Development and Application of Pik Locus-Specific Molecular Markers for Blast Resistance Genes in Yunnan Japonica Rice Cultivars
Abstract
:1. Introduction
2. Results
2.1. Development and Identification of Resistance Gene-Specific Molecular Markers
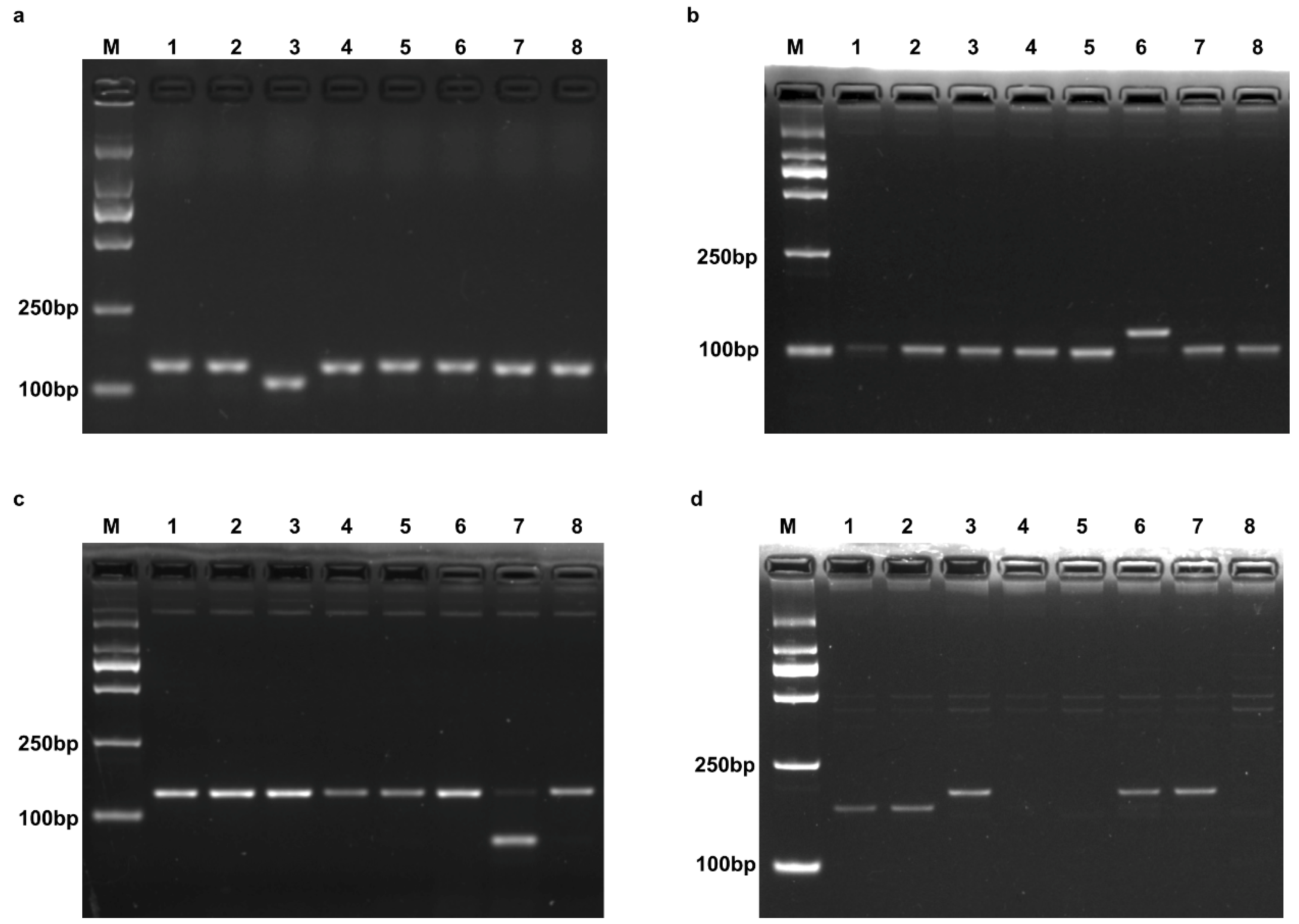
2.1.1. Pik
2.1.2. Pi1
2.1.3. Pikm
2.1.4. Piks
2.1.5. Pikp and Pikh
2.1.6. A Novel Haplotype of Pik Locus
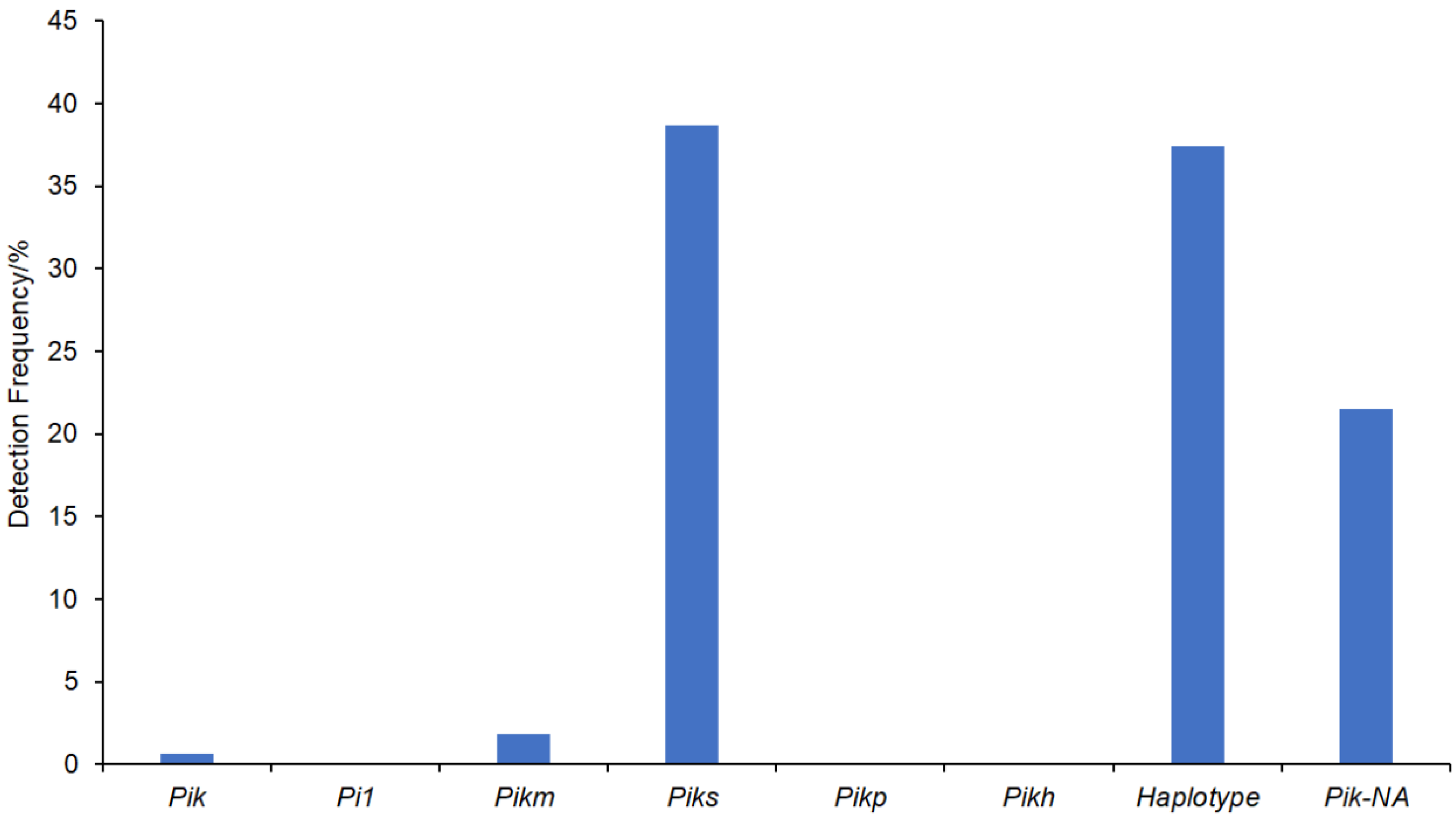
2.2. Determination of 163 Japonica Rice Cultivars with Pik Allele-Specific Molecular Markers
3. Discussion
4. Materials and Methods
4.1. Rice Cultivars
4.2. Development of Gene-Specific Molecular Markers
4.3. Genomic DNA Extraction
4.4. PCR Amplification and Electrophoresis
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Khush, G.S. What it will take to Feed 5.0 Billion Rice consumers in 2030. Plant Mol. Biol. 2005, 59, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Couch, B.C.; Kohn, L.M. A multilocus gene genealogy concordant with host preference indicates segregation of a new species, Magnaporthe oryzae, from M. grisea. Mycologia 2002, 94, 683–693. [Google Scholar] [CrossRef] [PubMed]
- Zhang, N.; Luo, J.; Rossman, A.Y.; Aoki, T.; Chuma, I.; Crous, P.W.; Dean, R.; de Vries, R.P.; Donofrio, N.; Hyde, K.D.; et al. Generic names in Magnaporthales. IMA Fungus 2016, 7, 155–159. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Wang, X.; Mitchell, T.; Hu, Y.; Liu, X.; Dai, L.; Wang, G.-L. Recent progress and understanding of the molecular mechanisms of the rice-Magnaporthe oryzae interaction. Mol. Plant Pathol. 2010, 11, 419–427. [Google Scholar] [CrossRef]
- Nalley, L.; Tsiboe, F.; Durand-Morat, A.; Shew, A.; Thoma, G. Economic and environmental impact of rice blast pathogen (Magnaporthe oryzae) alleviation in the United States. PLoS ONE 2016, 11, e0167295. [Google Scholar] [CrossRef]
- Dean, R.; Van Kan, J.A.; Pretorius, Z.A.; Hammond-Kosack, K.E.; Di Pietro, A.; Spanu, P.D.; Rudd, J.J.; Dickman, M.; Kahmann, R.; Ellis, J.; et al. The Top 10 fungal pathogens in molecular plant pathology. Mol. Plant Pathol. 2012, 13, 414–430. [Google Scholar] [CrossRef]
- Sharma, T.R.; Rai, A.K.; Gupta, S.K.; Vijayan, J.; Devanna, B.N.; Ray, S. Rice blast management through host-plant resistance: Retrospect and prospects. Agric. Res. 2012, 1, 37–52. [Google Scholar] [CrossRef]
- Savary, S.; Willocquet, L.; Pethybridge, S.J.; Esker, P.; McRoberts, N.; Nelson, A. The global burden of pathogens and pests on major food crops. Nat. Ecol. Evol. 2019, 3, 430–439. [Google Scholar] [CrossRef]
- Ashkani, S.; Rafii, M.Y.; Shabanimofrad, M.; Miah, G.; Sahebi, M.; Azizi, P.; Tanweer, F.A.; Akhtar, M.S.; Nasehi, A. Molecular breeding strategy and challenges towards improvement of blast disease resistance in rice crop. Front. Plant Sci. 2015, 6, 886. [Google Scholar] [CrossRef]
- Kalia, S.; Rathour, R. Current status on mapping of genes for resistance to leaf- and neck-blast disease in rice. 3 Biotech 2019, 9, 209. [Google Scholar] [CrossRef]
- Yang, T.; Song, L.; Hu, J.; Qiao, L.; Yu, Q.; Wang, Z.; Chen, X.; Lu, G.D. Magnaporthe oryzae effector AvrPik-D targets a transcription factor WG7 to suppress rice immunity. Rice 2024, 17, 14. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Qiu, J.; Shen, Z.; Wang, C.; Jiang, N.; Shi, H.; Kou, Y. The E3 ubiquitin ligase OsRGLG5 targeted by the Magnaporthe oryzae effector AvrPi9 confers basal resistance against rice blast. Plant Commun. 2023, 4, 100626. [Google Scholar] [CrossRef] [PubMed]
- Devanna, B.N.; Jain, P.; Solanke, A.U.; Das, A.; Thakur, S.; Singh, P.K.; Kumari, M.; Dubey, H.; Jaswal, R.; Pawar, D.; et al. Understanding the dynamics of blast resistance in rice-Magnaporthe oryzae interactions. J. Fungi 2022, 8, 584. [Google Scholar] [CrossRef] [PubMed]
- Fukuoka, S.; Saka, N.; Koga, H.; Ono, K.; Shimizu, T.; Ebana, K.; Hayashi, N.; Takahashi, A.; Hirochika, H.; Okuno, K.; et al. Loss of function of a proline-containing protein confers durable disease resistance in rice. Science 2009, 325, 998–1001. [Google Scholar] [CrossRef]
- He, X.; Liu, X.; Wang, L.; Wang, L.; Lin, F.; Cheng, Y.; Chen, Z.; Liao, Y.; Pan, Q. Identification of the novel recessive gene pi55(t) conferring resistance to Magnaporthe oryzae. Sci. China Life Sci. 2012, 55, 141–149. [Google Scholar] [CrossRef]
- Zhao, H.; Wang, X.; Jia, Y.; Minkenberg, B.; Wheatley, M.; Fan, J.; Jia, M.H.; Famoso, A.; Edwards, J.D.; Wamishe, Y.; et al. The rice blast resistance gene Ptr encodes an atypical protein required for broad-spectrum disease resistance. Nat. Commun. 2018, 9, 2039. [Google Scholar] [CrossRef]
- Chen, X.; Shang, J.; Chen, D.; Lei, C.; Zou, Y.; Zhai, W.; Liu, G.; Xu, J.; Ling, Z.; Cao, G.; et al. A B-lectin receptor kinase gene conferring rice blast resistance. Plant J. 2006, 46, 794–804. [Google Scholar] [CrossRef]
- Zhang, M.; Wang, S.; Yuan, M. An update on molecular mechanism of disease resistance genes and their application for genetic improvement of rice. Mol. Breed. 2019, 39, 154. [Google Scholar] [CrossRef]
- Tsunematsu, H.; Yanoria, M.J.T.; Ebron, L.A.; Hayashi, N.; Ando, I.; Kato, H.; Imbe, T.; Khush, G.S. Development of monogenic lines of rice for blast resistance. Breed. Sci. 2000, 50, 229–234. [Google Scholar] [CrossRef]
- Fang, W.W.; Liu, C.C.; Zhang, H.W.; Xu, H.; Zhou, S.; Fang, K.X.; Peng, Y.L.; Zhao, W.S. Selection of differential isolates of Magnaporthe oryzae for postulation of blast resistance genes. Phytopathology 2018, 108, 878–884. [Google Scholar] [CrossRef]
- Michelmore, R.W.; Paran, I.; Kesseli, R.V. Identification of markers linked to disease-resistance genes by bulked segregant analysis: A rapid method to detect markers in specific genomic regions by using segregating populations. Proc. Natl. Acad. Sci. USA 1991, 88, 9828–9832. [Google Scholar] [CrossRef]
- Jena, K.K.; Mackill, D.J. Molecular markers and their use in marker-assisted selection in rice. Crop Sci. 2008, 48, 1266–1276. [Google Scholar] [CrossRef]
- Li, J.; Zhang, H.; Yang, R.; Zeng, Q.; Han, G.; Du, Y.; Yang, J.; Yang, G.; Luo, Q. Identification of miRNAs contributing to the broad-spectrum and durable blast resistance in the Yunnan local rice germplasm. Front. Plant Sci. 2021, 12, 749919. [Google Scholar] [CrossRef] [PubMed]
- Zhai, C.; Lin, F.; Dong, Z.; He, X.; Yuan, B.; Zeng, X.; Wang, L.; Pan, Q. The isolation and characterization of Pik, a rice blast resistance gene which emerged after rice domestication. N. Phytol. 2011, 189, 321–334. [Google Scholar] [CrossRef]
- Hua, L.; Wu, J.; Chen, C.; Wu, W.; He, X.; Lin, F.; Wang, L.; Ashikawa, I.; Matsumoto, T.; Wang, L.; et al. The isolation of Pi1, an allele at the Pik locus which confers broad spectrum resistance to rice blast. Theor. Appl. Genet. 2012, 125, 1047–1055. [Google Scholar] [CrossRef]
- Ashikawa, I.; Hayashi, N.; Yamane, H.; Kanamori, H.; Wu, J.; Matsumoto, T.; Ono, K.; Yano, M. Two adjacent nucleotide-binding site-leucine-rich repeat class genes are required to confer Pikm-specific rice blast resistance. Genetics 2008, 180, 2267–2276. [Google Scholar] [CrossRef]
- Kiyosawa, S. Inheritance of blast resistance of a U.S. rice variety, Dawn. Jpn. J. Breed. 1974, 24, 117–124. [Google Scholar] [CrossRef]
- Fjellstrom, R.; Conaway-Bormans, C.A.; McClung, A.M.; Marchetti, M.A.; Shank, A.R.; Park, W.D. Development of DNA markers suitable for marker assisted selection of three Pi genes conferring resistance to multiple Pyricularia grisea pathotypes. Crop Sci. 2004, 44, 1790–1798. [Google Scholar] [CrossRef]
- Yuan, B.; Zhai, C.; Wang, W.; Zeng, X.; Xu, X.; Hu, H.; Lin, F.; Wang, L.; Pan, Q. The Pik-p resistance to Magnaporthe oryzae in rice is mediated by a pair of closely linked CC-NBS-LRR genes. Theor. Appl. Genetic. 2011, 122, 1017–1028. [Google Scholar] [CrossRef]
- Zhai, C.; Zhang, Y.; Yao, N.; Lin, F.; Liu, Z.; Dong, Z.; Wang, L.; Pan, Q. Function and interaction of the coupled genes responsible for Pik-h encoded rice blast resistance. PLoS ONE 2014, 9, e98067. [Google Scholar] [CrossRef]
- Chen, J.; Peng, P.; Tian, J.; He, Y.; Zhang, L.; Liu, Z.; Yin, D.; Zhang, Z. Pike, a rice blast resistance allele consisting of two adjacent NBS–LRR genes, was identified as a novel allele at the Pik locus. Mol. Biol. 2015, 35, 117. [Google Scholar] [CrossRef]
- Costanzo, S.; Jia, Y. Sequence variation at the rice blast resistance gene Pi-km locus: Implications for the development of allele specific markers. Plant Sci. 2010, 178, 523–530. [Google Scholar] [CrossRef]
- Sahu, P.K.; Sao, R.; Choudhary, D.K.; Thada, A.; Kumar, V.; Mondal, S.; Das, B.K.; Jankuloski, L.; Sharma, D. Advancement in the breeding, biotechnological and genomic tools towards development of durable genetic resistance against the rice blast disease. Plants 2022, 11, 2386. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.P.; Li, X.; Wang, C.Y.; Li, X.H.; He, Y.Q. Improvement of resistance to rice blast in Zhenshan 97 by molecular marker-aided selection. Acta Agron. Sin. 2003, 45, 1346–1350. [Google Scholar]
- Liu, W.; Liu, J.; Triplett, L.; Leach, J.E.; Wang, G.L. Novel insights into rice innate immunity against bacterial and fungal pathogens. Annu. Rev. Phytopathol. 2014, 52, 213–241. [Google Scholar] [CrossRef]
- Saleh, D.; Milazzo, J.; Adreit, H.; Fournier, E.; Tharreau, D. South-East Asia is the center of origin, diversity and dispersion of the rice blast fungus, Magnaporthe oryzae. N. Phytol. 2014, 201, 1440–1456. [Google Scholar] [CrossRef]
- Saleh, D.; Xu, P.; Shen, Y.; Li, C.Y.; Adreit, H.; Milazzo, J.; Ravigné, V.; Bazin, E.; Nottéghem, J.L.; Fournier, E.; et al. Sex at the origin: An Asian population of the rice blast fungus Magnaporthe oryzae reproduces sexually. Mol. Ecol. 2012, 21, 1330–1344. [Google Scholar] [CrossRef]
- Pink, D.A.C. Strategies using genes for non-durable disease resistance. Euphytica 2002, 124, 227–236. [Google Scholar] [CrossRef]
- Dong, L.; Liu, S.; Tian, W.; Zhou, W.; Zhang, X.; Li, X.; Yang, Q. Pathogenicity and mapping type of rice blast fungus Magnaporthe oryzae isolates in Yunnan Province. J. Plant Prot. 2023, 50, 316–324, (In Chinese with English Summary). [Google Scholar]
- Maksup, S.; Supaibulwatana, K.; Selvaraj, G. High-quality reference genes for quantifying the transcriptional responses of Oryza sativa L. (ssp. indica and japonica) to abiotic stress conditions. Chin. Sci. Bull. 2013, 58, 1919–1930. [Google Scholar]
- Neff, M.M.; Turk, E.; Kalishman, M. Web-based primer design for single nucleotide polymophism analysis. TRENDS Genet. 2002, 18, 613–615. [Google Scholar] [CrossRef] [PubMed]
- Warude, D.; Chavan, P.; Joshi, K.; Patwardhan, B. DNA isolation from fresh, dry plant samples with highly acidic tissue extracts. Plant Mol. Biol. Rep. 2003, 21, 467. [Google Scholar] [CrossRef]



| Target Gene | Marker Name | Sequence (5′-3′) | Fragment Length/bp | Enzyme Site | Annealing Temperature/°C | Marker Type |
|---|---|---|---|---|---|---|
| Pik | Pik-Fw | TCAGTACTCAGTAGTAGTGC | 111 a/133 b | - | 55 | InDel |
| Pik-Rv | TGAGAGAAAATAACCCGCTC | |||||
| Pi1 | Pi1-Fw | CTGTCAACTGATGAAGGC | 120/99 + 21 | Xba I | 55 | dCAPS |
| Pi1-Rv | AGAAAGGATTCTTATCtCtAG | |||||
| Pikm | Pikm-Fw | GCAATGTCATTGGTTGCAAG | 67 + 67/134 | BstN I | 58 | CAPS |
| Pikm-Rv | CCCACCTTCTTCCGGAG | |||||
| Piks | Piks-Fw | GCAATGTCATTGGTTGCAAG | 167 + 22/189 | Nde I | 53 | dCAPS |
| Piks-Rv | CGTTATCTCCTTCACATCTTCaTaT | |||||
| Pikp/Pikh | Pikp/kh-Fw | GTTGCCATGGAGGGCAATAATT | 122/- | - | 60 | dominant |
| Pikp/kh-Rv | CCATAACCGACCACCTCTATCTT | |||||
| TBC-Fw | TGGTCATGTTCCTTCAGCAC | 111 c | - | 60 | internal control | |
| TBC-Rv | GACTTGGCGAGCTTTTGAAC | |||||
| Pikp/kh-Fw1 | AGTTGCTGCAGGTCAGCCAAGCAAt | 51 + 25/76 | Mse I | 60 | dCAPS | |
| Pikp/kh-Rv1 | CATATGGATTTCACCGGCGCAA | |||||
| Piknew-Fw | GGGAGCAGTGAAAACATTGC | 89/80 | - | 55 | InDel | |
| Piknew-Rv | ACAGCAGGGAGATTATCATGC |
| No. | Name | Year of Registration | Pik | Pi1 | Pikm | Piks | Pikp | Pikh | Novel Haplotype |
|---|---|---|---|---|---|---|---|---|---|
| 1 | Chugeng 3 | 1983 | - | - | - | + | - | - | - |
| 2 | Chugeng 4 | 1985 | - | - | + | - | - | - | - |
| 3 | Chugeng 5 | 1986 | - | - | - | - | - | - | + |
| 4 | Chugeng 6 | 1990 | - | - | - | - | - | - | + |
| 5 | Chugeng 7 | 1991 | - | - | - | - | - | - | + |
| 6 | Chugeng 8 | 1990 | - | - | - | - | - | - | + |
| 7 | Chugeng 9 | - | - | - | - | - | - | - | + |
| 8 | Chugeng 13 | - | - | - | - | - | - | - | + |
| 9 | Chugeng 14 | 1995 | - | - | - | - | - | - | + |
| 10 | Chugeng 15 | - | - | - | - | - | - | - | + |
| 11 | Chugeng 17 | 1997 | - | - | - | + | - | - | - |
| 12 | Chugeng 18 | - | - | - | + | - | - | - | - |
| 13 | Chugeng 19 | - | - | - | - | + | - | - | - |
| 14 | Chugeng 21 | - | - | - | - | - | - | - | + |
| 15 | Chugeng 22 | 1999 | - | - | - | - | - | - | + |
| 16 | Chugeng 23 | 1999 | - | - | - | + | - | - | - |
| 17 | Chugeng 25 | 2002 | - | - | - | - | - | - | + |
| 18 | Chugeng 26 | 2005 | - | - | - | - | - | - | + |
| 19 | Chugeng 27 | 2005 | - | - | - | - | - | - | + |
| 20 | Chugeng 28 | 2007 | - | - | - | + | - | - | - |
| 21 | Chugeng 29 | 2007 | - | - | - | + | - | - | - |
| 22 | Chugeng 30 | 2007 | - | - | - | + | - | - | - |
| 23 | Chugeng 31 | 2010 | - | - | - | + | - | - | - |
| 24 | Chugeng 32 | 2011 | - | - | - | + | - | - | - |
| 25 | Chugeng 34 | - | - | - | - | + | - | - | - |
| 26 | Chugeng 35 | - | - | - | - | - | - | - | + |
| 27 | Chugeng 36 | - | - | - | - | + | - | - | - |
| 28 | Chugeng 37 | 2014 | - | - | - | - | - | - | + |
| 29 | Chugeng 38 | 2014 | - | - | - | - | - | - | + |
| 30 | Chugeng 40 | 2015 | - | - | - | - | - | - | + |
| 31 | Chugeng 41 | 2016 | - | - | - | - | - | - | + |
| 32 | Chugeng 42 | 2016 | - | - | - | + | - | - | - |
| 33 | Chugeng 45 | 2017 | - | - | - | - | - | - | + |
| 34 | Chugeng 48 | 2019 | - | - | - | - | - | - | - |
| 35 | Chugeng 53 | - | - | - | - | - | - | - | + |
| 36 | Chugeng 54 | - | - | - | - | + | - | - | - |
| 37 | Hongza 135 | 1989 | - | - | - | - | - | - | - |
| 38 | Yundao 1 | 2005 | - | - | - | + | - | - | - |
| 39 | DJY 5 | 2005 | - | - | - | - | - | - | - |
| 40 | Dian 4 | 2001 | - | - | - | - | - | - | + |
| 41 | Yinguang | 2001 | - | - | - | + | - | - | - |
| 42 | Yunzigeng 41 | 2012 | - | - | - | + | - | - | - |
| 43 | Yungeng 2 | - | - | - | - | + | - | - | - |
| 44 | Yungeng 3 | - | - | - | - | + | - | - | - |
| 45 | Yungeng 4 | 2001 | - | - | - | - | - | - | + |
| 46 | Yungeng 5 | - | - | - | - | + | - | - | - |
| 47 | Yungeng 6 | - | - | - | - | + | - | - | - |
| 48 | Yungeng 7 | - | - | - | - | + | - | - | - |
| 49 | Yungeng 10 | - | - | - | - | + | - | - | - |
| 50 | Yungeng 12 | 2005 | - | - | - | - | - | - | + |
| 51 | Yungeng 14 | - | - | - | - | - | - | - | + |
| 52 | Yungeng 16 | - | - | - | - | - | - | - | - |
| 53 | Yungeng 17 | - | - | - | - | - | - | - | + |
| 54 | Yungeng 18 | - | - | - | - | - | - | - | + |
| 55 | Yungeng 19 | 2010 | - | - | - | - | - | - | + |
| 56 | Yungeng 20 | 2011 | - | - | - | - | - | - | + |
| 57 | Yungeng 21 | - | - | - | - | - | - | - | + |
| 58 | Yungeng 24 | 2007 | - | - | - | + | - | - | - |
| 59 | Yungeng 25 | 2007 | - | - | - | - | - | - | + |
| 60 | Yungeng 26 | 2010 | - | - | - | - | - | - | + |
| 61 | Yungeng 29 | 2011 | - | - | - | - | - | - | + |
| 62 | Yungeng 30 | 2011 | - | - | - | - | - | - | + |
| 63 | Yungeng 31 | 2011 | - | - | - | - | - | - | + |
| 64 | Yungeng 32 | 2011 | - | - | - | - | - | - | + |
| 65 | Yungeng 35 | 2014 | - | - | - | + | - | - | - |
| 66 | Yungeng 37 | - | - | - | - | - | - | - | + |
| 67 | Yungeng 38 | 2014 | - | - | - | - | - | - | + |
| 68 | Yungeng 39 | 2014 | - | - | - | + | - | - | - |
| 69 | Yungeng 42 | 2016 | - | - | - | - | - | - | + |
| 70 | Yungeng 43 | 2016 | - | - | - | - | - | - | + |
| 71 | Yungeng 46 | 2018 | - | - | - | - | - | - | + |
| 72 | Yungeng 48 | - | - | - | - | - | - | - | + |
| 73 | Yungeng 135 | - | - | - | - | + | - | - | - |
| 74 | Yungeng 136 | 1983 | - | - | - | - | - | - | - |
| 75 | Yungengyou 1 | 2004 | - | - | - | + | - | - | - |
| 76 | Yungengyou 5 | 2005 | - | - | - | + | - | - | - |
| 77 | Hexi 1 | - | - | - | - | + | - | - | - |
| 78 | Hexi 2 | 1991 | - | - | - | + | - | - | - |
| 79 | Hexi 3 | - | - | - | - | - | - | - | + |
| 80 | Hexi 4 | 1990 | - | - | - | - | - | - | + |
| 81 | Hexi 5 | 1990 | - | - | - | + | - | - | - |
| 82 | Hexi 6 | - | - | - | - | + | - | - | - |
| 83 | Hexi 8 | - | - | - | - | - | - | - | + |
| 84 | Hexi 9 | - | - | - | - | + | - | - | - |
| 85 | Hexi 10 | 1990 | - | - | - | - | - | - | + |
| 86 | Hexi 12 | - | - | - | - | - | - | - | + |
| 87 | Hexi 13 | - | - | - | - | + | - | - | - |
| 88 | Hexi 14 | - | - | - | - | + | - | - | - |
| 89 | Hexi 15 | 1993 | - | - | - | - | - | - | - |
| 90 | Hexi 16 | - | - | - | - | + | - | - | - |
| 91 | Hexi 17 | - | - | - | - | - | - | - | + |
| 92 | Hexi 20 | - | - | - | - | - | - | - | + |
| 93 | Hexi 22 | 1991 | - | - | - | - | - | - | + |
| 94 | Hexi 23 | - | - | - | - | + | - | - | - |
| 95 | Hexi 24 | 1993 | - | - | - | - | - | - | + |
| 96 | Hexi 25 | 1993 | - | - | - | - | - | - | - |
| 97 | Hexi 28 | - | - | - | - | - | - | - | - |
| 98 | Hexi 30 | 1993 | - | - | - | - | - | - | + |
| 99 | Hexi 32 | - | - | - | - | - | - | - | + |
| 100 | Hexi 34 | 1997 | - | - | - | + | - | - | - |
| 101 | Hexi 35 | 1997 | - | - | - | - | - | - | - |
| 102 | Hexi 38 | - | - | - | - | + | - | - | - |
| 103 | Hexi 40 | 1999 | - | - | - | + | - | - | - |
| 104 | Hexi 41 | 1999 | - | - | - | - | - | - | + |
| 105 | Hexi 42 | 1999 | - | - | - | - | - | - | + |
| 106 | Fengdao 9 | 1997 | - | - | - | - | - | - | - |
| 107 | Fengdao 11 | 1999 | - | - | - | + | - | - | - |
| 108 | Fengdao 12 | - | - | - | - | + | - | - | - |
| 109 | Fengdao 14 | 2001 | - | - | - | - | - | - | - |
| 110 | Fengdao 15 | 2002 | - | - | - | - | - | - | - |
| 111 | Fengdao 16 | 2004 | - | - | - | - | - | - | - |
| 112 | Fengdao 17 | 2003 | - | - | - | + | - | - | - |
| 113 | Fengdao 18 | 2005 | - | - | - | - | - | - | - |
| 114 | Fengdao 19 | 2006 | - | - | - | - | - | - | - |
| 115 | Fengdao 20 | 2006 | - | - | - | - | - | - | - |
| 116 | Fengdao 21 | 2007 | - | - | - | + | - | - | - |
| 117 | Fengdao 23 | 2010 | - | - | - | - | - | - | + |
| 118 | Fengdao 29 | 2014 | - | - | - | - | - | - | - |
| 119 | Fengdao 30 | 2017 | - | - | - | + | - | - | - |
| 120 | Jinggeng 3 | 2005 | - | - | - | - | - | - | - |
| 121 | Jinggeng 6 | 2009 | - | - | - | - | - | - | - |
| 122 | Dianyuyi | 1983 | - | - | - | + | - | - | - |
| 123 | Jinggeng 8 | 2001 | - | - | - | + | - | - | - |
| 124 | Jinggeng 11 | 2007 | - | - | - | + | - | - | - |
| 125 | Jinggeng 12 | 2007 | - | - | - | + | - | - | - |
| 126 | Jinggeng 13 | 2007 | - | - | - | + | - | - | - |
| 127 | Jinggeng 14 | 2007 | - | - | - | + | - | - | - |
| 128 | Jinggeng 16 | 2010 | - | - | - | + | - | - | - |
| 129 | Jinggeng 17 | 2010 | - | - | - | + | - | - | - |
| 130 | Jinggeng 18 | 2010 | - | - | - | + | - | - | - |
| 131 | Jinggeng 26 | 2014 | + | - | - | - | - | - | - |
| 132 | Jingdao 1 | 2018 | - | - | - | - | - | - | - |
| 133 | Jingdao 5 | 2018 | - | - | - | - | - | - | + |
| 134 | Jinggengyou 1 | 2003 | - | - | - | - | - | - | - |
| 135 | Jinggengyou 2 | 2005 | - | - | - | - | - | - | - |
| 136 | Jinggengyou 3 | 2005 | - | - | - | - | - | - | + |
| 137 | Ligeng 4 | - | - | - | - | - | - | - | - |
| 138 | Ligeng 6 | 2004 | - | - | - | - | - | - | - |
| 139 | Ligeng 9 | 2012 | - | - | - | - | - | - | - |
| 140 | Ligeng 10 | 2009 | - | - | - | - | - | - | - |
| 141 | Ligeng 11 | 2010 | - | - | - | - | - | - | - |
| 142 | Ligeng 15 | 2014 | - | - | - | - | - | - | + |
| 143 | Ligeng 18 | 2018 | - | - | - | + | - | - | - |
| 144 | Ligeng 22 | 2019 | - | - | - | + | - | - | - |
| 145 | Ligeng 23 | 2019 | - | - | - | - | - | - | - |
| 146 | Ligeng 314 | 2007 | - | - | - | - | - | - | - |
| 147 | Xiugeng 12 | 2011 | - | - | - | - | - | - | - |
| 148 | Xiugeng 18 | 2015 | - | - | + | - | - | - | - |
| 149 | Xiugeng 22 | 2013 | - | - | - | - | - | - | - |
| 150 | Xiugeng 26 | 2018 | - | - | - | - | - | - | - |
| 151 | Xiugeng 28 | 2018 | - | - | - | + | - | - | - |
| 152 | Xiugeng 29 | 2020 | - | - | - | + | - | - | - |
| 153 | Xiu 87-15 | 2003 | - | - | - | + | - | - | - |
| 154 | Xiu 191-7 | 2011 | - | - | - | - | - | - | - |
| 155 | Changgeng 8 | 2004 | - | - | - | + | - | - | - |
| 156 | Changgeng 9 | 2007 | - | - | - | - | - | - | - |
| 157 | Longke 16 | 2015 | - | - | - | - | - | - | + |
| 158 | Yugeng 24 | 2018 | - | - | - | + | - | - | - |
| 159 | Yugeng 25 | 2019 | - | - | - | - | - | - | + |
| 160 | Tageng 3 | 2014 | - | - | - | + | - | - | - |
| 161 | Niangeng 7 | 1993 | - | - | - | + | - | - | - |
| 162 | Jinrui 4 | 2019 | - | - | - | - | - | - | + |
| 163 | Jinning 78-102 | - | - | - | - | - | - | - | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, P.; Zhou, W.; Dong, L.; Liu, S.; Nawaz, G.; Huang, L.; Yang, Q. Development and Application of Pik Locus-Specific Molecular Markers for Blast Resistance Genes in Yunnan Japonica Rice Cultivars. Plants 2025, 14, 592. https://doi.org/10.3390/plants14040592
Liu P, Zhou W, Dong L, Liu S, Nawaz G, Huang L, Yang Q. Development and Application of Pik Locus-Specific Molecular Markers for Blast Resistance Genes in Yunnan Japonica Rice Cultivars. Plants. 2025; 14(4):592. https://doi.org/10.3390/plants14040592
Chicago/Turabian StyleLiu, Pei, Wumin Zhou, Liying Dong, Shufang Liu, Gul Nawaz, Liyu Huang, and Qinzhong Yang. 2025. "Development and Application of Pik Locus-Specific Molecular Markers for Blast Resistance Genes in Yunnan Japonica Rice Cultivars" Plants 14, no. 4: 592. https://doi.org/10.3390/plants14040592
APA StyleLiu, P., Zhou, W., Dong, L., Liu, S., Nawaz, G., Huang, L., & Yang, Q. (2025). Development and Application of Pik Locus-Specific Molecular Markers for Blast Resistance Genes in Yunnan Japonica Rice Cultivars. Plants, 14(4), 592. https://doi.org/10.3390/plants14040592






