Combining Physiology and Transcriptome to Reveal Mechanisms of Hosta ‘Golden Cadet’ in Response to Alkali Stress
Abstract
:1. Introduction
2. Results
2.1. Screening for Alkali Tolerance Thresholds in Hosta ‘Golden Cadet’
2.2. Effect of NaHCO3 Stress on Osmoregulatory Substances and Antioxidant Activity of Hosta ‘Golden Cadet’
2.3. Principal Component and Correlation Analyses of the Response Pattern of Hosta ‘Golden Cadet’ to Alkali Stress
2.4. Transcriptome Sequencing, Assembly, and Functional Annotation Classification of Unigenes
2.5. Differential Gene Expression Analysis
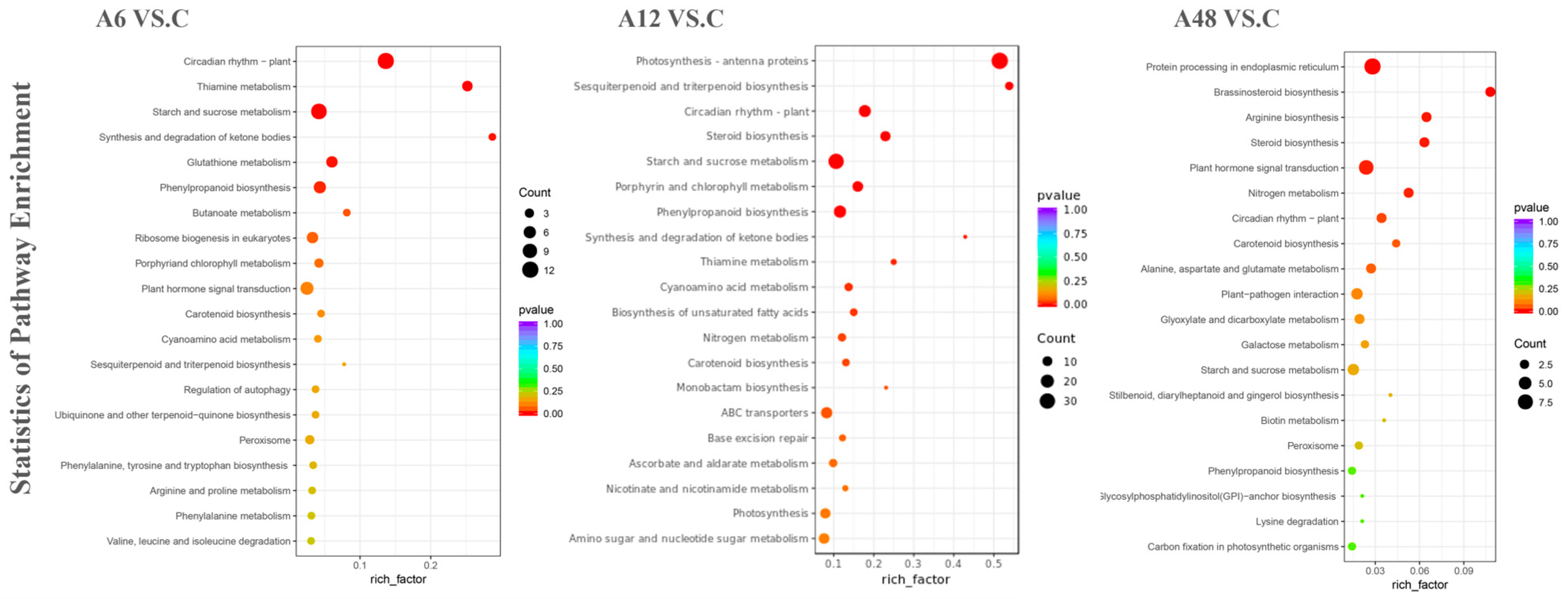
2.6. Functional Annotation and Enrichment Analysis of DEGs
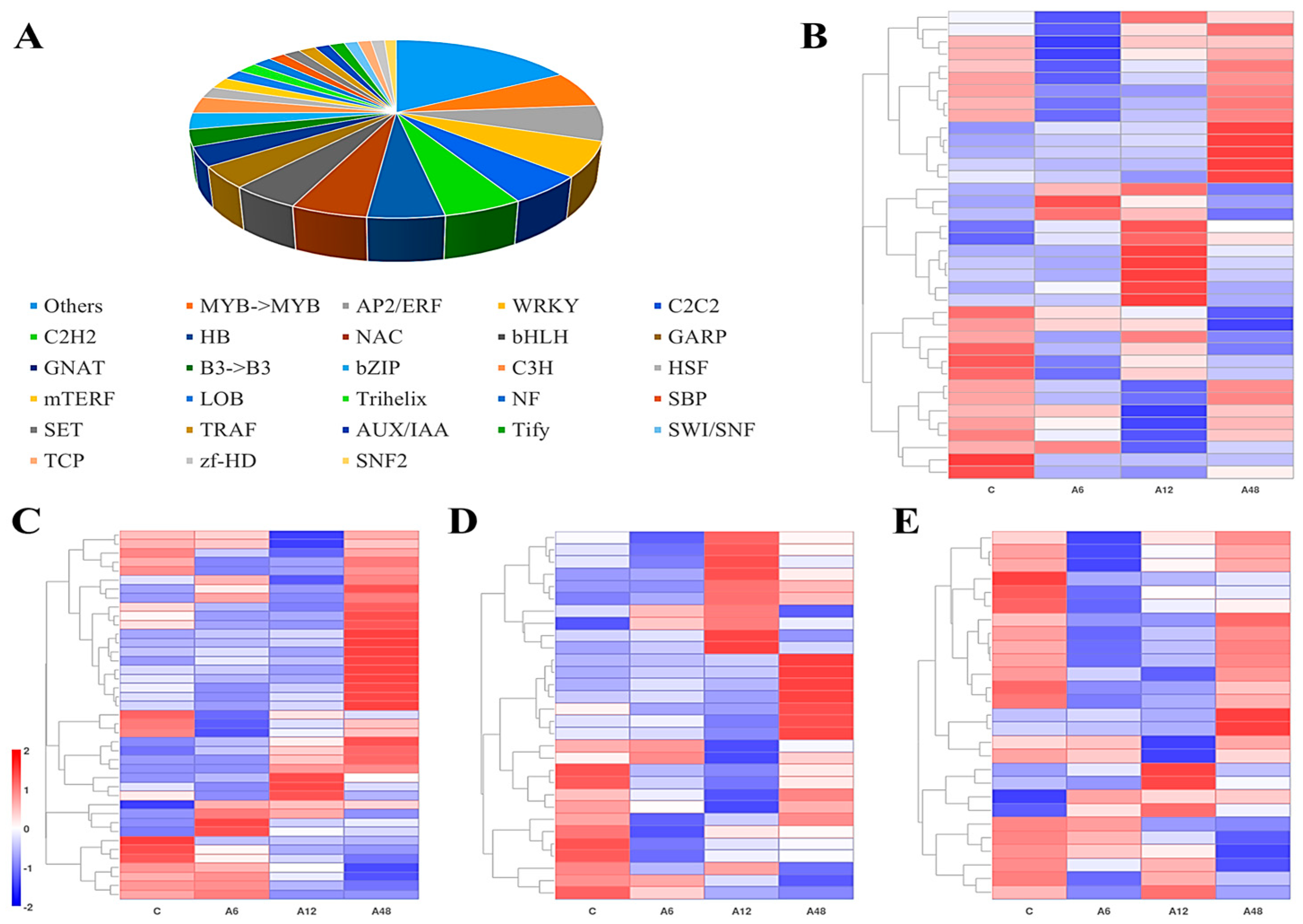
2.7. Potential Transcription Factors of Hosta ‘Golden Cadet ’in Response to NaHCO3 Stress
2.8. Validation of RNA-Seq Data with qRT-PCR
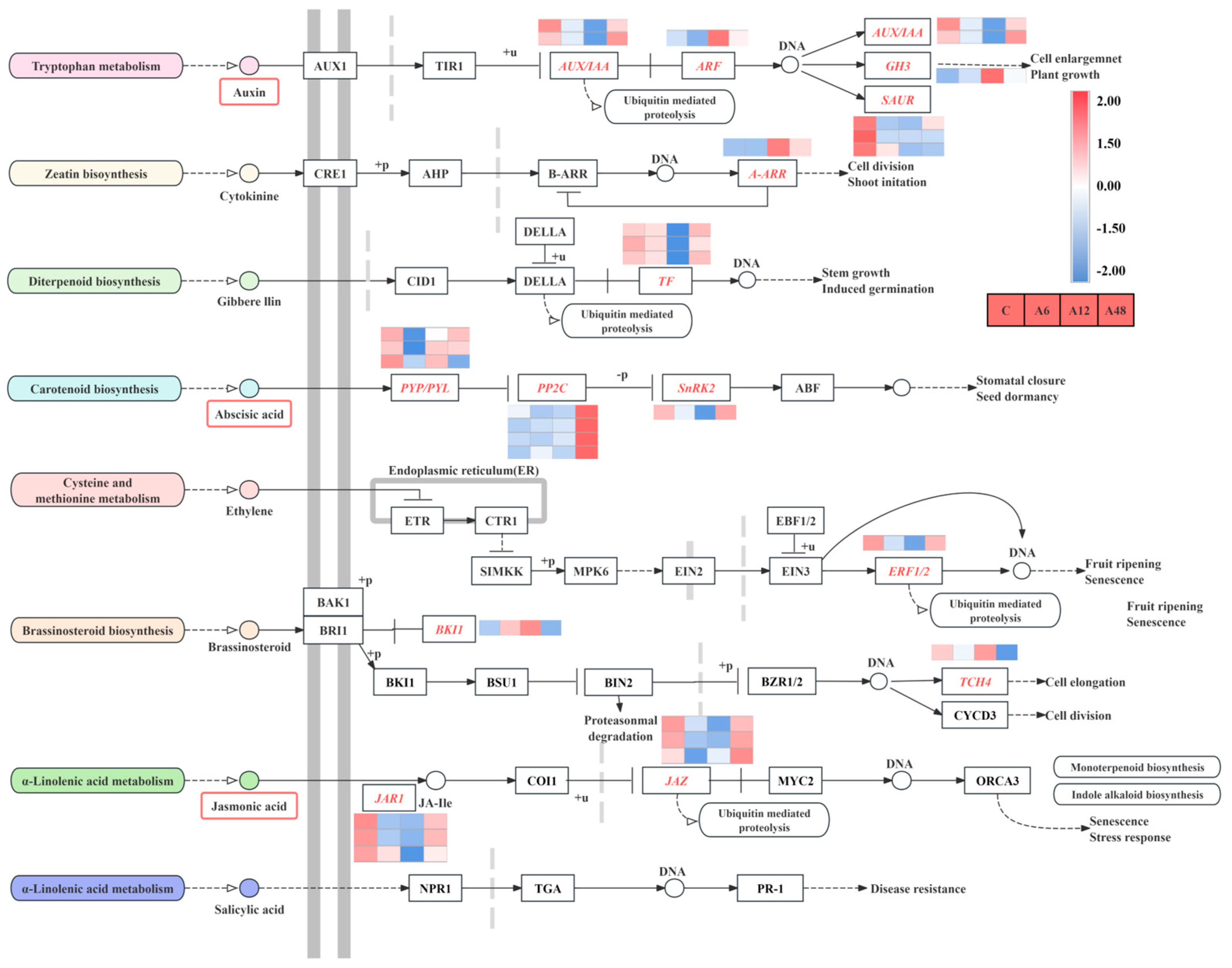
2.9. DEGs Associated with Plant Hormone Signal Transduction
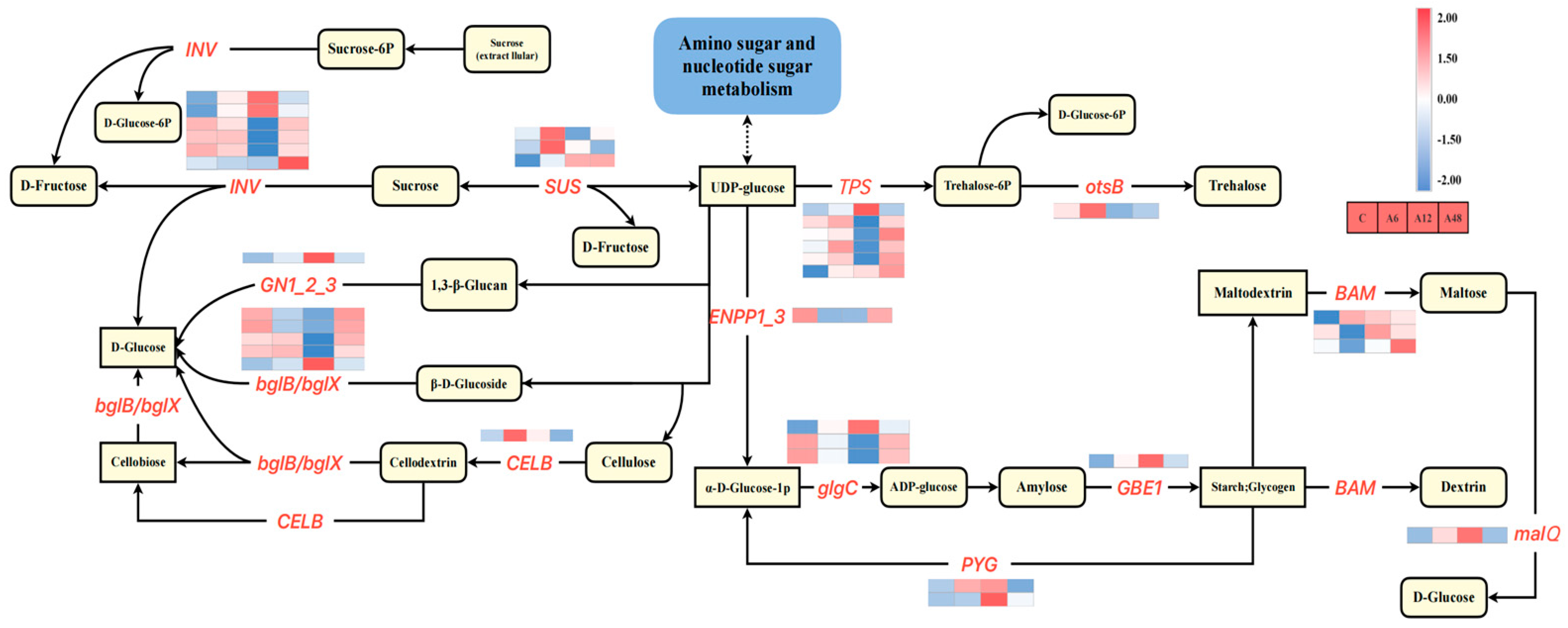
2.10. Starch and Sucrose Metabolism Pathways in Response to Alkali Stress
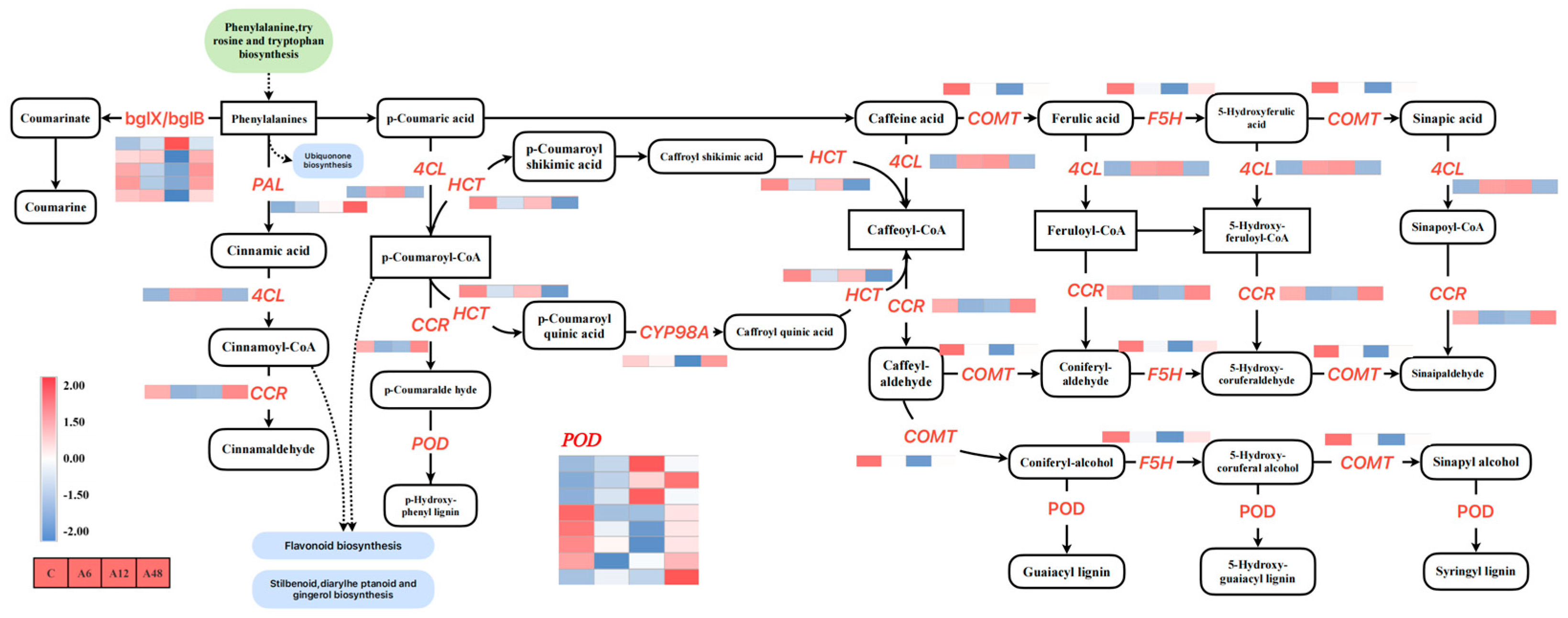
2.11. DEGs Associated with the Phenylpropanoid Biosynthesis Pathway
3. Discussion
3.1. Hosta ‘Golden Cadet’ Seedling Leaves Respond to Alkali Stress by Increasing Osmoregulatory and Antioxidant Capacity
3.2. Hosta ‘Golden Cadet’ Leaves Respond to Alkali Stress by Activating Transcription Factors
3.3. Hosta ‘Golden Cadet’ Responds to Alkali Stress by Activating Phytohormone Signaling
3.4. Hosta ‘Golden Cadet’ Responds to Alkali Stress by Increasing Starch and Sucrose Metabolizing Capacity
3.5. Hosta ‘Golden Cadet’ Responds to Alkali Stress by Regulating Phenylpropane Biosynthesis
4. Materials and Methods
4.1. Experimental Materials and Treatment
4.2. Physiological Trait Determination
4.3. RNA Extraction, Library Construction, and Sequencing
4.4. Analyses of Sequencing Data and Functional Annotation
4.5. Differential Expression and Functional Enrichment Analysis
4.6. Quantitative Real-Time PCR
4.7. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Abbreviations
| AUX/IAA | Auxin-responsive protein IAA |
| ARF | Auxin response factor |
| GH3 | Auxin responsive GH3 gene family |
| SAUR | SAUR family protein |
| ARR-A | Two-component response regulator ARR-A family |
| PLF4 | Phytochrome-interacting factor 4 |
| PYL | Abscisic acid receptor PYR/PYL family |
| PP2C | Protein phosphatase 2C |
| SnRK2 | Serine/Threonine-protein kinase SRK2 |
| ERF1 | Ethylene-responsive transcription factor 1 |
| BKI1 | BRI1 kinase inhibitor 1 |
| TCH4 | Xyloglucan:xyloglucosyl transferase TCH4 |
| JAR1 | Jasmonic acid-amino synthetase |
| JAZ | Jasmonate ZIM domain-containing protein |
| SUS | Sucrose synthase gene |
| bglB/bglX | beta-glucosidase gene |
| ENPP1_3 | Endoglucanase gene |
| TPS | Trehalose 6-phosphate synthase/phosphatase gene |
| PYG | Glycogen phosphorylase gene |
| malQ | 4-alpha-glucanotransferase gene |
| GBE1 | 1,4-alpha-glucan branching enzyme gene |
| BAM | beta-amylase gene |
| INV | beta-fructofuranosidase gene |
| GN1_2_3 | Glucan endo-1,3-beta-glucosidase 1/2/3 |
| glgC | Glucose-1-phosphate adenylyltransferase |
| otsB | Trehalose 6-phosphate phosphatase |
| PAL | Phenylalanine ammonia-lyase gene |
| 4CL | 4-coumarate--CoA ligase gene |
| CCR | Cinnamoyl-CoA reductase gene |
| POD | Peroxidase gene |
| HCT | Shikimate O-hydroxycinnamoyltransferase gene |
| CYP98A | 5-O-(4-coumaroyl)-D-quinate 3′-monooxygenase gene |
| COMT | Caffeic acid 3-O-methyltransferase/Acetylserotonin O-methyltransferase gene |
| CYP84A | Ferulate-5-hydroxylase gene |
References
- Zhang, P.; Dong, Y.; Guo, Y.; Wang, C.; Wang, G.; Ma, Z.; Zhou, W.; Zhang, D.; Ren, Z.; Wang, W. Urban Forest Soil Is Becoming Alkaline under Rapid Urbanization: A Case Study of Changchun, Northeast China. Catena 2023, 224, 106993. [Google Scholar] [CrossRef]
- Litalien, A.; Zeeb, B. Curing the Earth: A Review of Anthropogenic Soil Salinization and Plant-Based Strategies for Sustainable Mitigation. Sci. Total Environ. 2020, 698, 134235. [Google Scholar] [CrossRef] [PubMed]
- García-Caparrós, P.; Lao, M.T. The Effects of Salt Stress on Ornamental Plants and Integrative Cultivation Practices. Sci. Hortic. 2018, 240, 430–439. [Google Scholar] [CrossRef]
- Zhu, Y.; Yu, Q.; Luo, Q.; Zhang, H.; Zhao, J.; Ju, Z.; Du, Y.; Yang, Y. Impacts of Climate Change on Suitability Zonation for Potato Cultivation in Jilin Province, Northeast China. Sci. Rep. 2021, 11, 13103. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Wang, Q.; Lin, S.; Han, Y.; Cheng, S.; Wang, N. Relationship between the shear strength and microscopic pore parameters of saline soil with different freeze-thaw cycles and salinities. Symmetry 2020, 12, 1709. [Google Scholar] [CrossRef]
- Martirosyan, K.V.; Chernyshev, A.B.; Martirosyan, A.V.; Tatyana, K.V. Formation of the Anterior Heating Function under the Action of Uniformly Distributed Sources. In Proceedings of the 2020 IEEE Conference of Russian Young Researchers in Electrical and Electronic Engineering (EIConRus), St. Petersburg and Moscow, Russia, 27–30 January 2020; pp. 1–6. [Google Scholar]
- Sun, Q.; Yang, K.; Sun, Z.; Jia, L.; Tian, H.; Guo, X.; Li, X.; Zhu, W. Characteristics of groundwater quality in Changchun New Area and its evaluation on ecological health. Geol. China 2022, 49, 834–848. [Google Scholar] [CrossRef]
- Subramanian, S.; Ricci, E.; Souleimanov, A.; Smith, D.L. A proteomic approach to lipo-chitooligosaccharide and thuricin 17 effects on soybean germinationunstressed and salt stress. PLoS ONE 2016, 11, e0160660. [Google Scholar] [CrossRef]
- Sakamoto, H.; Matsuda, O.; Iba, K. ITN1, a Novel Gene Encoding an Ankyrin-Repeat Protein That Affects the ABA-Mediated Production of Reactive Oxygen Species and Is Involved in Salt-Stress Tolerance in Arabidopsis Thaliana. Plant J. 2008, 56, 411–422. [Google Scholar] [CrossRef] [PubMed]
- Xu, D.; Do, D.T. Genetic studies on saline and sodic tolerances in soybean. Breed. Sci. 2012, 61, 559–565. [Google Scholar] [CrossRef] [PubMed]
- Guo, R.; Yang, Z.; Li, F.; Yan, C.; Zhong, X.; Liu, Q.; Xia, X.; Li, H.; Zhao, L. Comparative Metabolic Responses and Adaptive Strategies of Wheat (Triticum Aestivum) to Salt and Alkali Stress. BMC Plant Biol. 2015, 15, 170. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Shan, C.; Zhang, Y.; Wang, X.; Tian, H.; Han, G.; Zhang, Y.; Wang, B. Mechanisms of Salt Tolerance and Molecular Breeding of Salt-Tolerant Ornamental Plants. Front. Plant Sci. 2022, 13, 854116. [Google Scholar] [CrossRef]
- Sabra, A.; Daayf, F.; Renault, S. Differential Physiological and Biochemical Responses of Three Echinacea Species to Salinity Stress. Sci. Hortic. 2012, 135, 23–31. [Google Scholar] [CrossRef]
- Acosta-Motos, J.-R.; Diaz-Vivancos, P.; Álvarez, S.; Fernández-García, N.; Sanchez-Blanco, M.J.; Hernández, J.A. Physiological and Biochemical Mechanisms of the Ornamental Eugenia Myrtifolia L. Plants for Coping with NaCl Stress and Recovery. Planta 2015, 242, 829–846. [Google Scholar] [CrossRef]
- Xu, W.; Jia, L.; Shi, W.; Balu¡ka, F.; Kronzucker, H.J.; Liang, J.; Zhang, J. The Tomato 14-3-3 Protein TFT4 Modulates H+ Efflux, Basipetal Auxin Transport, and the PKS5-J3 Pathway in the Root Growth Response to Alkaline Stress. Plant Physiol. 2013, 163, 1817–1828. [Google Scholar] [CrossRef] [PubMed]
- Higuchi, K.; Ono, K.; Araki, S.; Nakamura, S.; Uesugi, T.; Makishima, T.; Ikari, A.; Hanaoka, T.; Sue, M. Elongation of Barley Roots in High-pH Nutrient Solution Is Supported by Both Cell Proliferation and Differentiation in the Root Apex. Plant Cell Environ. 2017, 40, 1609–1617. [Google Scholar] [CrossRef]
- Nefissi Ouertani, R.; Arasappan, D.; Abid, G.; Ben Chikha, M.; Jardak, R.; Mahmoudi, H.; Mejri, S.; Ghorbel, A.; Ruhlman, T.A.; Jansen, R.K. Transcriptomic Analysis of Salt-Stress-Responsive Genes in Barley Roots and Leaves. Int. J. Mol. Sci. 2021, 22, 8155. [Google Scholar] [CrossRef] [PubMed]
- Guo, M.; Li, S.; Tian, S.; Wang, B.; Zhao, X. Transcriptome Analysis of Genes Involved in Defense against Alkaline Stress in Roots of Wild Jujube (Ziziphus Acidojujuba). PLoS ONE 2017, 12, e0185732. [Google Scholar] [CrossRef]
- Guan, Q.; Ma, H.; Wang, Z.; Wang, Z.; Bu, Q.; Liu, S. A Rice LSD1-like-Type ZFP Gene OsLOL5 Enhances Saline-Alkaline Tolerance in Transgenic Arabidopsis Thaliana, Yeast and Rice. BMC Genom. 2016, 17, 142. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; He, J. Traditional Uses, Phytochemistry, Pharmacology and Toxicological Aspects of the Genus Hosta (Liliaceae): A Comprehensive Review. J. Ethnopharmacol. 2021, 265, 113323. [Google Scholar] [CrossRef]
- Cui, P.; Liu, H.; Liu, S.; Chen, L. Research Progress of the Resistance and Cultivation of Hosta Plants. North. Hortic. 2013, 10, 185–189. [Google Scholar]
- Zhu, J.; Cai, Y.; Li, X.; Yang, L.; Zhang, Y. High-Nitrogen Fertilizer Alleviated Adverse Effects of Drought Stress on the Growth and Photosynthetic Characteristics of Hosta ‘Guacamole’. BMC Plant Biol. 2024, 24, 299. [Google Scholar] [CrossRef] [PubMed]
- Cheng, H.C.; Woo, S.Y.; Lee, S.H.; Kwak, M.J.; Kim, K.N. Physiological and Biochemical Responses of Sedum Kamtschaticum and Hosta Longipes to Ozone Stress. Korean J. Hortic. Sci. Technol. 2013, 31, 1–7. [Google Scholar] [CrossRef]
- Zhu, C.; Sun, X.; Sun, H.; Shi, J.; Song, J. Establishment and Optimisation of Tissue Culture Systems for Four Jade Hosta Varieties. North. Hortic. 2024, 49–58. [Google Scholar] [CrossRef]
- Sun, S.; Wang, Y.; Wang, J.; Tan, M.; Yan, H.; So, K.; Zhang, Y. Transcriptome Responses to Salt Stress in Roots and Leaves of Lilium pumilum. Sci. Hortic. 2023, 309, 111622. [Google Scholar] [CrossRef]
- Wang, J.; An, C.; Guo, H.; Yang, X.; Chen, J.; Zong, J.; Li, J.; Liu, J. Physiological and Transcriptomic Analyses Reveal the Mechanisms Underlying the Salt Tolerance of Zoysia Japonica Steud. BMC Plant Biol. 2020, 20, 114. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Rodríguez, P.; Riaño-Pachón, D.M.; Corrêa, L.G.G.; Rensing, S.A.; Kersten, B.; Mueller-Roeber, B. PlnTFDB: Updated Content and New Features of the Plant Transcription Factor Database. Nucleic Acids Res. 2009, 38, D822–D827. [Google Scholar] [CrossRef] [PubMed]
- Fang, X.; Mo, J.; Zhou, H.; Shen, X.; Xie, Y.; Xu, J.; Yang, S. Comparative Transcriptome Analysis of Gene Responses of Salt-Tolerant and Salt-Sensitive Rice Cultivars to Salt Stress. Sci. Rep. 2023, 13, 19065. [Google Scholar] [CrossRef] [PubMed]
- Lima, J.V.; Lobato, A.K.S. Brassinosteroids Improve Photosystem II Efficiency, Gas Exchange, Antioxidant Enzymes and Growth of Cowpea Plants Exposed to Water Deficit. Physiol. Mol. Biol. Plants 2017, 23, 59–72. [Google Scholar] [CrossRef]
- Demidchik, V.; Straltsova, D.; Medvedev, S.S.; Pozhvanov, G.A.; Sokolik, A.; Yurin, V. Stress-Induced Electrolyte Leakage: The Role of K+ Permeable Channels and Involvement in Programmed Cell Death and Metabolic Adjustment. J. Exp. Bot. 2014, 65, 1259–1270. [Google Scholar] [CrossRef] [PubMed]
- Sivankalyani, V.; Sela, N.; Feygenberg, O.; Zemach, H.; Maurer, D.; Alkan, N. Transcriptome Dynamics in Mango Fruit Peel Reveals Mechanisms of Chilling Stress. Front. Plant Sci. 2016, 7, 1579. [Google Scholar] [CrossRef] [PubMed]
- Zhao, D.; Zhang, X.; Fang, Z.; Wu, Y.; Tao, J. Physiological and Transcriptomic Analysis of Tree Peony (Paeonia Section Moutan DC.) in Response to Drought Stress. Forests 2019, 10, 135. [Google Scholar] [CrossRef]
- Gu, R.; Wan, Z.Q.; Tang, F.; Liu, X.T.; Yang, Y.t.; Shi, F. ling Physiological and Transcriptomic Analysis of Salt Tolerant Glaux Maritima Grown under High Saline Condition. Front. Plant Sci. 2023, 14, 1173191. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, P.; Abd_Allah, E.F.; Alyemeni, M.N.; Wijaya, L.; Alam, P.; Bhardwaj, R.; Siddique, K.H.M. Exogenous Application of Calcium to 24-Epibrassinosteroid Pre-Treated Tomato Seedlings Mitigates NaCl Toxicity by Modifying Ascorbate–Glutathione Cycle and Secondary Metabolites. Sci. Rep. 2018, 8, 13515. [Google Scholar] [CrossRef]
- Qiu, N.; Liu, Q.; Li, J.; Zhang, Y.; Wang, F.; Gao, J. Physiological and Transcriptomic Responses of Chinese Cabbage (Brassica Rapa L. Ssp. Pekinensis) to Salt Stress. Int. J. Mol. Sci. 2017, 18, 1953. [Google Scholar] [CrossRef]
- Doganlar, Z.B.; Demir, K.; Basak, H.; Gul, I. Effects of Salt Stress on Pigment and Total Soluble Protein Contents of Three Different Tomato Cultivars. Afr. J. Agric. Res. 2010, 5, 2056–2065. [Google Scholar]
- Wang, R.; Wang, X.; Liu, K.; Zhang, X.-J.; Zhang, L.-Y.; Fan, S.-J. Comparative Transcriptome Analysis of Halophyte Zoysia Macrostachya in Response to Salinity Stress. Plants 2020, 9, 458. [Google Scholar] [CrossRef]
- Wang, J.; Zhang, Y.; Yan, X.; Guo, J. Physiological and transcriptomic analyses of yellow horn (Xanthoceras sorbifolia) provide important insights into salt and saline-alkali stress tolerance. PLoS ONE 2020, 15, e0244365. [Google Scholar] [CrossRef]
- Harb, A.; Krishnan, A.; Ambavaram, M.M.R.; Pereira, A. Molecular and Physiological Analysis of Drought Stress in Arabidopsis Reveals Early Responses Leading to Acclimation in Plant Growth. Plant Physiol. 2010, 154, 1254–1271. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Jin, Y.; Huang, W.; Sun, Q.; Liu, F.; Huang, X. Full-Length Transcriptome Sequences of Ephemeral Plant Arabidopsis Pumila Provides Insight into Gene Expression Dynamics during Continuous Salt Stress. BMC Genom. 2018, 19, 717. [Google Scholar] [CrossRef]
- Panchy, N.L.; Azodi, C.B.; Winship, E.F.; O’Malley, R.C.; Shiu, S.-H. Expression and Regulatory Asymmetry of Retained Arabidopsis Thaliana Transcription Factor Genes Derived from Whole Genome Duplication. BMC Evol. Biol. 2019, 19, 77. [Google Scholar] [CrossRef]
- Golldack, D.; Li, C.; Mohan, H.; Probst, N. Tolerance to Drought and Salt Stress in Plants: Unraveling the Signaling Networks. Front. Plant Sci. 2014, 5, 151. [Google Scholar] [CrossRef] [PubMed]
- Jin, J.; Tian, F.; Yang, D.-C.; Meng, Y.-Q.; Kong, L.; Luo, J.; Gao, G. PlantTFDB 4.0: Toward a Central Hub for Transcription Factors and Regulatory Interactions in Plants. Nucleic Acids Res. 2016, 45, D1040–D1045. [Google Scholar] [CrossRef] [PubMed]
- An, Y.-M.; Song, L.-L.; Liu, Y.-R.; Shu, Y.-J.; Guo, C.-H. De Novo Transcriptional Analysis of Alfalfa in Response to Saline-Alkaline Stress. Front. Plant Sci. 2016, 7, 931. [Google Scholar] [CrossRef] [PubMed]
- Sun, S.; Song, H.; Li, J.; Chen, D.; Tu, M.; Jiang, G.; Yu, G.; Zhou, Z. Comparative Transcriptome Analysis Reveals Gene Expression Differences between Two Peach Cultivars under Saline-Alkaline Stress. Hereditas 2020, 157, 9. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Deyholos, M.K. Functional Characterization of Arabidopsis NaCl-Inducible WRKY25 and WRKY33 Transcription Factors in Abiotic Stresses. Plant Mol. Biol. 2008, 69, 91–105. [Google Scholar] [CrossRef]
- Devaiah, B.N.; Karthikeyan, A.S.; Raghothama, K.G. WRKY75 Transcription Factor Is a Modulator of Phosphate Acquisition and Root Development in Arabidopsis. Plant Physiol. 2007, 143, 1789–1801. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Wang, Y.; Zhang, Y.; Li, C.; Gong, S.; Yan, S.; Li, G.; Hu, G.; Ren, H.; Yang, J.; et al. Comparative Transcriptome Analysis of Salt-Sensitive and Salt-Tolerant Maize Reveals Potential Mechanisms to Enhance Salt Resistance. Genes Genom. 2019, 41, 781–801. [Google Scholar] [CrossRef] [PubMed]
- Xie, Z.; Nolan, T.M.; Jiang, H.; Yin, Y. AP2/ERF Transcription Factor Regulatory Networks in Hormone and Abiotic Stress Responses in Arabidopsis. Front. Plant Sci. 2019, 10, 228. [Google Scholar] [CrossRef]
- Ma, Z.; Hu, L.; Jiang, W. Understanding AP2/ERF Transcription Factor Responses and Tolerance to Various Abiotic Stresses in Plants: A Comprehensive Review. Int. J. Mol. Sci. 2024, 25, 893. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.; Ma, Z.; Hu, L.; Huang, K.; Zhang, M.; Zhang, S.; Jiang, W.; Wu, T.; Du, X. Involvement of Rice Transcription Factor OsERF19 in Response to ABA and Salt Stress Responses. Plant Physiol. Biochem. 2021, 167, 22–30. [Google Scholar] [CrossRef]
- Dubos, C.; Stracke, R.; Grotewold, E.; Weisshaar, B.; Martin, C.; Lepiniec, L. MYB Transcription Factors in Arabidopsis. Trends Plant Sci. 2010, 15, 573–581. [Google Scholar] [CrossRef]
- Liu, X.; Ma, D.; Zhang, Z.; Wang, S.; Du, S.; Deng, X.; Yin, L. Plant Lipid Remodeling in Response to Abiotic Stresses. Environ. Exp. Bot. 2019, 165, 174–184. [Google Scholar] [CrossRef]
- Sukumaran, S.; Lethin, J.; Liu, X.; Pelc, J.; Zeng, P.; Hassan, S.; Aronsson, H. Genome-Wide Analysis of MYB Transcription Factors in the Wheat Genome and Their Roles in Salt Stress Response. Cells 2023, 12, 1431. [Google Scholar] [CrossRef]
- Verma, V.; Ravindran, P.; Kumar, P.P. Plant Hormone-Mediated Regulation of Stress Responses. BMC Plant Biol. 2016, 16, 86. [Google Scholar] [CrossRef]
- Arif, Y.; Singh, P.; Siddiqui, H.; Bajguz, A.; Hayat, S. Salinity Induced Physiological and Biochemical Changes in Plants: An Omic Approach towards Salt Stress Tolerance. Plant Physiol. Biochem. 2020, 156, 64–77. [Google Scholar] [CrossRef] [PubMed]
- Aizaz, M.; Lubna; Jan, R.; Asaf, S.; Bilal, S.; Kim, K.-M.; AL-Harrasi, A. Regulatory Dynamics of Plant Hormones and Transcription Factors under Salt Stress. Biology 2024, 13, 673. [Google Scholar] [CrossRef] [PubMed]
- Finkelstein, R. Abscisic Acid Synthesis and Response. Arab. Book 2013, 11, e0166. [Google Scholar] [CrossRef]
- Tan, W.; Zhang, D.; Zhou, H.; Zheng, T.; Yin, Y.; Lin, H. Transcription Factor HAT1 is a Substrate of SnRK2.3 Kinase and Negatively Regulates ABA Synthesis and Signaling in Arabidopsis Responding to Drought. PLoS Genet. 2018, 14, e1007336. [Google Scholar] [CrossRef] [PubMed]
- Matsuoka, D.; Yasufuku, T.; Furuya, T.; Nanmori, T. An Abscisic Acid Inducible Arabidopsis MAPKKK, MAPKKK18 Regulates Leaf Senescence via Its Kinase Activity. Plant Mol. Biol. 2015, 87, 565–575. [Google Scholar] [CrossRef]
- Thalmann, M.; Santelia, D. Starch as a Determinant of Plant Fitness under Abiotic Stress. New Phytol. 2017, 214, 943–951. [Google Scholar] [CrossRef]
- Yin, J.; Li, A.; Wang, Y.; Li, X.; Ning, W.; Zhou, X.; Liu, J.; Sun, Z. Melatonin Improves Cadmium Tolerance in Salix Viminalis by Enhancing Amino Acid and Saccharide Metabolism, and Increasing Polyphenols. Ecotoxicol. Environ. Saf. 2024, 288, 117393. [Google Scholar] [CrossRef] [PubMed]
- Mathan, J.; Singh, A.; Ranjan, A. Sucrose Transport in Response to Drought and Salt Stress Involves ABA-Mediated Induction of OsSWEET13 and OsSWEET15 in Rice. Physiol. Plant. 2020, 171, 620–637. [Google Scholar] [CrossRef]
- Ghaffari, M.R.; Ghabooli, M.; Khatabi, B.; Hajirezaei, M.R.; Schweizer, P.; Salekdeh, G.H. Metabolic and Transcriptional Response of Central Metabolism Affected by Root Endophytic Fungus Piriformospora Indica under Salinity in Barley. Plant Mol. Biol. 2016, 90, 699–717. [Google Scholar] [CrossRef] [PubMed]
- Cesarino, I. Structural Features and Regulation of Lignin Deposited upon Biotic and Abiotic Stresses. Curr. Opin. Biotechnol. 2019, 56, 209–214. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Wang, Q.; Wang, Y.; Xu, Y.; Li, J.; Zhao, S.; Wang, D.; Ma, Z.; Yan, F.; Liu, Y. Combined Transcriptomic and Metabolomic Analysis Reveals the Role of Phenylpropanoid Biosynthesis Pathway in the Salt Tolerance Process of Sophora alopecuroides. Int. J. Mol. Sci. 2021, 22, 2399. [Google Scholar] [CrossRef] [PubMed]
- Shafi, A.; Chauhan, R.; Gill, T.; Swarnkar, M.K.; Sreenivasulu, Y.; Kumar, S.; Kumar, N.; Shankar, R.; Ahuja, P.S.; Singh, A.K. Expression of SOD and APX Genes Positively Regulates Secondary Cell Wall Biosynthesis and Promotes Plant Growth and Yield in Arabidopsis under Salt Stress. Plant Mol. Biol. 2015, 87, 615–631. [Google Scholar] [CrossRef]
- Pazhany, A.S.; Henry, R.J. Genetic Modification of Biomass to Alter Lignin Content and Structure. Ind. Eng. Chem. Res. 2019, 58, 16190–16203. [Google Scholar] [CrossRef]
- Long, G.; Wu, P.; Fu, J.; Lu, H.; Zhang, R. Research progress on regulation of peroxidase on lignin synthesis. Mod. Agric. Sci. Technol. 2021, 23, 47–54. [Google Scholar]
- Dong, W.; Pan, X.; Su, Y. Effect of planting time and density on four ground cover plants. Sci. Technol. Countrys. 2016, 30, 2. [Google Scholar] [CrossRef]
- Tan, H.; Cai, Y.; Sun, H.; Wang, J.; Wang, G.; Liu, Z. Effects of planting density on sucrose metabolism and activities of enzymes related to starch synthesis in maize hybrids with different plant types. Chin. J. Eco-Agric. 2010, 18, 1183–1188. [Google Scholar]
- Dionisio-Sese, M.L.; Tobita, S. Antioxidant Responses of Rice Seedlings to Salinity Stress. Plant Sci. 1998, 135, 1–9. [Google Scholar] [CrossRef]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Full-Length Transcriptome Assembly from RNA-Seq Data without a Reference Genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef] [PubMed]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor Package for Differential Expression Analysis of Digital Gene Expression Data. Bioinformatics 2009, 26, 139–140. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, X.; Zhu, C.; Li, B.; Ning, W.; Yin, J. Combining Physiology and Transcriptome to Reveal Mechanisms of Hosta ‘Golden Cadet’ in Response to Alkali Stress. Plants 2025, 14, 593. https://doi.org/10.3390/plants14040593
Sun X, Zhu C, Li B, Ning W, Yin J. Combining Physiology and Transcriptome to Reveal Mechanisms of Hosta ‘Golden Cadet’ in Response to Alkali Stress. Plants. 2025; 14(4):593. https://doi.org/10.3390/plants14040593
Chicago/Turabian StyleSun, Xiaogang, Chunyao Zhu, Baizhou Li, Wei Ning, and Jiahui Yin. 2025. "Combining Physiology and Transcriptome to Reveal Mechanisms of Hosta ‘Golden Cadet’ in Response to Alkali Stress" Plants 14, no. 4: 593. https://doi.org/10.3390/plants14040593
APA StyleSun, X., Zhu, C., Li, B., Ning, W., & Yin, J. (2025). Combining Physiology and Transcriptome to Reveal Mechanisms of Hosta ‘Golden Cadet’ in Response to Alkali Stress. Plants, 14(4), 593. https://doi.org/10.3390/plants14040593





