Phylogenetic Inferences and Historical Biogeography of Onocleaceae
Abstract
1. Introduction
2. Materials and Methods
2.1. Taxon Sampling, DNA Extraction and Sequencing
2.2. Plastome Assembly, Annotation, and Phylogenetic Analysis
| Taxon | Plastome Size (bp) | GC Content (%) | LSC Size (bp) | LSC GC Content (%) | SSC Size (bp) | SSC GC Content (%) | IR Size (bp) | IR GC Content (%) | Voucher | Herbarium Acronyms | Location | GenBank ID | Reference |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Matteuccia struthiopteris (L.) Tod. | 151,101 | 44.2 | 81,974 | 43.7 | 21,695 | 42.1 | 23,716 | 46.1 | YUS8847 | PYU | China, Sichuan | PP712888 | This study |
| Matteuccia struthiopteris (L.) Tod. | 151,078 | 44.3 | 82,020 | 43.8 | 21,672 | 42.2 | 23,693 | 46.1 | Wei Q. et al. FB854 | KUN | China, Yunnan | MT130666 | [37] |
| Matteuccia struthiopteris (L.) Tod. | 151,003 | 44.3 | 81,964 | 43.8 | 21,675 | 42.2 | 23,682 | 46.1 | WR0331 | PE | China, Beijing | NC035859 | [32] |
| Onoclea sensibilis L. | 148,604 | 44.4 | 81,588 | 44.1 | 21,730 | 42.5 | 22,643 | 45.9 | ZhouXM677 | PYU | USA, Cult. | PP712887 | This study |
| Onoclea sensibilis L. | 148,395 | 44.4 | 81,571 | 44.1 | 21,726 | 42.6 | 22,549 | 45.8 | WR0327 | PE | China, Beijing | NC035860 | [32] |
| Onoclea sensibilis var. interrupta Maxim. | 138,259 | 44.7 | 75,518 | 44.3 | 17,057 | 42.9 | 22,903 | 45.9 | ZhaoJingLN | PYU | China, Liaoning | PP712885 | This study |
| Onoclea sensibilis var. interrupta Maxim. | 141,826 | 44.6 | 77,871 | 44.3 | 19,089 | 42.9 | 22,039 | 45.9 | ZhaoJingLN3 | PYU | China, Liaoning | PP712886 | This study |
| Onoclea sensibilis var. interrupta Maxim. | 148,739 | 44.4 | 81,629 | 44.1 | 21,744 | 42.5 | 22,683 | 45.9 | Lu J.M. Lu472 | KUN | China, Jilin | MT130573 | [37] |
| Pentarhizidium intermedium (C.Chr.) Hayata | 151,168 | 44.2 | 82,554 | 43.7 | 21,620 | 42.2 | 23,497 | 46 | YUS7829 | PYU | China, Yunnan | PP712890 | This study |
| Pentarhizidium intermedium (C.Chr.) Hayata | 151,167 | 44.2 | 82,555 | 43.7 | 21,620 | 42.2 | 23,496 | 46 | YUS7844 | PYU | China, Yunnan | PP712889 | This study |
| Pentarhizidium intermedium (C.Chr.) Hayata | 151,175 | 44.2 | 82,562 | 43.7 | 21,619 | 42.2 | 23,497 | 46 | YUS9511 | PYU | China, Yunnan | PP712891 | This study |
| Pentarhizidium orientale (Hook.) Hayata | 151,279 | 44 | 82,646 | 43.5 | 21,591 | 41.9 | 23,521 | 45.9 | YUS6905 | PYU | China, Yunnan | PP712894 | This study |
| Pentarhizidium orientale (Hook.) Hayata | 151,333 | 44 | 82,628 | 43.5 | 21,591 | 41.9 | 23,557 | 45.9 | YUS8029 | PYU | China, Yunnan | PP712895 | This study |
| Pentarhizidium orientale (Hook.) Hayata | 151,203 | 44 | 82,571 | 43.5 | 21,590 | 41.9 | 23,521 | 45.9 | YUS9702 | PYU | China, Yunnan | PP712893 | This study |
| Pentarhizidium orientale (Hook.) Hayata | 151,202 | 44 | 82,570 | 43.5 | 21,590 | 41.9 | 23,521 | 45.9 | YUS10377 | PYU | China, Yunnan | PP712892 | This study |
| Pentarhizidium orientale (Hook.) Hayata | 151,243 | 44 | 82,610 | 43.5 | 21,591 | 41.9 | 23,521 | 45.9 | Lu J.M. Lu715 | KUN | China, Jiangxi | MT130641 | [37] |
| Woodwardia japonica (L.f.) Sm. | 153,708 | 43.2 | 82,377 | 42.4 | 21,559 | 40.5 | 24,886 | 45.8 | YUS8839 | PYU | China, Sichuan | PP712896 | This study |
2.3. Molecular Dating and Historical Biogeography
3. Results
3.1. Plastome Organization and Features
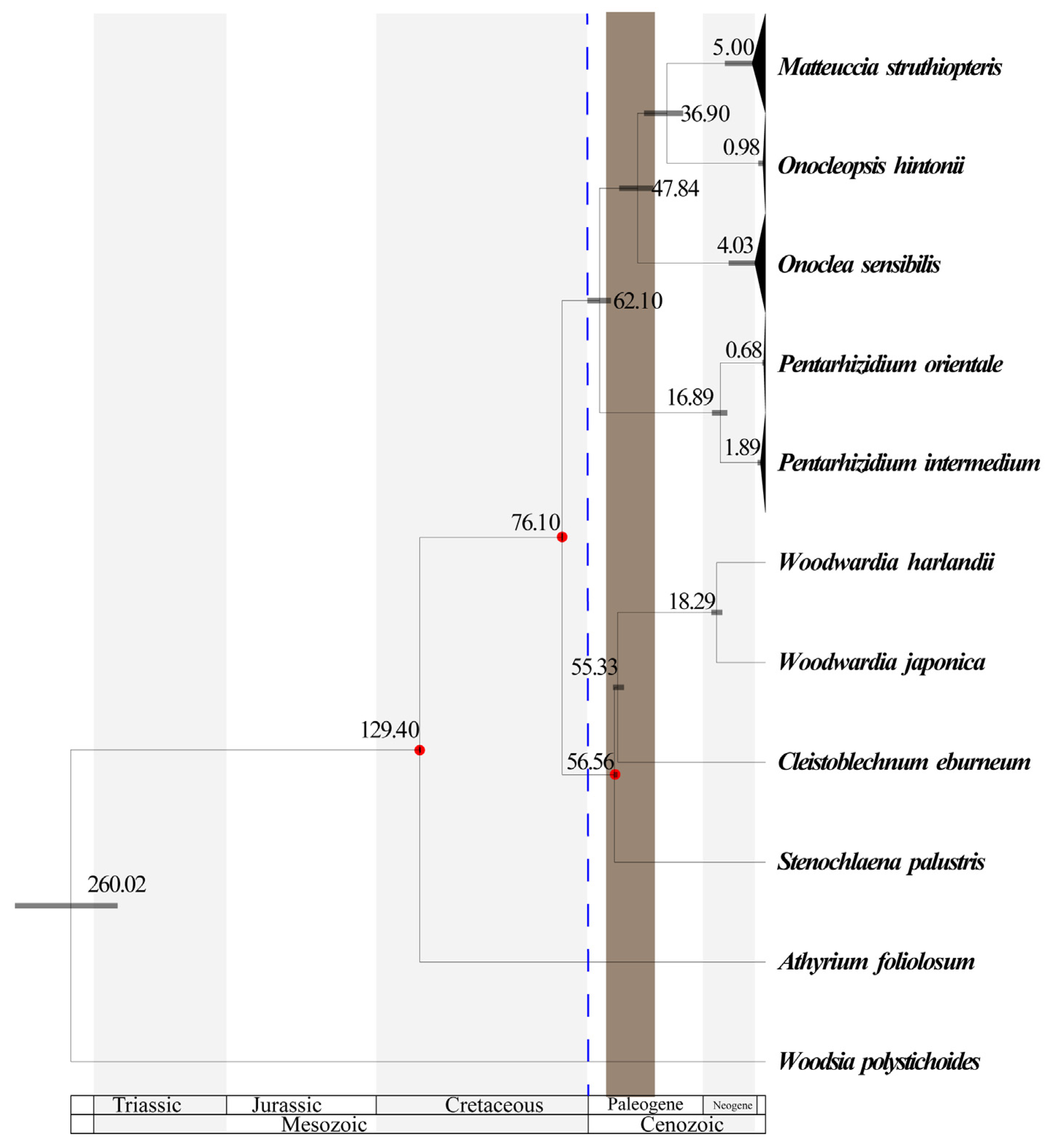
3.2. Phylogenetic Relationships
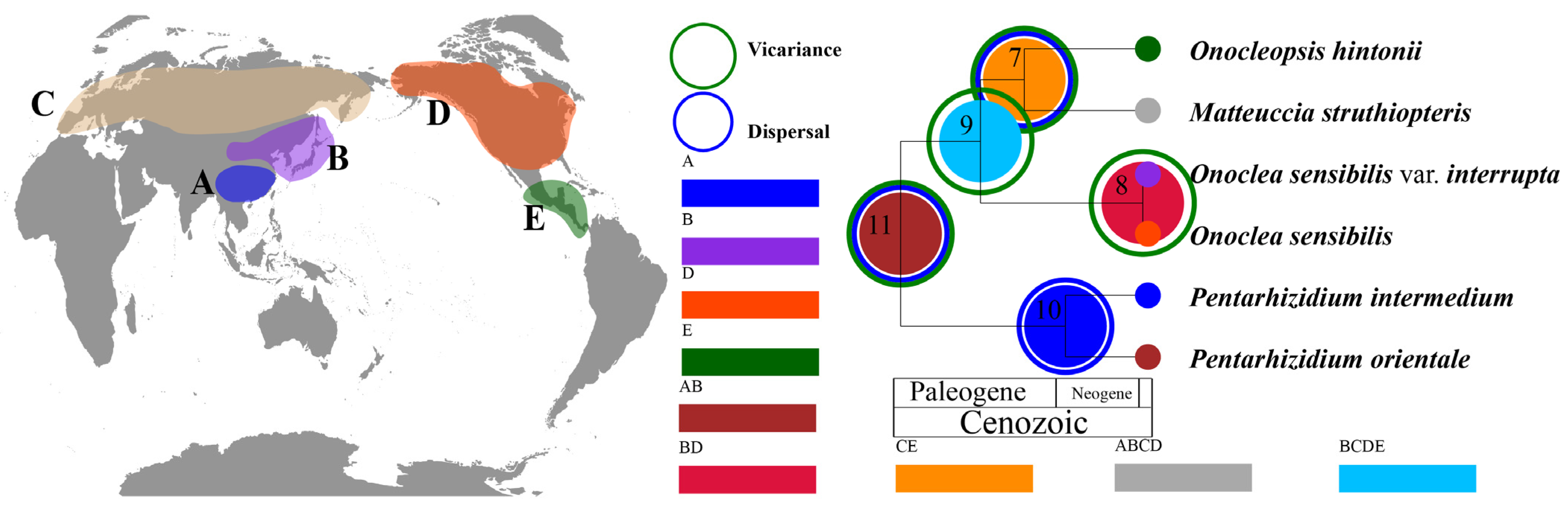
3.3. Biogeographic History and Ancestral Area Reconstruction
4. Discussion
4.1. Conservative Plastome Characteristics in Onocleaceae
4.2. Systematic Implications
4.3. Origin and Historical Biogeography of Onocleaceae
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Gastony, G.J.; Ungerer, M.C. Molecular systematics and a revised taxonomy of the onocleoid ferns (Dryopteridaceae: Onocleeae). Am. J. Bot. 1997, 84, 840–849. [Google Scholar] [CrossRef] [PubMed]
- Smith, A.R.; Pryer, K.M.; Schuettpelz, E.; Korall, P.; Schneider, H.; Wolf, P.G. A classification for extant ferns. Taxon 2006, 55, 705–731. [Google Scholar] [CrossRef]
- Rothfels, C.J.; Sundue, M.A.; Kuo, L.-Y.; Larsson, A.; Kato, M.; Schuettpelz, E.; Pryer, K.M. A revised family–level classification for eupolypod II ferns (Polypodiidae: Polypodiales). Taxon 2012, 61, 515–533. [Google Scholar] [CrossRef]
- PPGI. A community-derived classification for extant lycophytes and ferns. J. Syst. Evol. 2016, 54, 563–603. [Google Scholar] [CrossRef]
- Copeland, E.B. Genera filicum. In Chronica Botanica; Chronica Botanica Company: Waltham, MA, USA, 1947. [Google Scholar]
- Lloyd, R.M. Systematics of the Onocleoid Ferns, (University of California Publications in Botany); University of California Press: Berkeley, CA, USA, 1971; Volume 61, pp. 1–93. [Google Scholar]
- Tryon, R.M.; Tryon, A.F. Ferns and Allied Plants with Special Reference to Tropical America; Springer: New York, NY, USA, 1982. [Google Scholar]
- Kramer, K.U. Dryopteridaceae. In The Families and Genera of Vascular Plants, Pteridophytes and Gymnosperms; Kubitzki, K., Green, P.S., Eds.; Springer: Berlin, Germany, 1990; Volume 1, pp. 101–144. [Google Scholar]
- Hasebe, M.; Wolf, P.G.; Pryer, K.M.; Ueda, K.; Ito, M.; Samo, R.; Gastony, G.J.; Yokoyama, J.; Murakami, N.; Crane, E.H.; et al. Fern phylogeny based on rbcL nucleotide sequences. Am. Fern J. 1995, 85, 134–181. [Google Scholar] [CrossRef]
- Kato, M.; Sahashi, N. Affinities in the onocleoid ferns. Acta Phytotaxon. Geo. 1980, 31, 127–138. [Google Scholar]
- Kato, M.; Suzuki, T.; Nakato, N. The systematic status of Matteuccia intermedia (Woodsiaceae: Pteridophyta). Fern Gaz. 1991, 14, 65–71. [Google Scholar]
- Smith, A.R.; Pryer, K.M.; Schuettpelz, E.; Korall, P.; Schneider, H.; Wolf, P.G. Fern classification. In Biology and Evolution of Ferns and Lycophytes; Ranker, T.A., Haufler, C.H., Eds.; Cambridge University Press: Cambridge, UK, 2008; pp. 417–467. [Google Scholar]
- Christenhusz, M.J.M.; Zhang, X.-C.; Schneider, H. A linear sequence of extant families and genera of lycophytes and ferns. Phytotaxa 2011, 19, 7–54. [Google Scholar] [CrossRef]
- Rothfels, C.J.; Larsson, A.; Kuo, L.-Y.; Korall, P.; Chiou, W.-L.; Pryer, K.M. Overcoming deep roots, fast rates, and short internodes to resolve the ancient rapid radiation of eupolypod II ferns. Syst. Biol. 2012, 61, 490. [Google Scholar] [CrossRef]
- Schuettpelz, E.; Rouhan, G.; Pryer, K.M.; Rothfels, C.J.; Prado, J.; Sundue, M.A.; Windham, M.D.; Moran, M.D.; Smith, A.R. Are there too many fern genera? Taxon 2018, 67, 473–480. [Google Scholar] [CrossRef]
- Berry, K. East Asian-eastern North American disjunction in the Onoclea sensibilis species complex: Vicariance or peripatry? Palaeobotany 2021, 12, 88–94. [Google Scholar] [CrossRef]
- Xing, F.-W.; Wang, F.-G.; Kato, M. Onocleaceae. In Flora of China, (Pteridophytes); Wu, Z.Y., Raven, P.H., Hong, D.Y., Eds.; Science Press: Beijing, China; Missouri Botanical Garden Press: St. Louis, MO, USA, 2013; Volume 2–3, pp. 408–410. [Google Scholar]
- Koenemann, D.M. Broad-scale integrity and local divergence in the fiddlehead fern Matteuccia struthiopteris (L.) Todaro (Onocleaceae). Am. Fern J. 2011, 101, 213–230. [Google Scholar] [CrossRef]
- Ballard, F. Further Notes on Onocleopsis. Am. Fern J. 1948, 38, 125–132. [Google Scholar] [CrossRef]
- Lellinger, D.B. A Field Manual of the Ferns and Fern Allies of the United States and Canada; Smithson Institute Press: Washington, DC, USA, 1985; p. 389. [Google Scholar]
- Li, H.-L. Floristic relationships between eastern Asia and eastern North-America. Trans. Am. Phil. Soci. 1952, 42, 371–429. [Google Scholar] [CrossRef]
- Kato, M. Biogeography of ferns: Dispersal and vicariance. J. Biogeogr. 1993, 20, 265–274. [Google Scholar] [CrossRef]
- Xiang, J.-Y.; Wen, J.; Peng, H. Evolution of the eastern Asian–North American biogeographic disjunctions in ferns and lycophytes. J. Syst. Evol. 2015, 53, 2–32. [Google Scholar] [CrossRef]
- Boureau, E. Traité de Paléobotanique IV(I). Filicopside; Masson et Cie: Paris, France, 1970. [Google Scholar]
- Barrington, D.S. Ecological and historical factors in fern biogeography. J. Biogeogr. 1993, 20, 275–280. [Google Scholar] [CrossRef]
- Newberry, J.S. Notes on the latter extinct floras of North America, with descriptions of some new species of fossil plants from the Cretaceous and Tertiary strata. Lyceum Nat. Hist. N. Y. Ann. 1868, 9, 1–76. [Google Scholar]
- Rothwell, G.W.; Stockey, R.A. Fossil Ophioglossales in the Paleocene of Western North America. Am. J. Bot. 1989, 76, 637–644. [Google Scholar] [CrossRef]
- Brown, R.W. Paleocene flora of the Rockey Mountains and Great Plains. Geol. Surv. Prof. Pap. 1962, 375, 1–119. [Google Scholar]
- Gardner, J.S.; Ettingshausen, C. A Monograph of the British Eocene Flora; Filices Paleontological Society: London, UK, 1879; Volume 1, p. 283. [Google Scholar]
- Chen, S.-F.; Zhou, Y.-Q.; Chen, Y.-R.; Gu, J. Fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef] [PubMed]
- Jin, J.-J.; Yu, W.-B.; Yang, J.-B.; Song, Y.; de Pamphilis, C.W.; Yi, T.-S.; Li, D.-Z. GetOrganelle: A fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 2020, 21, 241. [Google Scholar] [CrossRef] [PubMed]
- Wei, R.; Yan, Y.-H.; Harris, A.J.; Kang, J.-S.; Shen, H.; Xiang, Q.-P.; Zhang, X.-C. Plastid phylogenomics resolve deep relationships among eupolypod II ferns with rapid radiation and rate heterogeneity. Genome Biol. Evol. 2017, 9, 1646–1657. [Google Scholar] [CrossRef]
- Tillich, M.; Lehwark, P.; Pellizzer, T.; Ulbricht-Jones, E.S.; Fischer, A.; Bock, R.; Greiner, S. GeSeq–versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 2017, 45, W6–W11. [Google Scholar] [CrossRef] [PubMed]
- Shi, L.; Chen, H.; Jiang, M.; Wang, L.; Wu, X.; Huang, L.; Liu, C. CPGAVAS2, an integrated plastome sequence annotator and analyzer. Nucleic Acids Res. 2019, 47, W65–W73. [Google Scholar] [CrossRef]
- Chan, P.-P.; Lowe, T.M. tRNAscan-SE: Searching for tRNA genes in genomic sequences. Methods Mol. Biol. 2019, 1962, 1–14. [Google Scholar]
- Miao, B.-B.; Dong, W.; Gu, Y.-X.; Han, Z.-F.; Luo, X.; Ke, C.-H.; You, W.-W. OmicsSuite: A customized and pipelined suite for analysis and visualization of multi-omics big data. Hortic. Res. 2023, 10, uhad195. [Google Scholar] [CrossRef]
- Du, X.-Y.; Lu, J.-M.; Zhang, L.-B.; Wen, J.; Kuo, L.-Y.; Mynsseng, C.M.; Schneider, H.; Li, D.-Z. Simultaneous diversification of Polypodiales and angiosperms in the Mesozoic. Cladistics 2021, 37, 518–539. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.F.; von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, L.T.; Schmidt, H.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef] [PubMed]
- Rambaut, A. FigTree-Version 1.4.3, A Graphical Viewer of Phylogenetic Trees. 2017. Available online: http://tree.bio.ed.ac.uk/software/figtree/ (accessed on 5 May 2023).
- Ronquist, F.; Teslenko, M.; van der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef]
- Minh, B.Q.; Hahn, M.W.; Lanfear, R. New methods to calculate concordance factors for phylogenomic datasets. Mol. Biol. Evol. 2020, 37, 2727–2733. [Google Scholar] [CrossRef]
- Mo, Y.-K.; Lanfear, R.; Hahn, M.W.; Minh, B.Q. Updated site concordance factors minimize effects of homoplasy and taxon sampling. Bioinformatics 2023, 39, btac741. [Google Scholar] [CrossRef]
- Maurin, J.L.K. An empirical guide for producing a dated phylogeny with treePL in a maximum likelihood framework. arXiv 2020, arXiv:2008.07054v2. [Google Scholar]
- Smith, S.A.; O’Meara, B.C. treePL: Divergence time estimation using penalized likelihood for large phylogenies. Bioinformatics 2012, 28, 2689–2690. [Google Scholar] [CrossRef]
- Drummond, A.J.; Suchard, M.A.; Xie, D.; Rambaut, A. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol. Biol. Evol. 2012, 29, 1969–1973. [Google Scholar] [CrossRef]
- Chen, F.; Deng, S.-H.; Sun, K.-Q. Early Cretaceous Athyrium Roth from northeastern China. Palaeobotanist 1997, 46, 117–133. [Google Scholar] [CrossRef]
- Upchurch, G.R.; Mack, G.H. Latest Cretaceous leaf megafloras from the Jose Creek Member, McRae Formation of New Mexico. In Fall Field Conference Guidebook-49: Las Cruces Country II; Mack, G.H., Austin, G.S., Barker, J.M., Eds.; New Mexico Geological Society: Socorro, NM, USA, 1998; pp. 209–222. [Google Scholar]
- Wilf, P.; Beard, K.C.; Daviesvollum, K.S.; Norejko, J.W. Portrait of a late Paleocene (early Clarkforkian) terrestrial ecosystem; big multi quarry and associated strata, Washakie Basin, southwestern Wyoming. Palaios 1998, 13, 514–532. [Google Scholar] [CrossRef]
- Schuettpelz, E.; Pryer, K.M. Evidence for a Cenozoic radiation of ferns in an angiosperm-dominated canopy. Proc. Natl. Acad. Sci. USA 2009, 106, 11200–11205. [Google Scholar] [CrossRef] [PubMed]
- Qi, X.-P.; Kuo, L.-Y.; Guo, C.; Li, H.; Li, Z.-Y.; Qi, J.; Wang, L.; Hu, Y.; Xiang, J.; Zhang, C.; et al. A well-resolved fern nuclear phylogeny reveals the evolution history of numerous transcription factor families. Mol. Phylogenet. Evol. 2018, 127, 961–977. [Google Scholar] [CrossRef] [PubMed]
- Pelosi, J.A.; Kim, E.H.; Barbazuk, W.B.; Sessa, E.B. Phylotranscriptomics illuminates the placement of whole genome duplications and gene retention in ferns. Front. Plant Sci. 2022, 13, 882441. [Google Scholar] [CrossRef]
- Ballard, F. A new fern genus from Mexico and Guatemala. Am. Fern J. 1945, 35, 1–3. [Google Scholar] [CrossRef]
- Kato, M. Woodsiaceae. In Flora of Japan, Pteridophyta and Gymnospermae; Iwatsuki, K., Yamazaki, T., Boufford, D.E., Ohba, H., Eds.; Kodansha Ltd.: Tokyo, Japan, 1995; Volume 1, pp. 195–231. [Google Scholar]
- GBIF.org. GBIF Occurrence Download. Available online: https://www.gbif.org/occurrence/download/0015267-231120084113126 (accessed on 1 December 2023).
- Matzke, N.J. BioGeoBEARS: BioGeography with Bayesian (and Likelihood) Evolutionary Analysis with R Scripts, Version 1.1.1. 2018. Available online: https://github.com/nmatzke/BioGeoBEARS (accessed on 6 November 2018).
- Buerki, S.; Forest, F.; Alvarez, N.; Nylander, J.A.A.; Arrigo, N.; Sanmartın, I. An evaluation of new parsimony-based versus parametric inference methods in biogeography: A case study using the globally distributed plant family Sapindaceae. J. Biogeogr. 2011, 38, 531–550. [Google Scholar] [CrossRef]
- Wei, R.; Xiang, Q.-P.; Schneider, H.; Sundue, M.A.; Kessler, M.; Kamau, P.W.; Hidayat, A.; Zhang, X.-C. Eurasian origin, boreotropical migration and transoceanic dispersal in the pantropical fern genus Diplazium (Athyriaceae). J. Biogeogr. 2015, 42, 1809–1819. [Google Scholar] [CrossRef]
- Matzke, N.J. Model selection in historical biogeography reveals that founder-event speciation is a crucial process in Island clades. Syst. Biol. 2014, 63, 951–970. [Google Scholar] [CrossRef]
- Yu, Y.; Blair, C.; He, X.-J. RASP 4: Ancestral state reconstruction tool for multiple genes and characters. Mol. Biol. Evol. 2020, 37, 604–606. [Google Scholar] [CrossRef]
- Matzke, N.J. Stochastic Mapping Under Biogeographical Models. 2015. Available online: http://phylo.wikidot.com/biogeobears#stochastic_mapping (accessed on 1 June 2015).
- Kuo, L.-Y.; Qi, X.; Ma, H.; Li, F.-W. Order-level fern plastome phylogenomics: New insights from Hymenophyllales. Am. J. Bot. 2018, 105, 1545–1555. [Google Scholar] [CrossRef]
- Mower, J.P.; Ma, P.-F.; Grewe, F.; Taylor, A.; Michael, T.P.; VanBuren, R.; Qiu, Y.-L. Lycophyte plastid genomics: Extreme variation in GC, gene and intron content and multiple inversions between a direct and inverted orientation of the rRNA repeat. New Phytol. 2019, 222, 1061–1075. [Google Scholar] [CrossRef]
- Du, X.-Y.; Kuo, L.-Y.; Zuo, Z.-Y.; Li, D.-Z.; Lu, J.-M. Structural variation of plastomes provides key insight into the deep phylogeny of ferns. Front. Plant Sci. 2022, 13, 862772. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.-M.; Zhao, J.; Yang, J.-J.; Le Péchonc, T.; Zhang, L.; He, Z.-R.; Zhang, L.-B. Plastome structure, evolution, and phylogeny of Selaginella. Mol. Phylogenet. Evol. 2022, 169, 107410. [Google Scholar] [CrossRef]
- Grewe, F.; Guo, W.H.; Gubbels, E.A.; Hansen, A.K.; Mower, J.P. Complete plastid genomes from Ophioglossum californicum, Psilotum nudum, and Equisetum hyemale reveal an ancestral land plant genome structure and resolve the position of Equisetales among monilophytes. BMC Evol. Biol. 2013, 13, 8. [Google Scholar] [CrossRef]
- Kim, H.T.; Chung, M.G.; Kim, K.J. Chloroplast genome evolution in early diverged leptosporangiate ferns. Mol. Cell. 2014, 37, 372–382. [Google Scholar] [CrossRef] [PubMed]
- Wolf, P.G.; Rowe, C.A.; Sinclair, R.B.; Hasebe, M. Complete nucleotide sequence of the chloroplast genome from a leptosporangiate fern, Adiantum capillus-veneris L. DNA Res. 2003, 10, 59–65. [Google Scholar] [CrossRef]
- Wolf, P.G.; Roper, J.M.; Duffy, A.M. The evolution of chloroplast genome structure in ferns. Genome 2010, 53, 731–738. [Google Scholar] [CrossRef]
- Lu, J.-M.; Zhang, N.; Du, X.-Y.; Wen, J.; Li, D.-Z. Chloroplast phylogenomics resolves key relationships in ferns. J. Syst. Evol. 2015, 53, 448–457. [Google Scholar] [CrossRef]
- Zhao, J.; Zhou, X.-M.; Fang, S.-L.; Zhu, Z.-M.; Li, Y.-X.; Yu, H.; He, Z.-R. Transcriptome-based study on the phylogeny and hybridization of Marattialean ferns (Marattiaceae). Plants 2023, 12, 2237. [Google Scholar] [CrossRef]
- Jiang, L.-J.; Zhao, J.; Wang, J.-G.; Landrein, S.; Shi, J.-P.; Huang, C.-J.; Luo, M.; Zhou, X.-M.; Niu, H.-B.; He, Z.-R. Deciphering the evolution and biogeography of ant-ferns Lecanopteris s.s. Mol. Phylogenet. Evol. 2024, 201, 108199. [Google Scholar] [CrossRef]
- Maximowicz, C.J. Primitiae florae Amuresis; Saraswati Press: St. Petersburg, Russia, 1859. [Google Scholar]
- Testo, W.; Sundue, M. A 4000-species dataset provides new insight into the evolution of ferns. Mol. Phylogenet. Evol. 2016, 105, 200–211. [Google Scholar] [CrossRef]
- Berry, K. Fern macroflora from the basal Danian of the Raton Formation supports palynological signal of a widespread “postdisaster” fern flora. Int. J. Plant Sci. 2023, 184, 1–13. [Google Scholar] [CrossRef]
- Ali, J.R.; Aitchison, J.C. Gondwana to Asia: Plate tectonics, paleogeography and the biological connectivity of the Indian subcontinent from the Middle Jurassic through latest Eocene (166–35 Ma). Earth-Sci. Rev. 2008, 88, 145–166. [Google Scholar] [CrossRef]
- Rothwell, G.W.; Stockey, R.A. Onoclea sensibilis in the Paleocene of North America, a dramatic example of structural and ecological stasis. Rev. Palaeobot. Palynol. 1991, 70, 113–124. [Google Scholar] [CrossRef]
- Fox, R.C. The succession of Paleocene mammals in western Canada. In Dawn of the Age of Mammals in the Northern Part of the Rocky Mountain Interior, North America; Brown, T.M., Rose, K.D., Eds.; Geological Society of America: Boulder, CO, USA, 1990; p. 243. [Google Scholar]
- Johnson, K. Megaflora of the Hell Creek Formation and lower Fort Union Formations in the western Dakotas: Vegetational response to climate change, the Cretaceous-Tertiary boundary event, and rapid marine transgression. In The Hell Creek Formation and the Cretaceous-Tertiary Boundary in the Northern Great Plains: An Integrated Continental Record of the End of the Cretaceous; Hartman, J., Johnson, K.R., Nichols, D.J., Eds.; Geological Society of America: Boulder, CO, USA, 2002; pp. 329–391. [Google Scholar]
- Wilf, P.; Johnson, K.R.; Huber, B.T. Correlated terrestrial and marine evidence for global climate changes before mass extinction at the Cretaceous-Paleogene boundary. Proc. Natl. Acad. Sci. USA 2003, 100, 599–604. [Google Scholar] [CrossRef] [PubMed]
- Aitchison, J.C.; Ali, J.R.; Davis, A.M. When and where did India and Asia collide? J. Geophys. Res. 2007, 112, B05423. [Google Scholar] [CrossRef]
- Wei, X.-P.; Qi, Y.-D.; Zhang, X.-C.; Luo, L.; Shang, H.; Wei, R.; Liu, H.-T.; Zhang, B.-G. Phylogeny, historical biogeography and characters evolution of the drought resistant fern Pyrrosia Mirbel (Polypodiaceae) inferred from plastid and nuclear markers. Sci. Rep. 2017, 7, 12757. [Google Scholar] [CrossRef]
- Zhao, J.; Huang, C.-J.; Jiang, L.-J.; He, Z.-R.; Yang, S.; Zhu, Z.-M.; Zhang, L.; Yu, H.; Zhou, X.-M.; Wang, J.-G. Phylogenomic analyses of the pantropical Platycerium Desv. (Platycerioideae) reveal their complex evolution and historical biogeography. Mol. Phylogenet. Evol. 2024, 201, 108213. [Google Scholar] [CrossRef]
- Davis, C.C.; Bell, C.D.; Mathews, S.; Donoghue, M.J. Laurasian migration explains Gondwanan disjunctions: Evidence from Malpighiaceae. Proc. Natl. Acad. Sci. USA 2002, 99, 6833–6837. [Google Scholar] [CrossRef]
- Testo, W.L.; de Gasper, A.L.; Molino, S.; Galán, J.M.G.Y.; Salino, A.; Dittrich, V.A.O.; Sessa, E.B. Deep vicariance and frequent transoceanic dispersal shape the evolutionary history of a globally distributed fern family. Am. J. Bot. 2022, 109, 1579–1595. [Google Scholar] [CrossRef]
- Zachos, J.; Pagani, M.; Sloan, L.; Thomas, E.; Billups, K. Trends, rhythms, and aberrations in global climate 65 Ma to present. Science 2001, 292, 686–693. [Google Scholar] [CrossRef]
- Morley, R.J. Cretaceous and Tertiary climate change and the past distribution of megathermal rainforests. In Tropical Rainforest Responses to Climatic Change; Springer Praxis Books; Bush, M.B., Flenley, J.R., Eds.; Springer: Berlin/Heidelberg, Germany, 2007; pp. 1–31. [Google Scholar]
- Wen, J. Evolution of eastern Asian and eastern North American disjunct distributions in flowering plants. Ann. Rev. Ecol. Evol. Syst. 1999, 30, 421–455. [Google Scholar] [CrossRef]
- Tiffney, B.H.; Manchester, S.R. The use of geological and paleontological evidence in evaluating plant phylogeographic hypotheses in the northern hemisphere Tertiary. Int. J. Plant Sci. 2001, 162, S3–S17. [Google Scholar] [CrossRef]
- Wen, J.; Ickert-Bond, S.; Nie, Z.-L.; Li, R. Timing and modes of evolution of eastern Asian–North American biogeographic disjunctions in seed plants. In Darwin′s Heritage Today: Proceedings of the Darwin 200 Beijing International Conference; Long, M., Gu, H., Zhou, Z., Eds.; Higher Education Press: Beijing, China, 2010; pp. 252–269. [Google Scholar]


| Genus | Matteuccia | Onoclea |
|---|---|---|
| Rhizomes | erect | creeping |
| Lamina | dimorphic | dimorphic |
| Trophophylls veins | open pinnate, free | reticulate |
| Chromosome number | x = 39, 40 | x = 37 |
| Glandular hairs of gametophytes | absent | present |
| Distribution | Disjunctly circumboreal | North America and Eastern Asia |
| Genus | Onoclepsis | Pentarhizidium |
| Rhizomes | erect | creeping |
| Lamina | dimorphic | dimorphic |
| Trophophylls veins | reticulate | open pinnate, free |
| Chromosome number | x = 40 | x = 40, 41 |
| Glandular hairs of gametophytes | absent | present |
| Distribution | Southern Mexico and Guatemala | Asiatic |
| Model | LnL | Numparams | d | e | j | AICc | AICc_wt |
|---|---|---|---|---|---|---|---|
| DEC | −14.12 | 2 | 0.018 | 0.05 | 0 | 36.24 | 0.2 |
| DEC + J | −13.04 | 3 | 0.013 | 0.037 | 0.23 | 44.07 | 0.004 |
| DIVALIKE | −13.22 | 2 | 0.016 | 0.04 | 0 | 34.45 | 0.49 |
| DIVALIKE + J | −12.89 | 3 | 0.015 | 0.045 | 0.28 | 43.77 | 0.0046 |
| BAYAREALIKE | −13.71 | 2 | 0.54 | 2.38 | 0 | 35.41 | 0.3 |
| BAYAREALIKE + J | −13.67 | 3 | 0.56 | 2.35 | 0.96 | 45.33 | 0.0021 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, J.; Wang, J.-G.; Hu, Y.-P.; Huang, C.-J.; Fang, S.-L.; Wan, Z.-Y.; Li, R.-J.; Yu, H.; He, Z.-R.; Zhou, X.-M. Phylogenetic Inferences and Historical Biogeography of Onocleaceae. Plants 2025, 14, 510. https://doi.org/10.3390/plants14040510
Zhao J, Wang J-G, Hu Y-P, Huang C-J, Fang S-L, Wan Z-Y, Li R-J, Yu H, He Z-R, Zhou X-M. Phylogenetic Inferences and Historical Biogeography of Onocleaceae. Plants. 2025; 14(4):510. https://doi.org/10.3390/plants14040510
Chicago/Turabian StyleZhao, Jing, Jia-Guan Wang, Yu-Ping Hu, Chuan-Jie Huang, Shao-Li Fang, Zi-Yue Wan, Rong-Juan Li, Hong Yu, Zhao-Rong He, and Xin-Mao Zhou. 2025. "Phylogenetic Inferences and Historical Biogeography of Onocleaceae" Plants 14, no. 4: 510. https://doi.org/10.3390/plants14040510
APA StyleZhao, J., Wang, J.-G., Hu, Y.-P., Huang, C.-J., Fang, S.-L., Wan, Z.-Y., Li, R.-J., Yu, H., He, Z.-R., & Zhou, X.-M. (2025). Phylogenetic Inferences and Historical Biogeography of Onocleaceae. Plants, 14(4), 510. https://doi.org/10.3390/plants14040510






