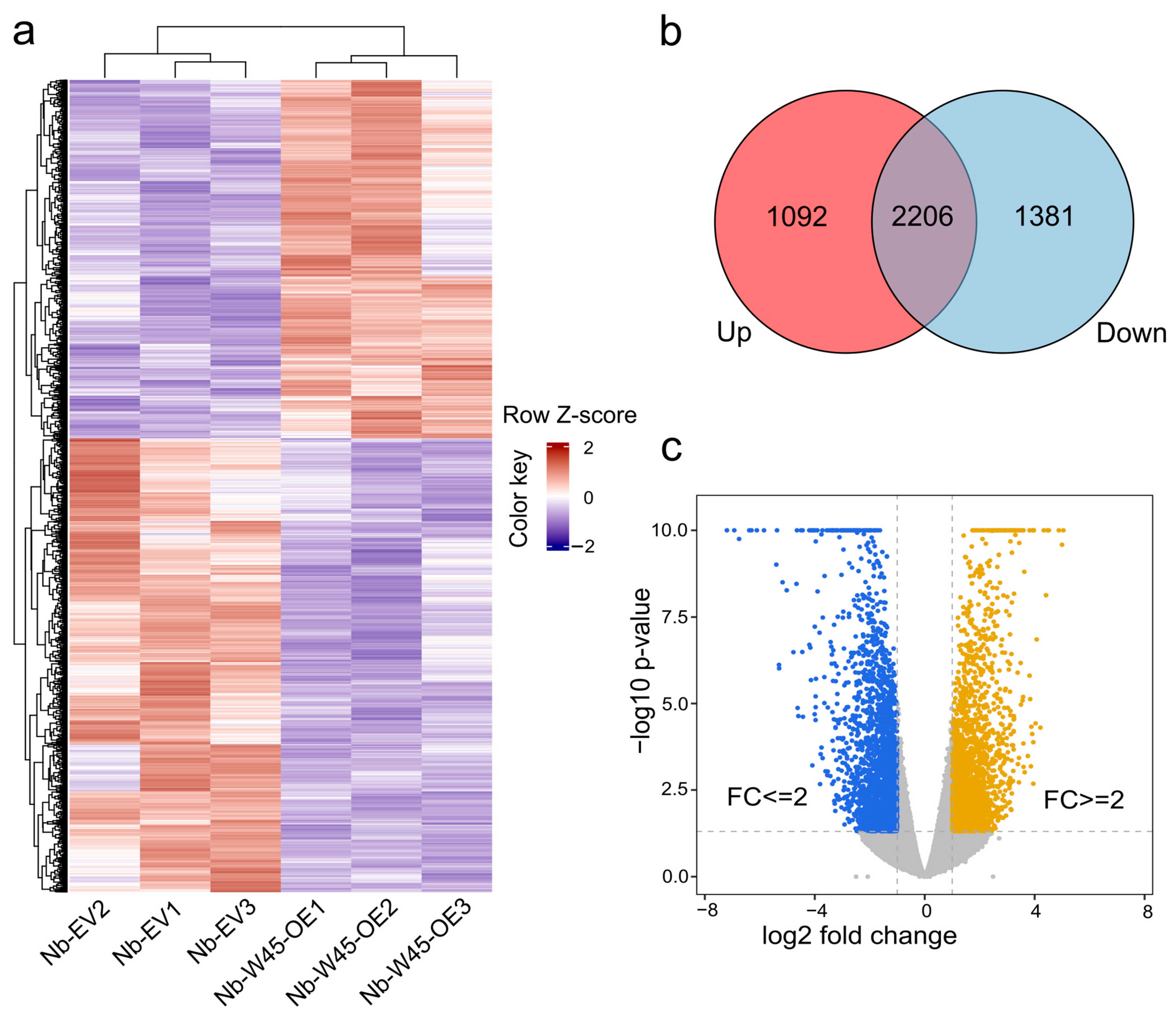
2.2. Global DEG Analysis of N. benthamiana Leaves Overexpressing MaWRKY45
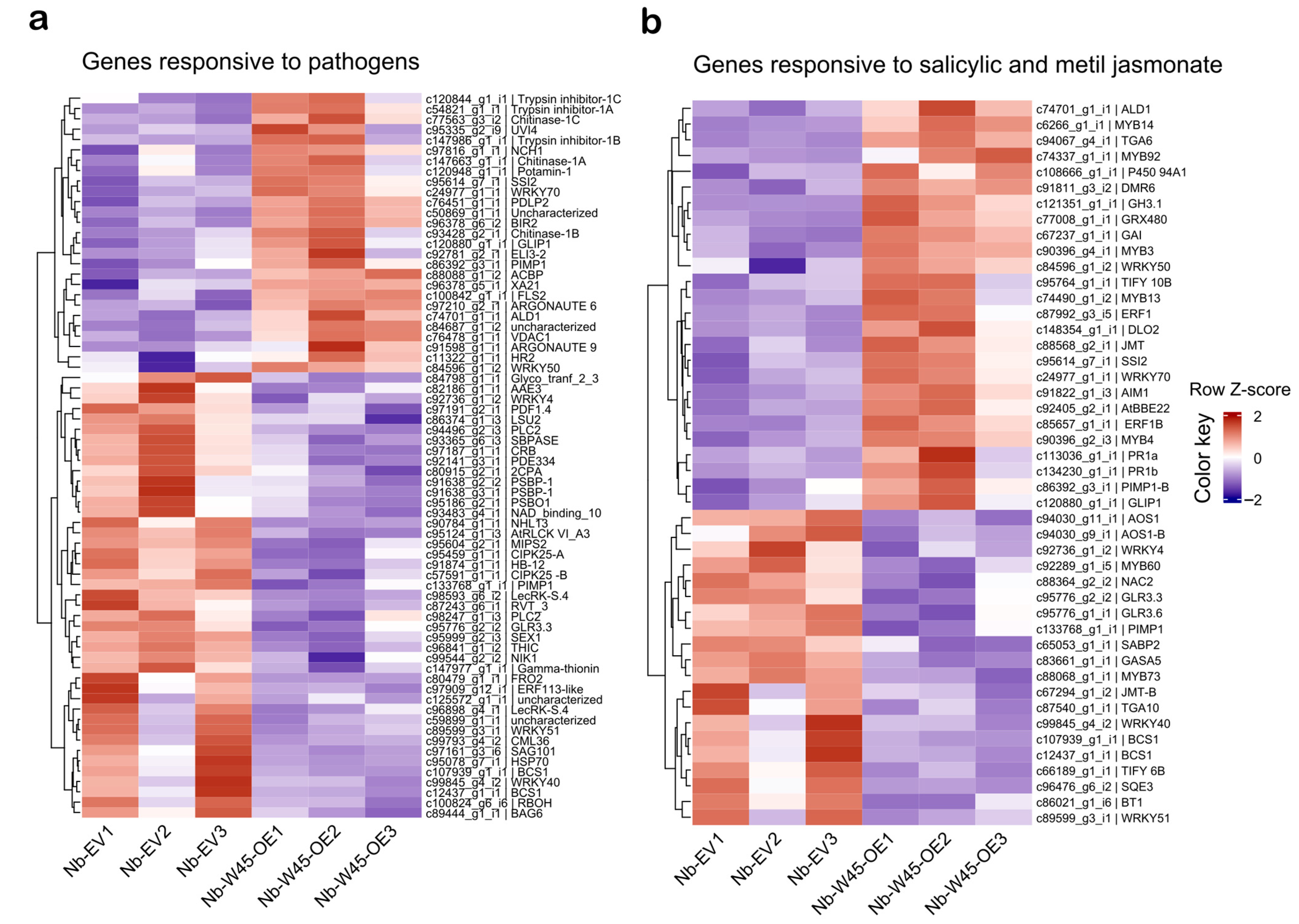
2.3. Transcriptional Regulation of Biotic Stress Responses in N. benthamiana Leaves Overexpressing MaWRKY45
2.4. MaWRKY45 Overexpression in N. benthamiana Leaves Induces Transcriptional Changes in Multiple Transcription Factor Genes
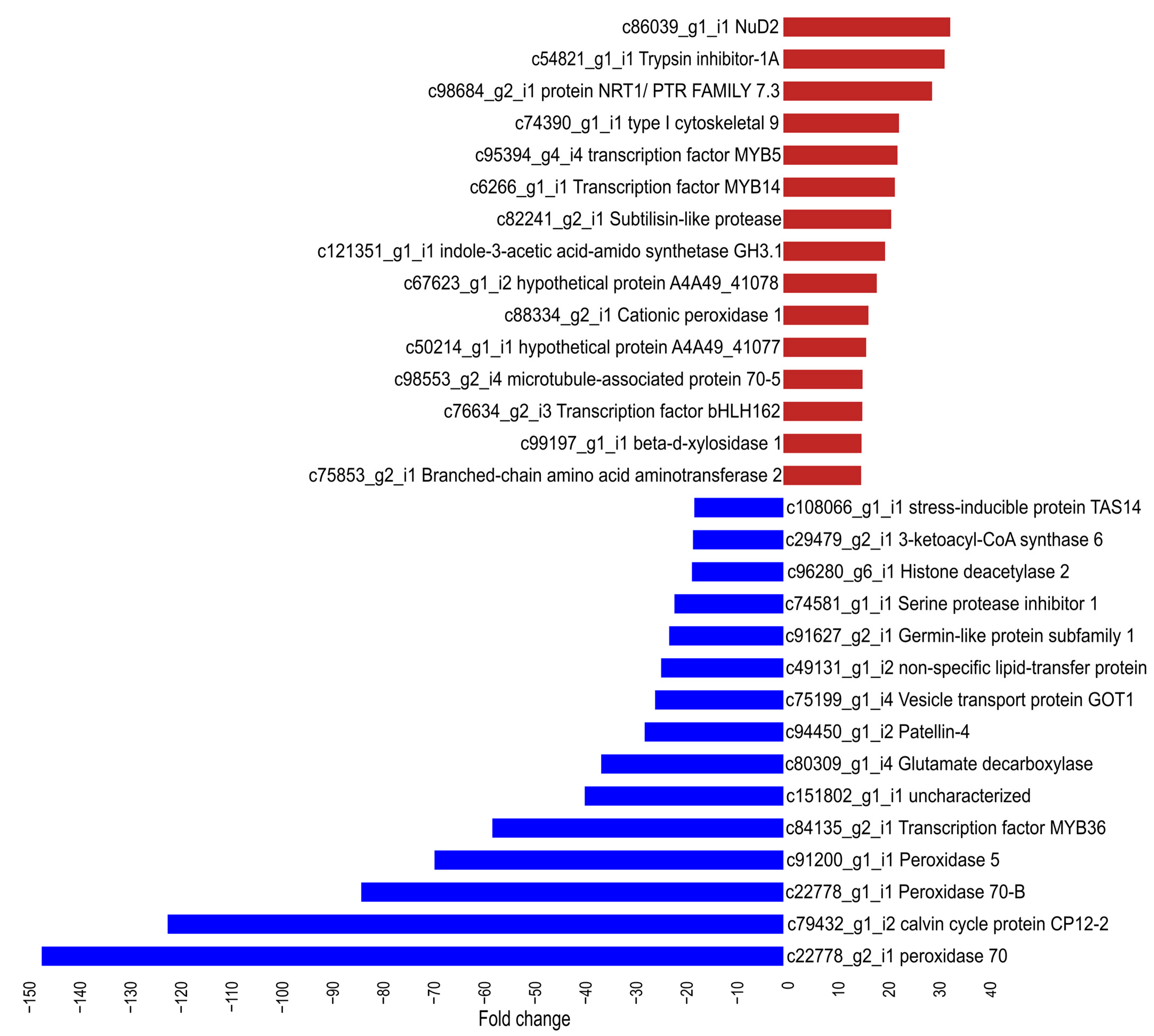
2.5. RT-qPCR Analysis of Genes Associated with MaWRKY45 Overexpression
3. Discussion
4. Materials and Methods
4.1. Biological Material
4.2. Amplification and Cloning of MaWRKY45
4.3. Agroinfiltration of N. benthamiana Leaves
4.4. RNA Isolation and cDNA Library Preparation for Sequencing
4.5. De Novo Transcriptome Assembly and Functional Annotation
4.6. Analysis of Differentially Expressed Genes (DEGs)
4.7. RT-qPCR
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Food and Agriculture Organization of the United Nations (FAO). FAOSTAT Database. Available online: https://www.fao.org/faostat/es/#data/QCL (accessed on 24 May 2023).
- Ploetz, R.C. Fusarium wilt of banana. Phytopathology 2015, 105, 1512–1521. [Google Scholar] [CrossRef]
- Ploetz, R.C.; Kema, G.H.; Ma, L.J. Impact of diseases on export and smallholder production of banana. Annu. Rev. Phytopathol. 2015, 53, 269–288. [Google Scholar] [CrossRef]
- D’Hont, A.; Denoeud, F.; Aury, J.-M.; Baurens, F.-C.; Carreel, F.; Garsmeur, O.; Noel, B.; Bocs, S.; Droc, G.; Rouard, M.; et al. The banana (Musa acuminata) genome and the evolution of monocotyledonous plants. Nature 2012, 488, 213–217. [Google Scholar] [CrossRef]
- Rouard, M.; Droc, G.; Martin, G.; Sardos, J.; Hueber, Y.; Guignon, V.; Cenci, A.; Geigle, B.; Hibbins, M.S.; Yahiaoui, N.; et al. Three new genome assemblies support a rapid radiation in Musa acuminata (wild banana). Genome Biol. Evol. 2018, 10, 3129–3140. [Google Scholar] [CrossRef]
- Belser, C.; Baurens, F.-C.; Noel, B.; Martin, G.; Cruaud, C.; Istace, B.; Yahiaoui, N.; Labadie, K.; Hřibová, E.; Doležel, J. Telomere-to-telomere gapless chromosomes of banana using nanopore sequencing. Commun. Biol. 2021, 4, 1047. [Google Scholar] [CrossRef]
- Dale, J.; James, A.; Paul, J.Y.; Khanna, H.; Smith, M.; Peraza-Echeverria, S.; Garcia-Bastidas, F.; Kema, G.; Waterhouse, P.; Mengersen, K.; et al. Transgenic Cavendish bananas with resistance to Fusarium wilt tropical race 4. Nat. Commun. 2017, 8, 1496. [Google Scholar] [CrossRef]
- Tripathi, J.N.; Ntui, V.O.; Tripathi, L. Precision genetics tools for genetic improvement of banana. Plant Genome 2024, 17, e20416. [Google Scholar] [CrossRef]
- Chen, X.; Li, C.; Wang, H.; Guo, Z. WRKY transcription factors: Evolution, binding, and action. Phytopathol. Res. 2019, 1, 13. [Google Scholar] [CrossRef]
- Dhatterwal, P.; Basu, S.; Mehrotra, S.; Mehrotra, R. Genome wide analysis of W-box element in Arabidopsis thaliana reveals TGAC motif with genes down regulated by heat and salinity. Sci. Rep. 2019, 9, 1681. [Google Scholar] [CrossRef] [PubMed]
- Maleck, K.; Levine, A.; Eulgem, T.; Morgan, A.; Schmid, J.; Lawton, K.A.; Dietrich, R.A. The transcriptome of Arabidopsis thaliana during systemic acquired resistance. Nat. Genet. 2000, 26, 403–410. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Cheng, X.; Yin, D.; Chen, D.; Luo, C.; Liu, H.; Huang, C. Advances in the research on plant WRKY transcription factors responsive to external stresses. Curr. Issues Mol. Biol. 2023, 45, 2861–2880. [Google Scholar] [CrossRef]
- Javed, T.; Gao, S.J. WRKY transcription factors in plant defense. Trends Genet. 2023, 39, 787–801. [Google Scholar] [CrossRef] [PubMed]
- Shimono, M.; Sugano, S.; Nakayama, A.; Jiang, C.J.; Ono, K.; Toki, S.; Takatsuji, H. Rice WRKY45 plays a crucial role in benzothiadiazole-inducible blast resistance. Plant Cell 2007, 19, 2064–2076. [Google Scholar] [CrossRef]
- Shimono, M.; Koga, H.; Akagi, A.; Hayashi, N.; Goto, S.; Sawada, M.; Kurihara, T.; Matsushita, A.; Sugano, S.; Jiang, C.J.; et al. Rice WRKY45 plays important roles in fungal and bacterial disease resistance. Mol. Plant Pathol. 2012, 13, 83–94. [Google Scholar] [CrossRef]
- Goto, S.; Sasakura-Shimoda, F.; Suetsugu, M.; Selvaraj, M.G.; Hayashi, N.; Yamazaki, M.; Takatsuji, H. Development of disease-resistant rice by optimized expression of WRKY45. Plant Biotechnol. J. 2015, 13, 753–765. [Google Scholar] [CrossRef] [PubMed]
- Goto, S.; Sasakura-Shimoda, F.; Yamazaki, M.; Hayashi, N.; Suetsugu, M.; Ochiai, H.; Takatsuji, H. Development of disease-resistant rice by pathogen-responsive expression of WRKY45. Plant Biotechnol. J. 2016, 14, 1127–1138. [Google Scholar] [CrossRef]
- García-Laynes, S.; Herrera-Valencia, V.A.; Tamayo-Torres, L.G.; Limones-Briones, V.; Barredo-Pool, F.A.; Baas-Espinola, F.M.; Alpuche-Solís, A.G.; Puch-Hau, C.; Peraza-Echeverria, S. The banana MaWRKY18, MaWRKY45, MaWRKY60 and MaWRKY70 genes encode functional transcription factors and display differential expression in response to defense phytohormones. Genes 2022, 13, 1891. [Google Scholar] [CrossRef]
- Vlot, A.C.; Sales, J.H.; Lenk, M.; Bauer, K.; Brambilla, A.; Sommer, A.; Nayem, S. Systemic propagation of immunity in plants. New Phytol. 2021, 229, 1234–1250. [Google Scholar] [CrossRef]
- Zhang, J.Z. Overexpression analysis of plant transcription factors. Curr. Opin. Plant Biol. 2003, 6, 430–440. [Google Scholar] [CrossRef]
- Tripathi, J.N.; Oduor, R.O.; Tripathi, L. A high-throughput regeneration and transformation platform for production of genetically modified banana. Front. Plant Sci. 2015, 6, 1025. [Google Scholar] [CrossRef] [PubMed]
- Scarpeci, T.E.; Frea, V.S.; Zanor, M.I.; Valle, E.M. Overexpression of AtERF019 delays plant growth and senescence, and improves drought tolerance in Arabidopsis. J. Exp. Bot. 2017, 68, 673–685. [Google Scholar]
- Bond, D.M.; Albert, N.W.; Lee, R.H.; Gillard, G.B.; Brown, C.M.; Hellens, R.P.; Macknight, R.C. Infiltration-RNAseq: Transcriptome profiling of Agrobacterium-mediated infiltration of transcription factors to discover gene function and expression networks in plants. Plant Methods 2016, 12, 41. [Google Scholar] [CrossRef][Green Version]
- Bally, J.; Jung, H.; Mortimer, C.; Naim, F.; Philips, J.G.; Hellens, R.; Bombarely, A.; Goodin, M.M.; Waterhouse, P.M. The rise and rise of Nicothiana benthamiana: A plant for all reasons. Annu. Rev. Phytopathol. 2018, 56, 405–426. [Google Scholar] [CrossRef]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef]
- Kaufmann, K.; Airoldi, C.A. Master regulatory transcription factors in plant development: A blooming perspective. Plant Transcr. Factors Methods Protoc. 2018, 3, 3–22. [Google Scholar]
- Mhamdi, A. NPR1 has everything under control. Plant Physiol. 2019, 181, 6–7. [Google Scholar] [CrossRef]
- Nakayama, A.; Fukushima, S.; Goto, S.; Matsushita, A.; Shimono, M.; Sugano, S.; Takatsuji, H. Genome-wide identification of WRKY45-regulated genes that mediate benzothiadiazole-induced defense responses in rice. BMC Plant Biol. 2013, 13, 150. [Google Scholar] [CrossRef]
- Liu, Y.; Tikunov, Y.; Schouten, R.E.; Marcelis, L.F.; Visser, R.G.; Bovy, A. Anthocyanin biosynthesis and degradation mechanisms in Solanaceous vegetables: A review. Front. Chem. 2018, 6, 52. [Google Scholar] [CrossRef]
- Wang, W.; Wang, Z.Y. At the intersection of plant growth and immunity. Cell Host Microbe 2014, 15, 400–402. [Google Scholar] [CrossRef] [PubMed]
- Slavokhotova, A.; Korostyleva, T.; Shelenkov, A.; Pukhalskiy, V.; Korottseva, I.; Slezina, M.; Odintsova, T. Transcriptomic analysis of genes involved in plant defense response to the cucumber green mottle mosaic virus infection. Life 2021, 11, 1064. [Google Scholar] [CrossRef]
- Zhang, X.; Guo, W.; Du, D.; Pu, L.; Zhang, C. Overexpression of a maize BR transcription factor ZmBZR1 in Arabidopsis enlarges organ and seed size of the transgenic plants. Plant Sci. 2020, 292, 110378. [Google Scholar] [CrossRef] [PubMed]
- Rochon, A.; Boyle, P.; Wignes, T.; Fobert, P.R.; Després, C. The coactivator function of Arabidopsis NPR1 requires the core of its BTB/POZ domain and the oxidation of C-terminal cysteines. Plant Cell 2006, 18, 3670–3685. [Google Scholar] [CrossRef]
- Boyle, P.; Després, C. Dual-function transcription factors and their entourage: Unique and unifying themes governing two pathogenesis-related genes. Plant Signal Behav. 2010, 5, 629–634. [Google Scholar] [CrossRef]
- Birkenbihl, R.P.; Kracher, B.; Roccaro, M.; Somssich, I.E. Induced genome-wide binding of three Arabidopsis WRKY transcription factors during early MAMP-triggered immunity. Plant Cell 2017, 29, 20–38. [Google Scholar] [CrossRef] [PubMed]
- Ullah, C.; Schmidt, A.; Reichelt, M.; Tsai, C.J.; Gershenzon, J. Lack of antagonism between salicylic acid and jasmonate signalling pathways in poplar. New Phytol. 2022, 235, 701–717. [Google Scholar] [CrossRef]
- Coolen, S.; Proietti, S.; Hickman, R.; Davila Olivas, N.H.; Huang, P.-P.; Van Verk, M.C.; Van Pelt, J.A.; Wittenberg, A.H.J.; De Vos, M.; Prins, M.; et al. Transcriptome dynamics of Arabidopsis during sequential biotic and abiotic stresses. Plant J. 2016, 86, 249–267. [Google Scholar] [CrossRef]
- Swaminathan, S.; Lionetti, V.; Zabotina, O.A. Plant cell wall integrity perturbations and priming for defense. Plants 2022, 11, 3539. [Google Scholar] [CrossRef]
- Ercoli, M.F.; Luu, D.D.; Rim, E.Y.; Shigenaga, A.; Teixeira de Araujo, A., Jr.; Chern, M.; Ronald, P. Plant immunity: Rice XA21-mediated resistance to bacterial infection. Proc. Natl. Acad. Sci. USA 2022, 119, e2121568119. [Google Scholar] [CrossRef]
- Zhang, N.; Zhou, S.; Yang, D.; Fan, Z. Revealing shared and distinct genes responding to JA and SA signaling in Arabidopsis by meta-analysis. Front. Plant Sci. 2020, 11, 908. [Google Scholar]
- Caillaud, M.C.; Wirthmueller, L.; Sklenar, J.; Findlay, K.; Piquerez, S.J.; Jones, A.M.; Faulkner, C. The plasmodesmal protein PDLP1 localises to haustoria-associated membranes during downy mildew infection and regulates callose deposition. PLoS Pathog. 2014, 10, e1004496. [Google Scholar] [CrossRef]
- Xia, Y.; Yu, K.; Gao, Q.M.; Wilson, E.V.; Navarre, D.; Kachroo, P.; Kachroo, A. Acyl CoA binding proteins are required for cuticle formation and plant responses to microbes. Front. Plant Sci. 2012, 3, 224. [Google Scholar] [CrossRef]
- Shah, J.; Kachroo, P.; Nandi, A.; Klessig, D.F. A recessive mutation in the Arabidopsis SSI2 gene confers SA- and NPR1-independent expression of PR genes and resistance against bacterial and oomycete pathogens. Plant J. 2001, 25, 563–574. [Google Scholar] [CrossRef]
- Hong, J.K.; Choi, D.S.; Kim, S.H.; Yi, S.Y.; Kim, Y.J.; Hwang, B.K. Distinct roles of the pepper pathogen-induced membrane protein gene CaPIMP1 in bacterial disease resistance and oomycete disease susceptibility. Planta 2008, 228, 485–497. [Google Scholar] [CrossRef]
- Lee, D.H.; Lee, H.S.; Choi, M.S.; Parys, K.; Honda, K.; Kondoh, Y.; Belkhadir, Y. Reprogramming of flagellin receptor responses with surrogate ligands. Nat. Commun. 2024, 15, 9811. [Google Scholar] [CrossRef]
- Halter, T.; Imkampe, J.; Mazzotta, S.; Wierzba, M.; Postel, S.; Bücherl, C.; Kiefer, C.; Stahl, M.; Chinchilla, D.; Wang, X.; et al. The leucine-rich repeat receptor kinase BIR2 is a negative regulator of BAK1 in plant immunity. Curr. Biol. 2014, 24, 134–143. [Google Scholar] [CrossRef]
- Yang, X.M.; Zhao, J.H.; Xiong, X.Y.; Hu, Z.W.; Sun, J.F.; Su, H.; Li, Y. Broad-spectrum resistance gene RPW8.1 balances immunity and growth via feedback regulation of WRKYs. Plant Biotechnol. J. 2024, 22, 116–130. [Google Scholar] [CrossRef]
- Almagro, L.; Gómez Ros, L.V.; Belchi-Navarro, S.; Bru, R.; Ros Barceló, A.; Pedreño, M.A. Class III peroxidases in plant defense reactions. J. Exp. Bot. 2009, 60, 377–390. [Google Scholar] [CrossRef]
- Kim, H.G.; Kwon, S.J.; Jang, Y.J.; Nam, M.H.; Chung, J.H.; Na, Y.C.; Park, O.K. GDSL LIPASE1 modulates plant immunity through feedback regulation of ethylene signaling. Plant Physiol. 2013, 163, 1776–1791. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.J.; Lim, J.H.; Kim, M.J.; Kim, T.; Chung, H.M.; Paek, K.H. GDSL-lipase1 (CaGL1) contributes to wound stress resistance by modulation of CaPR-4 expression in hot pepper. Biochem. Biophys. Res. Commun. 2008, 374, 693–698. [Google Scholar] [CrossRef] [PubMed]
- de Souza Nascimento, A.M.; de Oliveira Segundo, V.H.; Felipe Camelo Aguiar, A.J.; Piuvezam, G.; Souza Passos, T.; Florentino da Silva Chaves Damasceno, K.S.F.; de Araújo Morais, A.H. Antibacterial action mechanisms and mode of trypsin inhibitors: A systematic review. J. Enzym. Inhib. Med. Chem. 2022, 37, 749–759. [Google Scholar] [CrossRef] [PubMed]
- Tzean, Y.; Hou, B.H.; Tsao, S.M.; Chen, H.M.; Cheng, A.P.; Chen, E.G.; Chou, W.Y.; Chao, C.P.; Shen, W.C.; Chen, C.C.; et al. Identification of MaWRKY40 and MaDLO1 as effective marker genes for tracking the salicylic acid-mediated immune response in bananas. Rev. Phytopathol. 2021, 111, 1800–1810. [Google Scholar] [CrossRef]
- Gao, Q.M.; Zhu, S.; Kachroo, P.; Kachroo, A. Signal regulators of systemic acquired resistance. Front. Plant Sci. 2015, 6, 228. [Google Scholar] [CrossRef] [PubMed]
- Kong, W.; Zhang, Y.; Deng, X.; Li, S.; Zhang, C.; Li, Y. Comparative genomic and transcriptomic analysis suggests the evolutionary dynamic of GH3 genes in Gramineae crops. Front. Plant Sci. 2019, 10, 1297. [Google Scholar] [CrossRef]
- Zhang, H.; Liu, Y.; Zhang, X.; Ji, W.; Kang, Z. A necessary considering factor for breeding: Growth-defense tradeoff in plants. Stress Biol. 2023, 3, 6. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Jun, J.H.; Dixon, R.A. MYB5 and MYB14 play pivotal roles in seed coat polymer biosynthesis in Medicago truncatula. Front. Plant Sci. 2014, 5, 169. [Google Scholar] [CrossRef]
- Luo, Y.; Wang, Q.; Bai, R.; Li, R.; Chen, L.; Xu, Y.; Duan, D. The effect of transcription factor MYB14 on defense mechanisms in Vitis quinquangularis-Pingyi. Int. J. Mol. Sci. 2020, 21, 706. [Google Scholar] [CrossRef]
- To, A.; Joubès, J.; Thueux, J.; Kazaz, S.; Lepiniec, L.; Baud, S. AtMYB92 enhances fatty acid synthesis and suberin deposition in leaves of Nicotiana benthamiana. Plant J. 2020, 103, 660–676. [Google Scholar] [CrossRef]
- Llorca, C.M.; Potschin, M.; Zentgraf, U. bZIPs and WRKYs: Two large transcription factor families executing two different functional strategies. Front. Plant Sci. 2014, 5, 169. [Google Scholar] [CrossRef]
- Thieme, C.J.; Rojas-Triana, M.; Stecyk, E.; Schudoma, C.; Zhang, W.; Yang, L.; Miñambres, M.; Walther, D.; Schulze, W.X.; Paz-Ares, J.; et al. Endogenous Arabidopsis messenger RNAs transported to distant tissues. Nat. Plants 2015, 1, 15025. [Google Scholar] [CrossRef]
- Kato, H.; Onai, K.; Abe, A.; Shimizu, M.; Takagi, H.; Tateda, C.; Utsushi, H.; Singkarabanit-Ogawa, S.; Kitakura, S.; Ono, E.; et al. Lumi-Map, a real-time luciferase bioluminescence screen of mutants combined with MutMap, reveals Arabidopsis genes involved in PAMP-triggered immunity. Mol. Plant-Microbe Interact. 2020, 33, 1366–1380. [Google Scholar] [CrossRef] [PubMed]
- Gkizi, D.; Lehmann, S.; L’Haridon, F.; Serrano, M.; Paplomatas, E.J.; Métraux, J.P.; Tjamos, S.E. The innate immune signaling system as a regulator of disease resistance and induced systemic resistance activity against Verticillium dahliae. Mol. Plant-Microbe Interact. 2016, 29, 313–323. [Google Scholar] [CrossRef]
- Li, J.; Brader, G.; Palva, E.T. The WRKY70 transcription factor: A node of convergence for jasmonate-mediated and salicylate-mediated signals in plant defense. Plant Cell 2004, 16, 319–331. [Google Scholar] [CrossRef]
- Mao, G.; Meng, X.; Liu, Y.; Zheng, Z.; Chen, Z.; Zhang, S. Phosphorylation of a WRKY transcription factor by two pathogen-responsive MAPKs drives phytoalexin biosynthesis in Arabidopsis. Plant Cell 2011, 23, 1639–1653. [Google Scholar] [CrossRef] [PubMed]
- Han, G.Z. Origin and evolution of the plant immune system. New Phytol. 2018, 222, 70–83. [Google Scholar] [CrossRef] [PubMed]
- Jones, J.D.G.; Staskawicz, B.J.; Dangl, J.L. The plant immune system: From discovery to deployment. Cell 2024, 187, 2095–2116. [Google Scholar] [CrossRef]
- Norkunas, K.; Harding, R.; Dale, J.; Dugdale, B. Improving agroinfiltration-based transient gene expression in Nicotiana benthamiana. Plant Methods 2018, 14, 71. [Google Scholar] [CrossRef]
- Assefa, A.T.; Vandesompele, J.; Thas, O. On the utility of RNA sample pooling to optimize cost and statistical power in RNA sequencing experiments. BMC Genom. 2020, 21, 312. [Google Scholar] [CrossRef] [PubMed]
- Andrews, S. 2010 FastQC: A Quality Control Tool for High Throughput Sequence Data. Available online: http://www.bioinformatics.babraham.ac.uk/projects/fastqc (accessed on 13 August 2023).
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef]
- Li, W.; Godzik, A. Cd-hit: A fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 2006, 22, 1658–1659. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S. Fast gapped-read alignment with bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef]
- Kanehisa, M.; Sato, Y.; Kawashima, M.; Furumichi, M.; Tanabe, M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 2016, 44, D457–D462. [Google Scholar] [CrossRef] [PubMed]
- Sayers, E.W.; Beck, J.; Bolton, E.E.; Bourexis, D.; Brister, J.R.; Canese, K.; Comeau, D.C.; Funk, K.; Kim, S.; Klimke, W.; et al. Database resources of the national center for biotechnology information. Nucleic Acids Res. 2021, 49, D10–D17. [Google Scholar] [CrossRef]
- Mistry, J.; Chuguransky, S.; Williams, L.; Qureshi, M.; Salazar, G.A.; Sonnhammer, E.L.L.; Tosatto, S.C.E.; Paladin, L.; Raj, S.; Richardson, L.J.; et al. Pfam: The protein families database in 2021. Nucleic Acids Res. 2021, 49, D412–D419. [Google Scholar] [CrossRef]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene ontology: Tool for the unification of biology. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef]
- Pruitt, K.D.; Tatusova, T.; Maglott, D.R. NCBI reference sequences (RefSeq): A curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2007, 35, D61–D65. [Google Scholar] [CrossRef] [PubMed]
- UniProt Consortium, 2023. UniProt: The Universal Protein knowledgebase in 2023. Nucleic Acids Res. 2023, 51, D523–D531. [Google Scholar] [CrossRef]
- Hernández-Plaza, A.; Szklarczyk, D.; Botas, J.; Cantalapiedra, C.P.; Giner-Lamia, J.; Mende, D.R.; Kirsch, R.; Rattei, T.; Letunic, I.; Jensen, L.J.; et al. eggNOG 6.0: Enabling comparative genomics across 12 535 organisms. Nucleic Acids Res. 2023, 51, D389–D394. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Buchfink, B.; Xie, C.; Huson, D.H. Fast and sensitive protein alignment using DIAMOND. Nat. Methods 2015, 12, 59–60. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Wickham, H. Data Analysis. In ggplot2: Elegant Graphics for Data Analysis, 2nd ed.; Gentleman, R., Hornik, K., Parmigiani, G., Eds.; Springer: Houston, TX, USA, 2016; pp. 189–201. ISBN 978-3-319-24277-4. [Google Scholar]
- Gu, Z.; Eils, R.; Schlesner, M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 2016, 32, 2847–2849. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.; Zhang, Z.; Wan, Q.; Zhou, H.; Jiao, M.; Zheng, H.; Lu, Y.; Rao, S.; Wu, G.; Chen, J.; et al. Selection and validation of reference genes for RT-qPCR analysis of gene expression in Nicotiana benthamiana upon single infections by 11 positive-sense single-stranded RNA viruses from four genera. Plants 2023, 12, 857. [Google Scholar] [CrossRef]






| Metric | Transcript Contig | Only Longest Isoform per Gene |
|---|---|---|
| N50 (bp) | 1289 | 898 |
| Average contig length (bp) | 782 | 620 |
| Maximum contig length (bp) | 14,833 | 14,833 |
| Minimum contig length (bp) | 201 | 201 |
| GC content (%) | 38 | 38 |
| Total transcripts | 273,793 | 189,020 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
García-Laynes, S.; Calderón-Vázquez, C.L.; Puch-Hau, C.; Herrera-Valencia, V.A.; Peraza-Echeverria, S. Infiltration-RNAseq Reveals Enhanced Defense Responses in Nicothiana benthamiana Leaves Overexpressing the Banana Gene MaWRKY45. Plants 2025, 14, 483. https://doi.org/10.3390/plants14030483
García-Laynes S, Calderón-Vázquez CL, Puch-Hau C, Herrera-Valencia VA, Peraza-Echeverria S. Infiltration-RNAseq Reveals Enhanced Defense Responses in Nicothiana benthamiana Leaves Overexpressing the Banana Gene MaWRKY45. Plants. 2025; 14(3):483. https://doi.org/10.3390/plants14030483
Chicago/Turabian StyleGarcía-Laynes, Sergio, Carlos Ligne Calderón-Vázquez, Carlos Puch-Hau, Virginia Aurora Herrera-Valencia, and Santy Peraza-Echeverria. 2025. "Infiltration-RNAseq Reveals Enhanced Defense Responses in Nicothiana benthamiana Leaves Overexpressing the Banana Gene MaWRKY45" Plants 14, no. 3: 483. https://doi.org/10.3390/plants14030483
APA StyleGarcía-Laynes, S., Calderón-Vázquez, C. L., Puch-Hau, C., Herrera-Valencia, V. A., & Peraza-Echeverria, S. (2025). Infiltration-RNAseq Reveals Enhanced Defense Responses in Nicothiana benthamiana Leaves Overexpressing the Banana Gene MaWRKY45. Plants, 14(3), 483. https://doi.org/10.3390/plants14030483








