Harnessing Jasmonate Pathways: PgJAR1’s Impact on Ginsenoside Accumulation in Ginseng
Abstract
:1. Introduction
2. Results
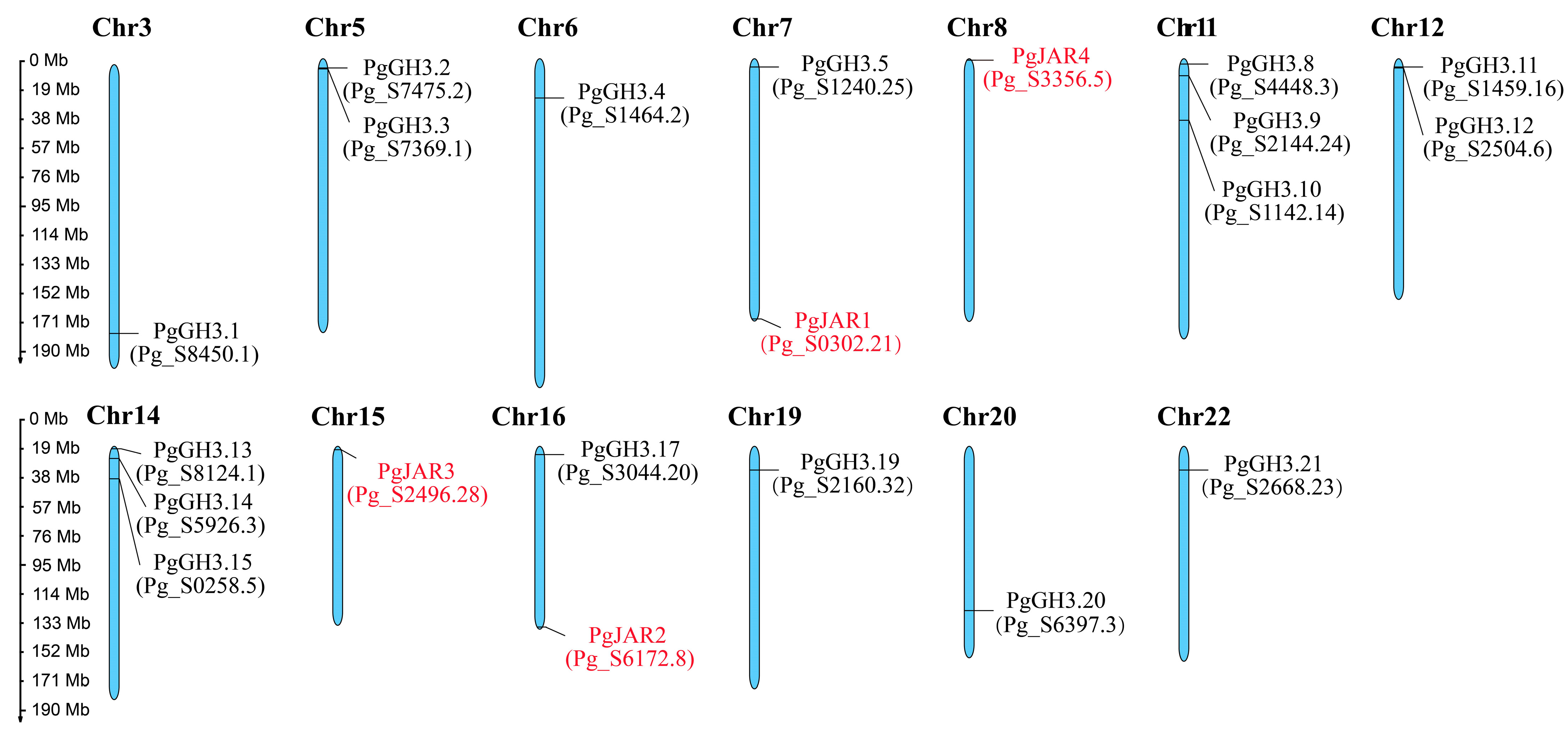
2.1. Chromosomal Distribution Analysis of the GH3 Family in P. ginseng
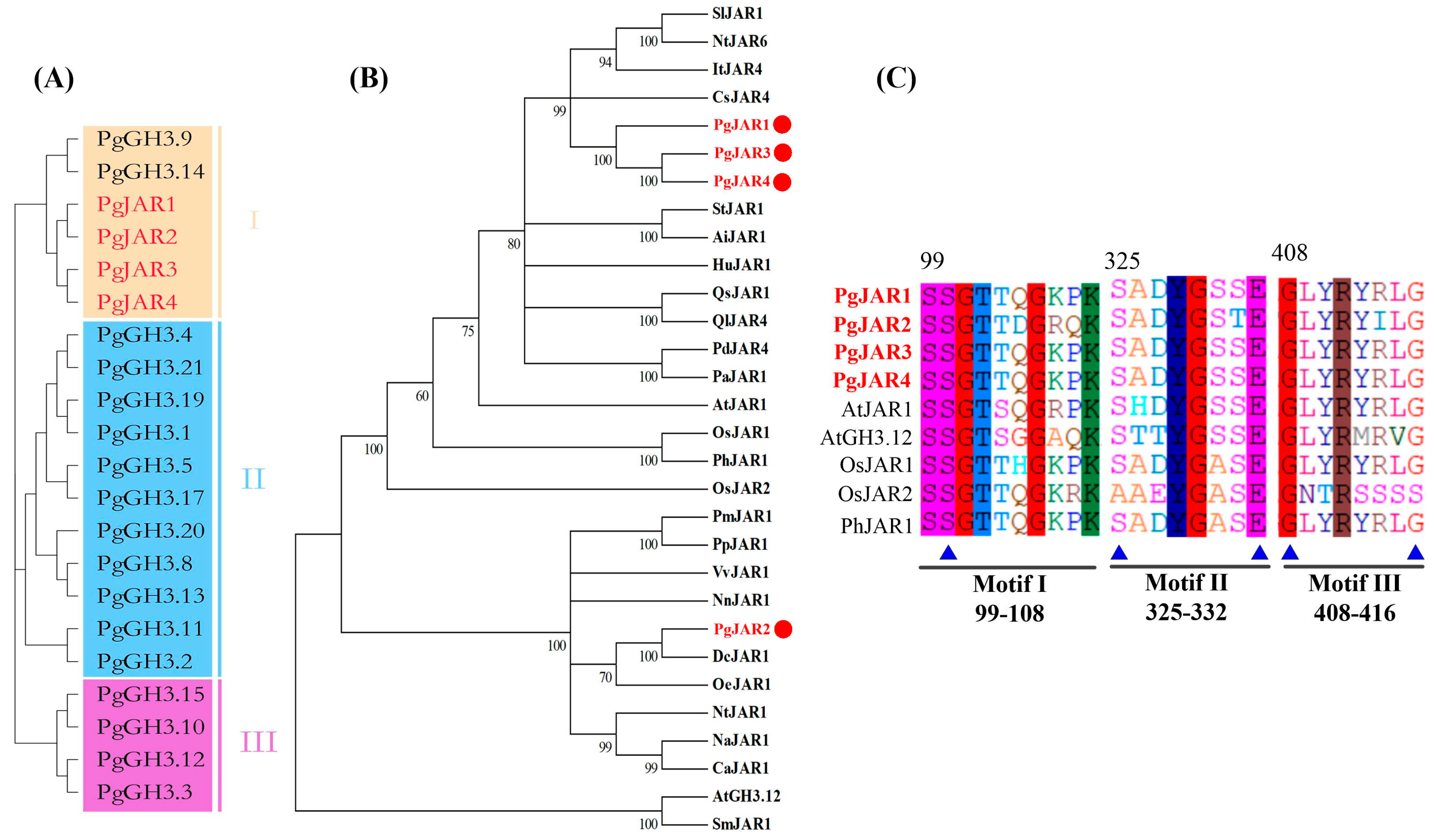
2.2. Cloning of the PgJAR1 Gene from P. ginseng and Its Sequence Analysis
2.3. Characterization of Cis Elements in the PgJAR1 Gene Promoter
2.4. Expression Pattern of PgJAR1 in P. ginseng
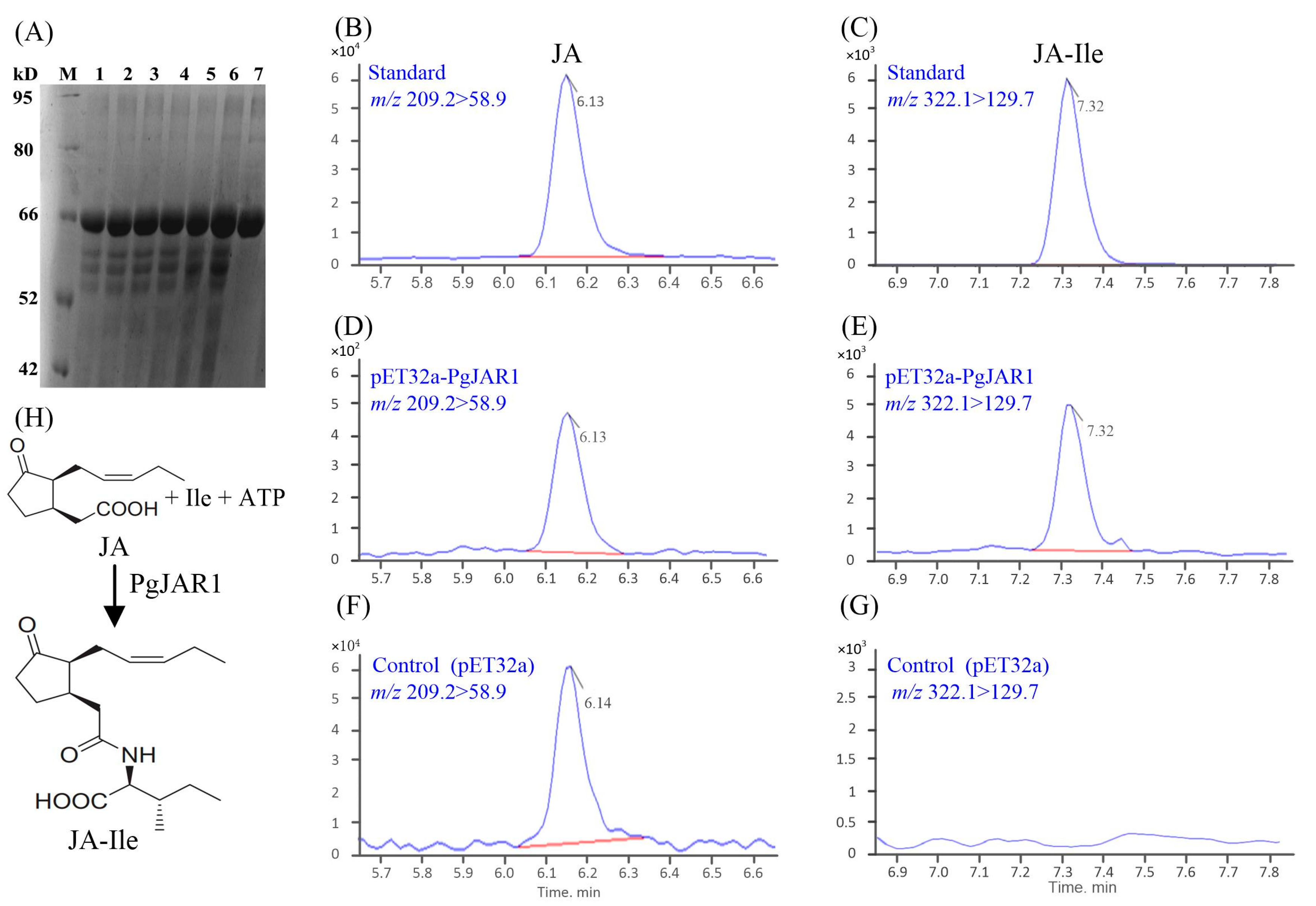
2.5. Enzymatic Production of JA-Ile by PgJAR1
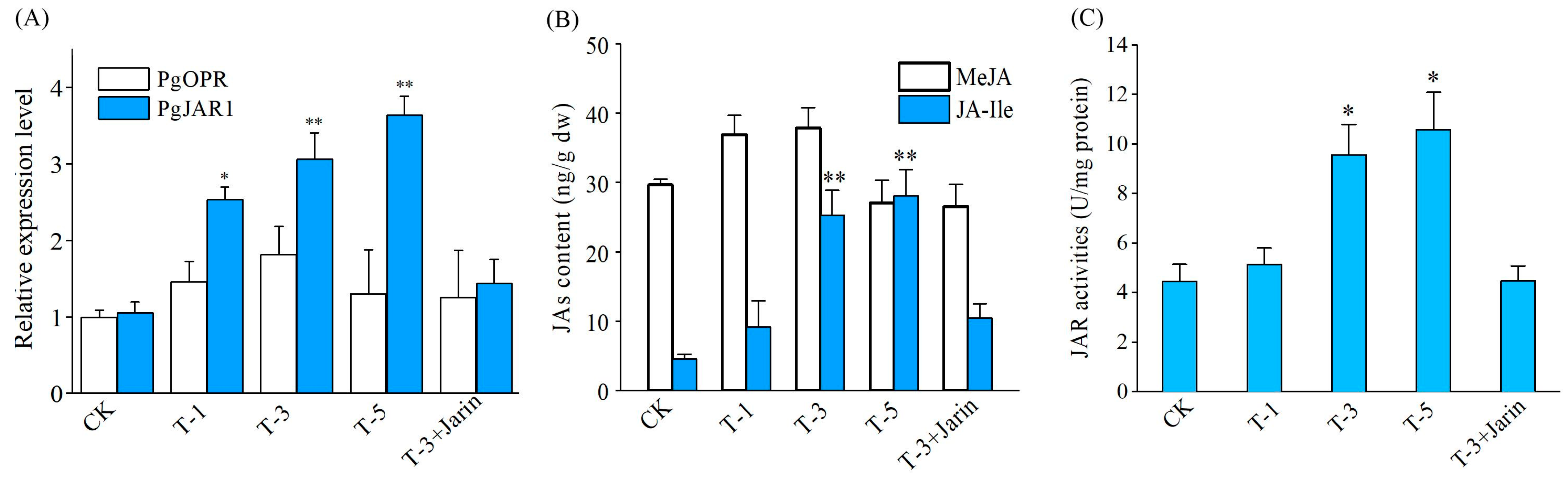
2.6. PgJAR1-Mediated JA Biosynthesis and Biosynthesis Enzyme Gene Expression
2.7. PgJAR1-Mediated PgSS, PgDDS, CYP, and UGT Expression and Ginsenoside Biosynthesis
3. Discussion
4. Materials and Methods
4.1. Chemicals, Materials, and Treatment
4.2. Chromosome Localization Analysis of Ginseng GH3 Genes
4.3. Gene Cloning and Bioinformatic Analysis
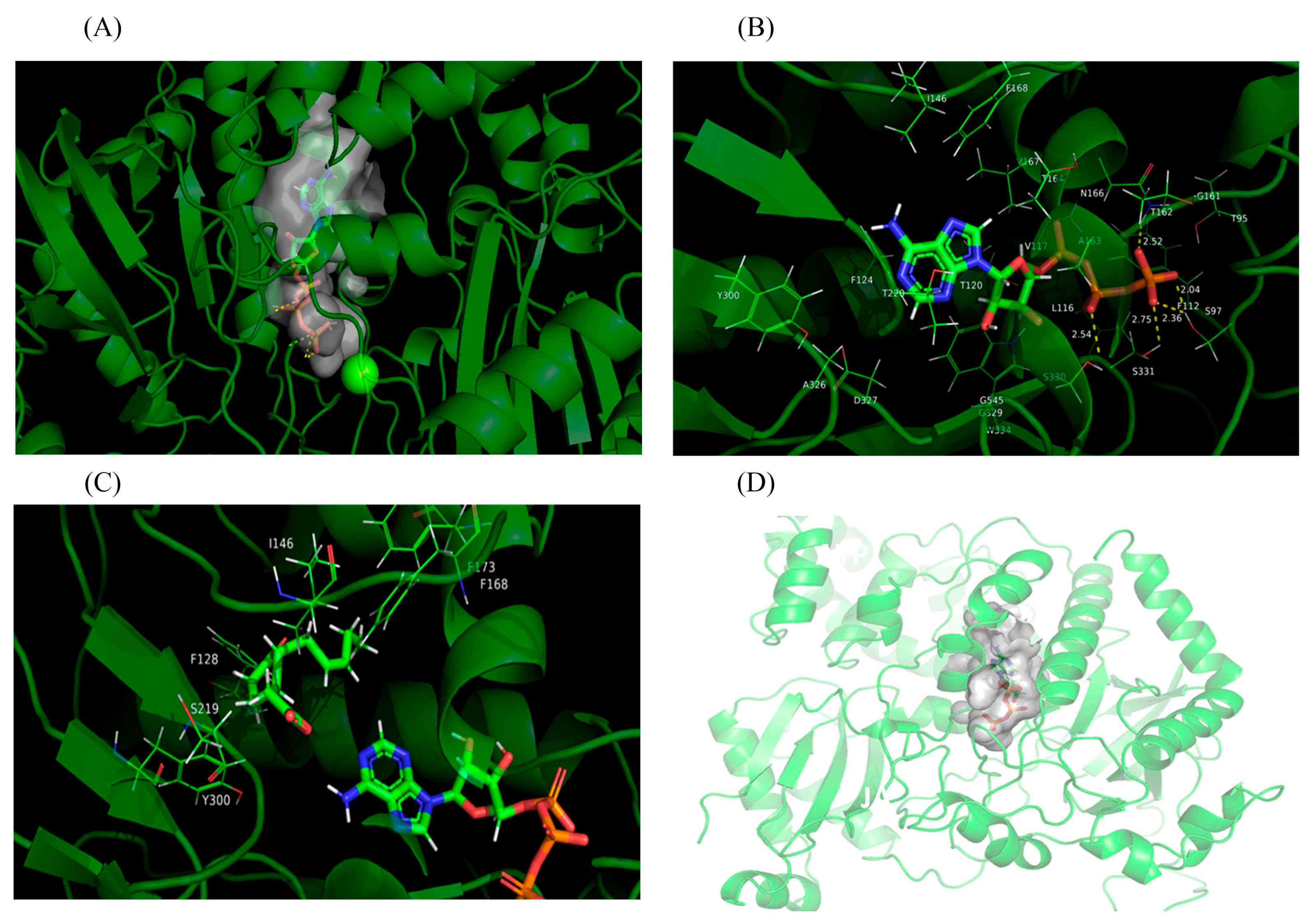
4.4. Homology Modeling and Molecular Docking
4.5. Prokaryotic Expression of PgJAR1 and Enzyme Assay
4.6. Transient Expression Analysis of PgJAR1
4.7. Measurement of JA-Ile and MeJA Levels
4.8. Determination of Ginsenoside Content
4.9. Gene Expression Analysis of Key Enzymes of JA and Ginsenoside Biosynthesis
4.10. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Jung, J.H.; Kim, H.Y.; Kim, H.S.; Jung, S.H. Transcriptome analysis of Panax ginseng response to high light stress. J. Ginseng Res. 2020, 44, 312–320. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Huang, X.M.; Yan, H.J.; Liu, X.Y.; Zhou, Q.; Luo, Z.Y.; Tan, X.N.; Zhang, B.L. Highly Selective Production of Compound K from Ginsenoside Rd by Hydrolyzing Glucose at C-3 Glycoside Using beta-Glucosidase of Bifidobacterium breve ATCC 15700. J. Microbiol. Biotechnol. 2019, 29, 410–418. [Google Scholar] [CrossRef]
- Jiang, T.; Zhang, Y.; Zuo, G.; Luo, T.; Wang, H.; Zhang, R.; Luo, Z. Transcription factor PgNAC72 activates DAMMARENEDIOL SYNTHASE expression to promote ginseng saponin biosynthesis. Plant Physiol. 2024, 195, 2952–2969. [Google Scholar] [CrossRef]
- Kim, H.M.; Song, Y.; Hyun, G.H.; Long, N.P.; Park, J.H.; Hsieh, Y.S.Y.; Kwon, S.W. Characterization and antioxidant activity determination of neutral and acidic polysaccharides from Panax ginseng C. A. Meyer. Molecules 2020, 25, 791. [Google Scholar] [CrossRef] [PubMed]
- Nakhjavani, M.; Smith, E.; Townsend, A.R.; Price, T.J.; Hardingham, J.E. Anti-angiogenic properties of ginsenoside Rg3. Molecules 2020, 25, 4905. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Liu, J.; Deng, J.; Wang, J.; Weng, W.; Chu, H.; Meng, Q. Advances in the chemistry, pharmacological diversity, and metabolism of 20(R)-ginseng saponins. J. Ginseng Res. 2020, 44, 14–23. [Google Scholar] [CrossRef]
- Chen, C.; Lv, Q.; Li, Y.; Jin, Y.H. The anti-tumor effect and underlying apoptotic mechanism of ginsenoside Rk1 and Rg5 in human liver cancer cells. Molecules 2021, 26, 3926. [Google Scholar] [CrossRef]
- Hou, M.; Wang, R.; Zhao, S.; Wang, Z. Ginsenosides in Panax genus and their biosynthesis. Acta Pharm. Sin. B 2021, 11, 1813–1834. [Google Scholar] [CrossRef]
- Jiang, Y.; Liu, S.; Li, L.; Zang, K.; Wang, Y.; Zhao, M.; Wang, K.; Zhu, L.; Chen, P.; Lei, J.; et al. Transcriptome and Phenotype Integrated Analysis Identifies Genes Controlling Ginsenoside Rb1 Biosynthesis and Reveals Their Interactions in the Process in Panax ginseng. Int. J. Mol. Sci. 2022, 23, 14016. [Google Scholar] [CrossRef]
- Zhang, S.; Shen, S.; Ma, P.; Fan, D. Biochemical Targets and Molecular Mechanism of Ginsenoside Compound K in Treating Osteoporosis Based on Network Pharmacology. Int. J. Mol. Sci. 2022, 23, 13921. [Google Scholar] [CrossRef]
- Jang, W.Y.; Hwang, J.Y.; Cho, J.Y. Ginsenosides from Panax ginseng as Key Modulators of NF-kappaB Signaling Are Powerful Anti-Inflammatory and Anticancer Agents. Int. J. Mol. Sci. 2023, 24, 6119. [Google Scholar] [CrossRef] [PubMed]
- Liang, W.; Wang, S.; Yao, L.; Wang, J.; Gao, W. Quality evaluation of Panax ginseng adventitious roots based on ginsenoside constituents, functional genes, and ferric-reducing antioxidant power. J. Food Biochem. 2019, 43, e12901. [Google Scholar] [CrossRef]
- Deng, L.; Luo, L.; Li, Y.; Wang, L.; Zhang, J.; Zi, B.; Ye, C.; Liu, Y.; Huang, H.; Mei, X.; et al. Autotoxic ginsenoside stress induces changes in root exudates to recruit the beneficial Burkholderia strain B36 as revealed by transcriptomic and metabolomic approaches. J. Agric. Food Chem. 2023, 71, 4536–4549. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, R.; Smith, S.A.; Nwangwa, E.E.; Arivett, B.A.; Bryant, D.L.; Fuller, M.L.; Hayes, D.; Bowling, J.L.; Nelson, D.E.; DuBois, J.D.; et al. Panax quinquefolius (North American ginseng) cell suspension culture as a source of bioactive polysaccharides: Immunostimulatory activity and characterization of a neutral polysaccharide AGC1. Int. J. Biol. Macromol. 2019, 139, 221–232. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Qiang, B.; Miao, J.; Phillips, N.; Wei, K. Recent advances in the tissue culture of American ginseng (Panax quinquefolius). Chem. Biodivers 2020, 17, e2000366. [Google Scholar] [CrossRef]
- Xu, P.; Zhao, P.X.; Cai, X.T.; Mao, J.L.; Miao, Z.Q.; Xiang, C.B. Integration of Jasmonic Acid and Ethylene into Auxin Signaling in Root Development. Front. Plant Sci. 2020, 11, 271. [Google Scholar] [CrossRef]
- Yan, J.; Li, S.; Gu, M.; Yao, R.; Li, Y.; Chen, J.; Yang, M.; Tong, J.; Xiao, L.; Nan, F.; et al. Endogenous bioactive jasmonate is composed of a set of (+)-7-iso-JA-amino acid conjugates. Plant Physiol. 2016, 172, 2154–2164. [Google Scholar] [CrossRef]
- Breeze, E. Master MYCs: MYC2, the jasmonate signaling “master switch”. Plant Cell 2019, 31, 9–10. [Google Scholar] [CrossRef]
- Yue, P.; Jiang, Z.; Sun, Q.; Wei, R.; Yin, Y.; Xie, Z.; Larkin, R.M.; Ye, J.; Chai, L.; Deng, X. Jasmonate activates a CsMPK6-CsMYC2 module that regulates the expression of beta-citraurin biosynthetic genes and fruit coloration in orange (Citrus sinensis). Plant Cell 2023, 35, 1167–1185. [Google Scholar] [CrossRef]
- Zhang, F.; Yao, J.; Ke, J.; Zhang, L.; Lam, V.Q.; Xin, X.F.; Zhou, X.E.; Chen, J.; Brunzelle, J.; Griffin, P.R.; et al. Structural basis of JAZ repression of MYC transcription factors in jasmonate signalling. Nature 2015, 525, 269–273. [Google Scholar] [CrossRef]
- Pei, T.; Ma, P.; Ding, K.; Liu, S.; Jia, Y.; Ru, M.; Dong, J.; Liang, Z. SmJAZ8 acts as a core repressor regulating JA-induced biosynthesis of salvianolic acids and tanshinones in Salvia miltiorrhiza hairy roots. J. Exp. Bot. 2018, 69, 1663–1678. [Google Scholar] [CrossRef] [PubMed]
- Shimizu, T.; Miyamoto, K.; Miyamoto, K.; Minami, E.; Nishizawa, Y.; Iino, M.; Nojiri, H.; Yamane, H.; Okada, K. OsJAR1 contributes mainly to biosynthesis of the stress-induced jasmonoyl-isoleucine involved in defense responses in rice. Biosci. Biotechnol. Biochem. 2013, 77, 1556–1564. [Google Scholar] [CrossRef]
- Fukumoto, K.; Alamgir, K.; Yamashita, Y.; Mori, I.C.; Matsuura, H.; Galis, I. Response of rice to insect elicitors and the role of OsJAR1 in wound and herbivory-induced JA-Ile accumulation. J. Integr. Plant Biol. 2013, 55, 775–784. [Google Scholar] [CrossRef] [PubMed]
- Deepika; Singh, A. Expression dynamics indicate the role of Jasmonic acid biosynthesis pathway in regulating macronutrient (N, P and K(+)) deficiency tolerance in rice (Oryza sativa L.). Plant Cell Rep. 2021, 40, 1495–1512. [Google Scholar] [CrossRef]
- Miyamoto, K.; Enda, I.; Okada, T.; Sato, Y.; Watanabe, K.; Sakazawa, T.; Yumoto, E.; Shibata, K.; Asahina, M.; Iino, M.; et al. Jasmonoyl-l-isoleucine is required for the production of a flavonoid phytoalexin but not diterpenoid phytoalexins in ultraviolet-irradiated rice leaves. Biosci. Biotechnol. Biochem. 2016, 80, 1934–1938. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Yu, G.; Cao, C.; Liu, P. Metabolism, signaling, and transport of jasmonates. Plant Commun. 2021, 2, 100231. [Google Scholar] [CrossRef] [PubMed]
- Suza, W.P.; Staswick, P.E. The role of JAR1 in Jasmonoyl-L: -isoleucine production during Arabidopsis wound response. Planta 2008, 227, 1221–1232. [Google Scholar] [CrossRef]
- Kitaoka, N.; Matsubara, T.; Sato, M.; Takahashi, K.; Wakuta, S.; Kawaide, H.; Matsui, H.; Nabeta, K.; Matsuura, H. Arabidopsis CYP94B3 encodes jasmonyl-L-isoleucine 12-hydroxylase, a key enzyme in the oxidative catabolism of jasmonate. Plant Cell Physiol. 2011, 52, 1757–1765. [Google Scholar] [CrossRef]
- Saito, R.; Morikawa, M.; Muto, T.; Saito, S.; Kaji, T.; Ueda, M. SlCYP94B18 and SlCYP94B19 monooxygenases for the catabolic turnover of jasmonates in tomato leaves. Phytochemistry 2024, 223, 114141. [Google Scholar] [CrossRef]
- Bozorov, T.A.; Dinh, S.T.; Baldwin, I.T. JA but not JA-Ile is the cell-nonautonomous signal activating JA mediated systemic defenses to herbivory in Nicotiana attenuata. J. Integr. Plant Biol. 2017, 59, 552–571. [Google Scholar] [CrossRef]
- Basso, V.; Veneault-Fourrey, C. Role of jasmonates in beneficial microbe-root interactions. Methods Mol. Biol. 2020, 2085, 43–67. [Google Scholar] [CrossRef]
- Bottcher, C.; Burbidge, C.A.; di Rienzo, V.; Boss, P.K.; Davies, C. Jasmonic acid-isoleucine formation in grapevine (Vitis vinifera L.) by two enzymes with distinct transcription profiles. J. Integr. Plant Biol. 2015, 57, 618–627. [Google Scholar] [CrossRef] [PubMed]
- Hazman, M.; Suhnel, M.; Schafer, S.; Zumsteg, J.; Lesot, A.; Beltran, F.; Marquis, V.; Herrgott, L.; Miesch, L.; Riemann, M.; et al. Characterization of Jasmonoyl-Isoleucine (JA-Ile) Hormonal Catabolic Pathways in Rice upon Wounding and Salt Stress. Rice 2019, 12, 45. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Cheng, L.; Li, R.; Cai, Y.; Wang, X.; Fu, X.; Dong, X.; Qi, M.; Jiang, C.Z.; Xu, T.; et al. The HD-Zip transcription factor SlHB15A regulates abscission by modulating jasmonoyl-isoleucine biosynthesis. Plant Physiol. 2022, 189, 2396–2412. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Allmann, S.; Wu, J.; Baldwin, I.T. Comparisons of LIPOXYGENASE3- and JASMONATE-RESISTANT4/6-silenced plants reveal that jasmonic acid and jasmonic acid-amino acid conjugates play different roles in herbivore resistance of Nicotiana attenuata. Plant Physiol. 2008, 146, 904–915. [Google Scholar] [CrossRef]
- Yao, L.; Zheng, Y.; Zhu, Z. Jasmonate suppresses seedling soil emergence in Arabidopsis thaliana. Plant Signal Behav. 2017, 12, e1330239. [Google Scholar] [CrossRef]
- Heitz, T.; Widemann, E.; Lugan, R.; Miesch, L.; Ullmann, P.; Desaubry, L.; Holder, E.; Grausem, B.; Kandel, S.; Miesch, M.; et al. Cytochromes P450 CYP94C1 and CYP94B3 catalyze two successive oxidation steps of plant hormone Jasmonoyl-isoleucine for catabolic turnover. J. Biol. Chem. 2012, 287, 6296–6306. [Google Scholar] [CrossRef]
- Westfall, C.S.; Zubieta, C.; Herrmann, J.; Kapp, U.; Nanao, M.H.; Jez, J.M. Structural basis for prereceptor modulation of plant hormones by GH3 proteins. Science 2012, 336, 1708–1711. [Google Scholar] [CrossRef]
- Wasternack, C.; Strnad, M. Jasmonate signaling in plant stress responses and development-active and inactive compounds. New Biotechnol. 2016, 33, 604–613. [Google Scholar] [CrossRef]
- Koo, A.J.; Thireault, C.; Zemelis, S.; Poudel, A.N.; Zhang, T.; Kitaoka, N.; Brandizzi, F.; Matsuura, H.; Howe, G.A. Endoplasmic reticulum-associated inactivation of the hormone jasmonoyl-L-isoleucine by multiple members of the cytochrome P450 94 family in Arabidopsis. J. Biol. Chem. 2014, 289, 29728–29738. [Google Scholar] [CrossRef]
- Widemann, E.; Miesch, L.; Lugan, R.; Holder, E.; Heinrich, C.; Aubert, Y.; Miesch, M.; Pinot, F.; Heitz, T. The amidohydrolases IAR3 and ILL6 contribute to jasmonoyl-isoleucine hormone turnover and generate 12-hydroxyjasmonic acid upon wounding in Arabidopsis leaves. J. Biol. Chem. 2013, 288, 31701–31714. [Google Scholar] [CrossRef]
- Rahimi, S.; Kim, Y.J.; Sukweenadhi, J.; Zhang, D.; Yang, D.C. PgLOX6 encoding a lipoxygenase contributes to jasmonic acid biosynthesis and ginsenoside production in Panax ginseng. J. Exp. Bot. 2017, 68, 4725. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Zhou, T.; Wang, Y.; Dong, J.; Bai, Y.; Huang, X.; Chen, C. Exploring the allelopathic autotoxicity mechanism of ginsenosides accumulation under ginseng decomposition based on integrated analysis of transcriptomics and metabolomics. Front. Bioeng Biotechnol. 2024, 12, 1365229. [Google Scholar] [CrossRef] [PubMed]
- Gomi, K. Jasmonic Acid: An Essential Plant Hormone. Int. J. Mol. Sci. 2020, 21, 1261. [Google Scholar] [CrossRef]
- Meesters, C.; Monig, T.; Oeljeklaus, J.; Krahn, D.; Westfall, C.S.; Hause, B.; Jez, J.M.; Kaiser, M.; Kombrink, E. A chemical inhibitor of jasmonate signaling targets JAR1 in Arabidopsis thaliana. Nat. Chem. Biol. 2014, 10, 830–836. [Google Scholar] [CrossRef] [PubMed]
- Wakuta, S.; Suzuki, E.; Saburi, W.; Matsuura, H.; Nabeta, K.; Imai, R.; Matsui, H. OsJAR1 and OsJAR2 are jasmonyl-L-isoleucine synthases involved in wound- and pathogen-induced jasmonic acid signalling. Biochem. Biophys Res. Commun. 2011, 409, 634–639. [Google Scholar] [CrossRef]
- Gulick, A.M. Conformational dynamics in the Acyl-CoA synthetases, adenylation domains of non-ribosomal peptide synthetases, and firefly luciferase. ACS Chem. Biol. 2009, 4, 811–827. [Google Scholar] [CrossRef]
- Pratiwi, P.; Tanaka, G.; Takahashi, T.; Xie, X.; Yoneyama, K.; Matsuura, H.; Takahashi, K. Identification of Jasmonic Acid and Jasmonoyl-Isoleucine, and Characterization of AOS, AOC, OPR and JAR1 in the Model Lycophyte Selaginella moellendorffii. Plant Cell Physiol. 2017, 58, 789–801. [Google Scholar] [CrossRef]
- Zhang, R.; Zhu, J.; Cao, H.Z.; An, Y.R.; Huang, J.J.; Chen, X.H.; Mohammed, Z.; Afrin, S.; Luo, Z.Y. Molecular cloning and expression analysis of PDR1-like gene in ginseng subjected to salt and cold stresses or hormonal treatment. Plant Physiol. Biochem. 2013, 71, 203–211. [Google Scholar] [CrossRef]
- Wasternack, C.; Song, S. Jasmonates: Biosynthesis, metabolism, and signaling by proteins activating and repressing transcription. J. Exp. Bot. 2017, 68, 1303–1321. [Google Scholar] [CrossRef]
- Liu, Z.; Zhang, S.; Sun, N.; Liu, H.; Zhao, Y.; Liang, Y.; Zhang, L.; Han, Y. Functional diversity of jasmonates in rice. Rice 2015, 8, 42. [Google Scholar] [CrossRef]
- Zeng, M.; Krajinski, F.; van Dam, N.M.; Hause, B. Jarin-1, an inhibitor of JA-Ile biosynthesis in Arabidopsis thaliana, acts differently in other plant species. Plant Signal. Behav. 2023, 18, 2273515. [Google Scholar] [CrossRef] [PubMed]
- Shi, M.; Zhou, W.; Zhang, J.; Huang, S.; Wang, H.; Kai, G. Methyl jasmonate induction of tanshinone biosynthesis in Salvia miltiorrhiza hairy roots is mediated by JASMONATE ZIM-DOMAIN repressor proteins. Sci. Rep. 2016, 6, 20919. [Google Scholar] [CrossRef] [PubMed]
- Sheard, L.B.; Tan, X.; Mao, H.; Withers, J.; Ben-Nissan, G.; Hinds, T.R.; Kobayashi, Y.; Hsu, F.F.; Sharon, M.; Browse, J.; et al. Jasmonate perception by inositol-phosphate-potentiated COI1-JAZ co-receptor. Nature 2010, 468, 400–405. [Google Scholar] [CrossRef] [PubMed]
- Bomer, M.; O’Brien, J.A.; Perez-Salamo, I.; Krasauskas, J.; Finch, P.; Briones, A.; Daudi, A.; Souda, P.; Tsui, T.L.; Whitelegge, J.P.; et al. COI1-dependent jasmonate signalling affects growth, metabolite production and cell wall protein composition in arabidopsis. Ann. Bot. 2018, 122, 1117–1129. [Google Scholar] [CrossRef]
- Ortigosa, A.; Fonseca, S.; Franco-Zorrilla, J.M.; Fernandez-Calvo, P.; Zander, M.; Lewsey, M.G.; Garcia-Casado, G.; Fernandez-Barbero, G.; Ecker, J.R.; Solano, R. The JA-pathway MYC transcription factors regulate photomorphogenic responses by targeting HY5 gene expression. Plant J. 2020, 102, 138–152. [Google Scholar] [CrossRef]
- Yan, L.; Zhai, Q.; Wei, J.; Li, S.; Wang, B.; Huang, T.; Du, M.; Sun, J.; Kang, L.; Li, C.B.; et al. Role of tomato lipoxygenase D in wound-induced jasmonate biosynthesis and plant immunity to insect herbivores. PLoS Genet. 2013, 9, e1003964. [Google Scholar] [CrossRef]
- Zhang, R.; Tan, S.; Zhang, B.; Hu, P.; Li, L. Cerium-promoted ginsenosides accumulation by regulating endogenous methyl jasmonate biosynthesis in hairy roots of Panax ginseng. Molecules 2021, 26, 5623. [Google Scholar] [CrossRef]
- Kazan, K.; Manners, J.M. JAZ repressors and the orchestration of phytohormone crosstalk. Trends Plant Sci 2012, 17, 22–31. [Google Scholar] [CrossRef]
- Han, X.; Kui, M.; He, K.; Yang, M.; Du, J.; Jiang, Y.; Hu, Y. Jasmonate-regulated root growth inhibition and root hair elongation. J. Exp. Bot. 2023, 74, 1176–1185. [Google Scholar] [CrossRef]
- Patil, R.A.; Lenka, S.K.; Normanly, J.; Walker, E.L.; Roberts, S.C. Methyl jasmonate represses growth and affects cell cycle progression in cultured Taxus cells. Plant Cell Rep. 2014, 33, 1479–1492. [Google Scholar] [CrossRef] [PubMed]
- Van der Does, D.; Leon-Reyes, A.; Koornneef, A.; Van Verk, M.C.; Rodenburg, N.; Pauwels, L.; Goossens, A.; Korbes, A.P.; Memelink, J.; Ritsema, T.; et al. Salicylic acid suppresses jasmonic acid signaling downstream of SCFCOI1-JAZ by targeting GCC promoter motifs via transcription factor ORA59. Plant Cell 2013, 25, 744–761. [Google Scholar] [CrossRef] [PubMed]
- Oh, J.Y.; Kim, Y.J.; Jang, M.G.; Joo, S.C.; Kwon, W.S.; Kim, S.Y.; Jung, S.K.; Yang, D.C. Investigation of ginsenosides in different tissues after elicitor treatment in Panax ginseng. J. Ginseng Res. 2014, 38, 270–277. [Google Scholar] [CrossRef] [PubMed]
- Chung, H.S.; Howe, G.A. A critical role for the TIFY motif in repression of jasmonate signaling by a stabilized splice variant of the JASMONATE ZIM-domain protein JAZ10 in Arabidopsis. Plant Cell 2009, 21, 131–145. [Google Scholar] [CrossRef]
- Yi, M.; Zhao, L.; Wu, K.; Liu, C.; Deng, D.; Zhao, K.; Li, J.; Deng, A. Simultaneous detection of plant growth regulators jasmonic acid and methyl jasmonate in plant samples by a monoclonal antibody-based ELISA. Analyst 2020, 145, 4004–4011. [Google Scholar] [CrossRef]
- Mielke, K.; Forner, S.; Kramell, R.; Conrad, U.; Hause, B. Cell-specific visualization of jasmonates in wounded tomato and Arabidopsis leaves using jasmonate-specific antibodies. New Phytol. 2011, 190, 1069–1080. [Google Scholar] [CrossRef]
- Zhang, Y.C.; Li, G.; Jiang, C.; Yang, B.; Yang, H.J.; Xu, H.Y.; Huang, L.Q. Tissue-specific distribution of ginsenosides in different aged ginseng and antioxidant activity of ginseng leaf. Molecules 2014, 19, 17381–17399. [Google Scholar] [CrossRef]
- Kim, Y.J.; Lee, O.R.; Oh, J.Y.; Jang, M.G.; Yang, D.C. Functional analysis of 3-hydroxy-3-methylglutaryl coenzyme a reductase encoding genes in triterpene saponin-producing ginseng. Plant Physiol. 2014, 165, 373–387. [Google Scholar] [CrossRef]
- Guo, W.; Zhao, Y.; Xu, H.; Xia, Y.; Tao, L.; You, X. PgDDS Changes the Plant Growth of Transgenic Aralia elata and Improves the Production of Re and Rg(3) in Its Leaves. Int. J. Mol. Sci. 2024, 25, 1945. [Google Scholar] [CrossRef]
- Han, J.Y.; Kim, H.J.; Kwon, Y.S.; Choi, Y.E. The Cyt P450 enzyme CYP716A47 catalyzes the formation of protopanaxadiol from dammarenediol-II during ginsenoside biosynthesis in Panax ginseng. Plant Cell Physiol. 2011, 52, 2062–2073. [Google Scholar] [CrossRef]
- Lu, C.; Zhao, S.; Wei, G.; Zhao, H.; Qu, Q. Functional regulation of ginsenoside biosynthesis by RNA interferences of a UDP-glycosyltransferase gene in Panax ginseng and Panax quinquefolius. Plant Physiol. Biochem. 2017, 111, 67–76. [Google Scholar] [CrossRef] [PubMed]
- Gwak, Y.S.; Han, J.Y.; Choi, Y.E. Production of ginsenoside aglycone (protopanaxatriol) and male sterility of transgenic tobacco co-overexpressing three Panax ginseng genes: PgDDS, CYP716A47, and CYP716A53v2. J. Ginseng Res. 2019, 43, 261–271. [Google Scholar] [CrossRef]
- Park, S.B.; Chun, J.H.; Ban, Y.W.; Han, J.Y.; Choi, Y.E. Alteration of Panax ginseng saponin composition by overexpression and RNA interference of the protopanaxadiol 6-hydroxylase gene (CYP716A53v2). J. Ginseng Res. 2016, 40, 47–54. [Google Scholar] [CrossRef]
- Ma, R.; Yang, P.; Jing, C.; Fu, B.; Teng, X.; Zhao, D.; Sun, L. Comparison of the metabolomic and proteomic profiles associated with triterpene and phytosterol accumulation between wild and cultivated ginseng. Plant Physiol. Biochem. 2023, 195, 288–299. [Google Scholar] [CrossRef]
- Zhang, R.; Zhang, B.L.; Li, G.C.; Xie, T.; Hu, T.; Luo, Z.Y. Enhancement of ginsenoside Rg1 in Panax ginseng hairy root by overexpressing the alpha-L-rhamnosidase gene from Bifidobacterium breve. Biotechnol. Lett. 2015, 37, 2091–2096. [Google Scholar] [CrossRef] [PubMed]
- Krieger, E.; Joo, K.; Lee, J.; Lee, J.; Raman, S.; Thompson, J.; Tyka, M.; Baker, D.; Karplus, K. Improving physical realism, stereochemistry, and side-chain accuracy in homology modeling: Four approaches that performed well in CASP8. Proteins 2009, 77 (Suppl. S9), 114–122. [Google Scholar] [CrossRef] [PubMed]
- Studer, G.; Tauriello, G.; Bienert, S.; Biasini, M.; Johner, N.; Schwede, T. ProMod3-A versatile homology modelling toolbox. PLoS Comput. Biol. 2021, 17, e1008667. [Google Scholar] [CrossRef]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef]
- Kim, S.; Chen, J.; Cheng, T.; Gindulyte, A.; He, J.; He, S.; Li, Q.; Shoemaker, B.A.; Thiessen, P.A.; Yu, B.; et al. PubChem in 2021: New data content and improved web interfaces. Nucleic Acids Res. 2021, 49, D1388–D1395. [Google Scholar] [CrossRef]
- Liu, T.; Luo, T.; Guo, X.; Zou, X.; Zhou, D.; Afrin, S.; Li, G.; Zhang, Y.; Zhang, R.; Luo, Z. PgMYB2, a MeJA-Responsive Transcription Factor, Positively Regulates the Dammarenediol Synthase Gene Expression in Panax Ginseng. Int. J. Mol. Sci. 2019, 20, 2219. [Google Scholar] [CrossRef]






Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, R.; Li, C.; Guo, R.; Li, Z.; Zhang, B. Harnessing Jasmonate Pathways: PgJAR1’s Impact on Ginsenoside Accumulation in Ginseng. Plants 2025, 14, 847. https://doi.org/10.3390/plants14060847
Zhang R, Li C, Guo R, Li Z, Zhang B. Harnessing Jasmonate Pathways: PgJAR1’s Impact on Ginsenoside Accumulation in Ginseng. Plants. 2025; 14(6):847. https://doi.org/10.3390/plants14060847
Chicago/Turabian StyleZhang, Ru, Chao Li, Rui Guo, Zhaoying Li, and Bianling Zhang. 2025. "Harnessing Jasmonate Pathways: PgJAR1’s Impact on Ginsenoside Accumulation in Ginseng" Plants 14, no. 6: 847. https://doi.org/10.3390/plants14060847
APA StyleZhang, R., Li, C., Guo, R., Li, Z., & Zhang, B. (2025). Harnessing Jasmonate Pathways: PgJAR1’s Impact on Ginsenoside Accumulation in Ginseng. Plants, 14(6), 847. https://doi.org/10.3390/plants14060847




