Genetic Assessment in the Andean Tropical Fruits Solanum quitoense Lam. and S. betaceum Cav.: Efforts Towards a Molecular Breeding Approach
Abstract
1. Introduction
2. Materials and Methods
2.1. DNA Library and NGS Sequencing
2.2. Microsatellite Search and Primer Screening
2.3. Polymorphism Screening and DNA Genotyping
2.4. Statistical Analysis
3. Results
3.1. NGS Sequencing Analysis and In Silico SSR Identification
3.2. SSR Variability
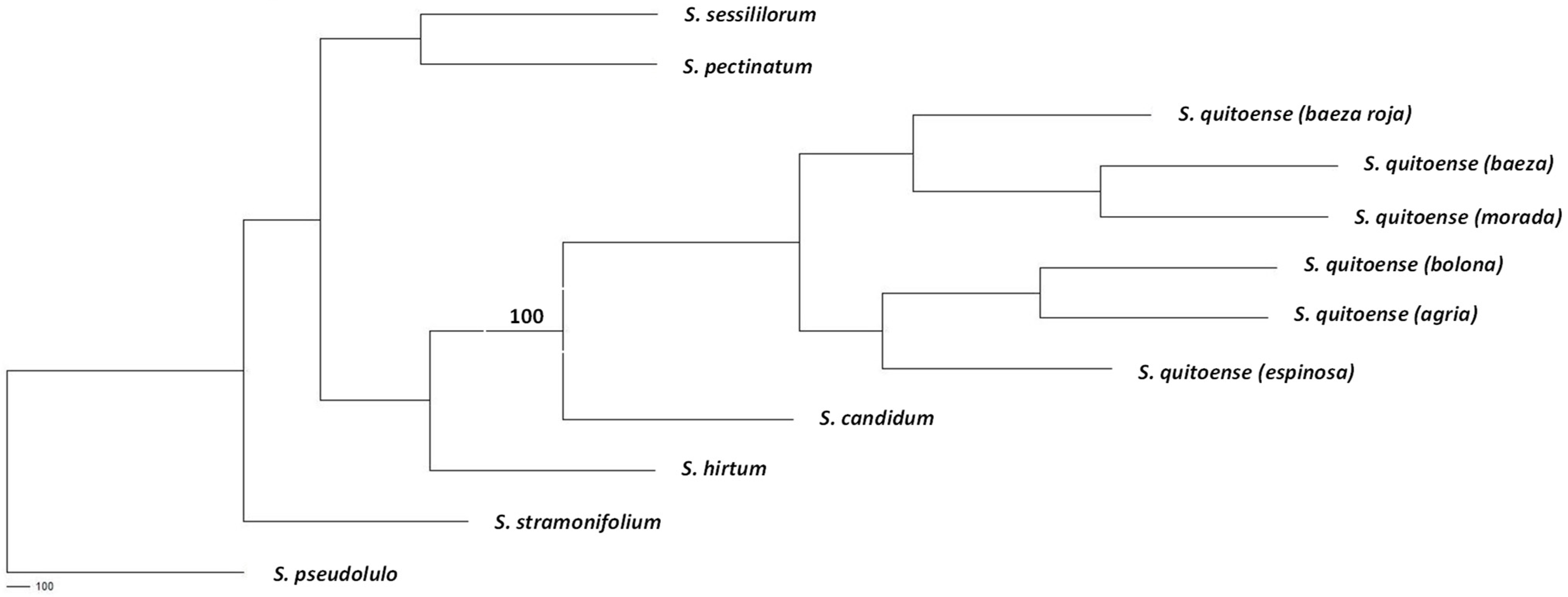
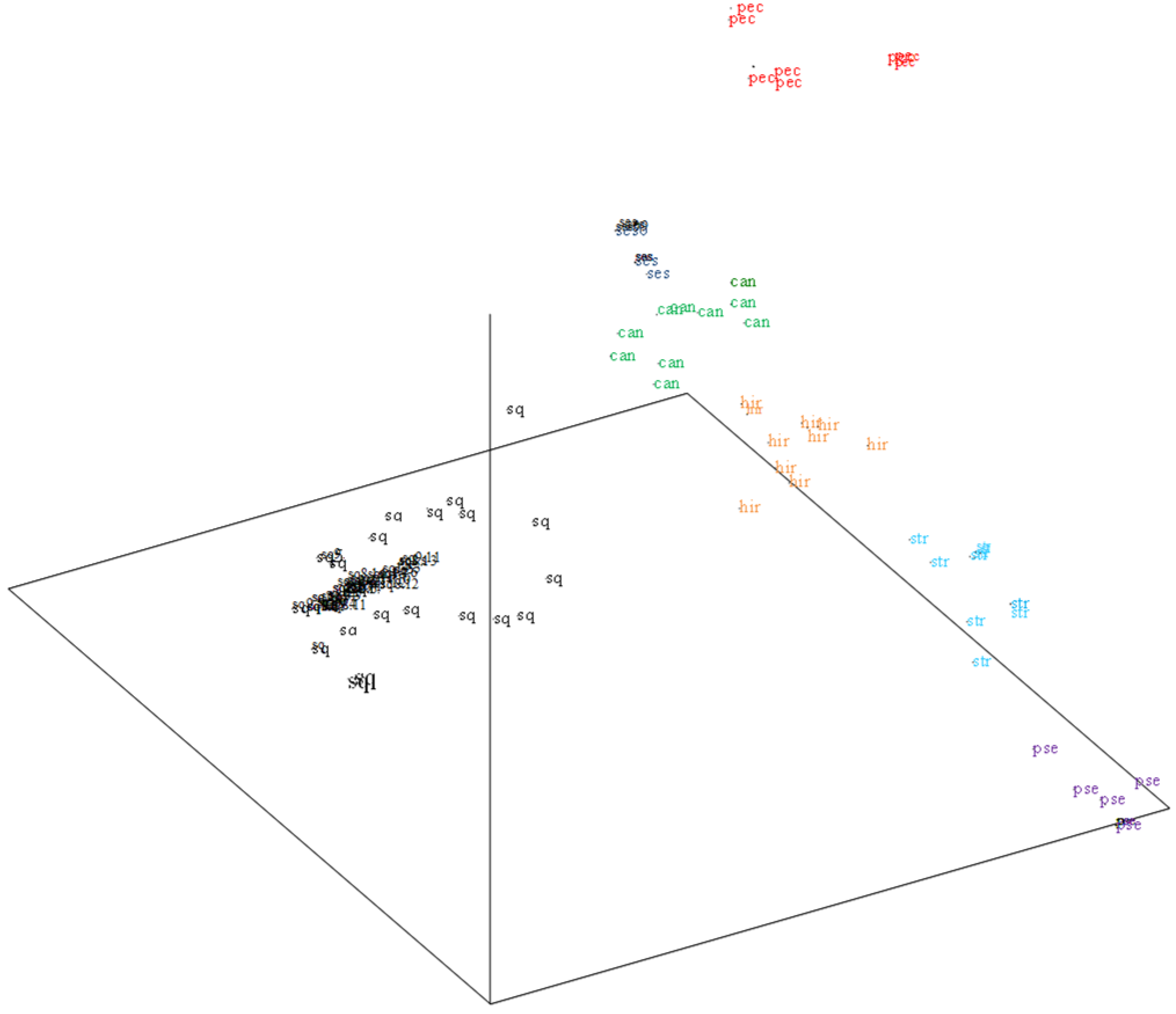
3.3. Genetic Diversity Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Heiser, C.B., Jr. The relationships of the naranjilla, Solanum quitoense. Biotropica 1972, 4, 77–84. [Google Scholar] [CrossRef]
- Silva, W.; Gómez, P.; Sotomayor, A.; Viteri, D.; Ron, L. Selección de líneas promisorias de naranjilla para mejorar la calidad de la fruta. Ecuad. Calidad. 2016, 3, 23–30. [Google Scholar]
- Carrera, K.; Isla, H.; Lidcay, F.; Cupull Santana, R. Identificación de aislados de Fusarium spp. asociados a Solanum quitoense Lam en Pastaza, Ecuador. Centro Agrícola. 2018, 45, 5–11. [Google Scholar]
- Salazar-González, C.; Betancourth-García, C. Reaction of genotypes of lulo (Solanum quitoense Lam.) to Meloidogyne spp. under field conditions. Cienc. Tecnol. Agropecu. 2017, 18, 295–306. [Google Scholar] [CrossRef]
- Lobo, M.; Medina, C.I.; Delgado Paz, O.A.; Bermeo Giraldo, A. Variabilidad morfológica de la colección colombiana de lulo (Solanum quitoense Lam.) y especies relacionadas de la sección Lasiocarpa. Rev. Fac. Nac. Agron. Medellin 2007, 60, 3939–3964. [Google Scholar]
- Torres, A.F.; Arias, A.S.; Arahana, V.; Torres, M.L. Preliminary Assessment of Genetic Diversity and Phenetic Relations for Section by Means of Heterologous SSR Markers. Crop Sci. 2008, 48, 2289–2297. [Google Scholar] [CrossRef]
- Soria, J. Genetic improvement of the “naranjilla” (Solanum quitoense Lam.) through interspecific crosses. In Utilization and Management of Phyto-Resources; Abya-Yala: Quito, Ecuador, 1997. (In Spanish) [Google Scholar]
- Fory Sánchez, P.A.; Sánchez Mosquera, I.; Bohórquez Cháux, A.; Ramírez, H.; Medina Cano, C.I.; Lobo Arias, M. Genetic variability of the Colombian collection of lulo (Solanum quitoense Lam.) and related species of section Lasiocarpa. Rev. Fac. Nac. Agron. Medellín 2010, 63, 5465–5476. [Google Scholar]
- Díaz Granda, L.; Canto Sáenz, M.; Alegre Orihuela, J.; Camarena Mayta, F.; Julca Otiniano, A. Sostenibilidad social de los subsistemas productivos de tomate de árbol (Solanum betaceum Cav) en el Cantón Guachapala, Provincia de Azuay-Ecuador. Ecol. Apl. 2017, 16, 99–104. [Google Scholar] [CrossRef][Green Version]
- Villares, M.; Sánchez, J.; Viera, W.; Soria, N.; Sotomayor, A.; Yanez, D.; Martínez, E. Caracterización morfológica de frutos de tomate de árbol (Solanum betaceum Cav.) de una población segregante. Rev. Investig. Talent. 2018, 5, 9–19. [Google Scholar]
- Moreno-Miranda, C.; Molina, J.I.; Ortiz, J.; Peñafiel, C.; Moreno, R. Cadena de valor en la red de tomate de árbol (Solanum betaceum) en Ecuador. Agron. Mesoam. 2020, 31, 13–29. [Google Scholar] [CrossRef]
- Criollo, H.; Insuasti, K.; Delgado, W. Regeneración in vitro de plántulas de tomate de árbol (Solanum betaceum Cav. Sendt.). Rev. Colomb. Cienc. Hortícolas 2016, 10, 252–261. [Google Scholar] [CrossRef]
- Caruso, G. Diversidad genética. Importancia y aplicaciones en el mejoramiento vegetal. Lhawet 2015, 4, 45–50. [Google Scholar]
- Sari, D.; Sari, H.; Ikten, C.; Toker, C. Genome-wide discovery of di-nucleotide SSR markers based on whole genome re-sequencing data of Cicer arietinum L. and Cicer reticulatum Ladiz. Sci. Rep. 2023, 13, 10351. [Google Scholar] [CrossRef] [PubMed]
- Peñafiel, N.L.; Arahana, V.S.; Torres, M. Evaluación de la variabilidad genética del tomate de árbol (Solanum betaceum Cav) en los cultivos de tres provincias del Ecuador por medio de marcadores microsatélites. ACI Av. Cienc. Ing. 2009, 1, 1. [Google Scholar] [CrossRef]
- Rodríguez, E.F.; Martínez, R.; Lobo, M.; Barrero, L.E. Genetic variation in the Solanaceae fruit bearing species lulo and tree tomato revealed by Conserved Ortholog (COSII) markers. Genet. Mol. Biol. 2010, 33, 271–278. [Google Scholar] [CrossRef][Green Version]
- Acosta-Quezada, P.G.; Raigón, M.D.; Riofrío-Cuenca, T.; García-Martínez, M.D.; Plazas, M.; Burneo, J.I.; Prohens, J. Diversity for chemical composition in a collection of different varietal types of tree tomato (Solanum betaceum Cav.), an Andean exotic fruit. Food Chem. 2015, 169, 327–335. [Google Scholar] [CrossRef]
- Souza, H.; Muller, L.; Brandão, R.; Lovato, M. Isolation of high quality and polysaccharide-free DNA from leaves of Dimor-384 phandra mollis (Leguminosae), a tree from the Brazilian Cerrado. Genet. Mol. Res. 2012, 11, 756–764. [Google Scholar] [CrossRef]
- Simpson, J.T.; Wong, K.; Jackman, S.D.; Schein, J.E.; Jones, S.J.; Birol, I. ABySS: A parallel assembler for short read sequence data. Genome Res. 2009, 19, 1117–1123. [Google Scholar] [CrossRef]
- Thiel, T. MISA-MIcroSAtellite Identification Tool, Version 1.0. MISA-MIcroSAtellite Identification Tool; Leibniz Institute of Plant. Genetics and Crop Plant Research: Gatersleben, Germany, 2001. [Google Scholar]
- Kang, H.; Cho, Y.; Yoon, U.; Eun, M. Rapid Genotypes Analysis Using DNA Extracted from Single Half Seed of Rice Plant. Mol. Biol. Reporter. 1988, 16, 90. [Google Scholar]
- Morillo, E.; Miño, G. Marcadores Moleculares en Biotecnología Agrícola: Manual de Procedimientos y Técnicas en INIAP; Manual técnico No. 91; Instituto Nacional Autónomo de Investigaciones Agropecuarias, Estación Experimental Santa Catalina: Quito, Ecuador, 2011; 121p. [Google Scholar]
- Liu, K.; Muse, S.V. PowerMarker: An integrated analysis environment for genetic marker analysis. Bioinformatics 2005, 21, 2128–2129. [Google Scholar] [CrossRef]
- Rohlf, F.J. NTSYS-pc. Numerical Taxonomy and Multivariate Analysis System; Applied Biostattistics: New York, NY, USA, 1992. [Google Scholar]
- Felsenstein, J. PHYLIP (Phylogeny Inference Package), version 3.5 c; Department of Genetics, University of Washington: Seatle, DC, USA, 1993.
- Taheri, S.; Lee Abdullah, T.; Yusop, M.R.; Hanafi, M.M.; Sahebi, M.; Azizi, P.; Shamshiri, R.R. Mining and development of novel SSR markers using next generation sequencing (NGS) data in plants. Molecules 2018, 23, 399. [Google Scholar] [CrossRef]
- Zhu, J.; Zhang, J.; Jiang, M.; Wang, W.; Jiang, J.; Li, Y.; Zhou, X. Development of genome-wide SSR markers in rapeseed by Next Generation Sequencing. Gene 2021, 798, 145798. [Google Scholar] [CrossRef]
- Botstein, D.; White, R.L.; Skolnick, M.; Davis, R.W. Construction of a genetic linkage map in man using restriction fragment length polymorphisms. Am. J. Hum. Genet. 1980, 32, 314. [Google Scholar]
- Otzen, T.; Manterola, C. Técnicas de Muestreo sobre una Población a Estudio. Int. J. Morphol. 2017, 35, 227–232. [Google Scholar] [CrossRef]
- Gepts, P. A comparison between crop domestication, classical plant breeding, and genetic engineering. Crop Sci. 2002, 42, 1780–1790. [Google Scholar] [CrossRef]
- Bernal, N.; Ocampo, J.; Hernández, J. Caracterización y análisis de la variabilidad Genética de la granadilla (Passiflora ligularis juss.) en Colombia empleando marcadores microsatélites. Rev. Bras. Frutic. 2014, 36, 586–597. [Google Scholar] [CrossRef][Green Version]
- Sosa, P.A.; Batista Hernández, F.; Bouza Carrelo, N.; González Pérez, M.Á. La conservación Genética de las Especies Vegetales Amenazadas; 2002; Available online: https://accedacris.ulpgc.es/bitstream/10553/1285/1/82.pdf (accessed on 30 December 2024).
- Zambrano-Fierro, P. Caracterización Molecular de un Grupo de Híbridos Provenientes del Cruzamiento (Solanum betaceum Cav. x Solanum unilobum) del CADET. Tumbaco; Tesis Ingeniero Agrónomo, Carrera de Ingeniería Agronómica, UCE: Quito, Ecuador, 2020; 94p. [Google Scholar]
- Leiva, L.E. Caracterización de dos Poblaciones Segregantes de Tomate de Árbol (Solanum betaceum) Mediante Marcadores Moleculares AFLP. Bachelor’s Thesis, Universidad San Francisco de Quito, Quito, Ecuador, 2008. Available online: http://repositorio.usfq.edu.ec/handle/23000/1314 (accessed on 1 December 2024).
| Species | Type | Variety or Group | INIAP ID or Source | Origin | Lab. Code |
|---|---|---|---|---|---|
| S. quitoense | Cultivated | Espinosa | INIAP ECU-3567 | Pichincha-Ecuador | Sq1 |
| S. quitoense | Cultivated | Naranjilla agria | INIAP ECU-3817 | Bolívar-Ecuador | Sq2 |
| S. quitoense | Cultivated | Naranjilla bolona | INIAP ECU-6235 | Morona Santiago-Ecuador | Sq5 |
| S. quitoense | Cultivated | Morada | INIAP Breeding program | Morona Santiago-Ecuador | Sq8 |
| S. quitoense | Cultivated | Baeza | INIAP Breeding program | Morona Santiago-Ecuador | Sq9 |
| S. quitoense | Cultivated | Baeza roja | INIAP Breeding program | Morona Santiago-Ecuador | Sq14 |
| S. candidum | Wild | Lasiocarpa | INIAP ECU-13242 | Not available | Sc |
| S. hirtum | Wild | Lasiocarpa | INIAP ECU-6242 | Morona Santiago-Ecuador | Sh |
| S. pectinatum | Wild | Lasiocarpa | INIAP ECU-7875 | Pastaza-Ecuador | Sp |
| S. pseudolulo | Wild | Lasiocarpa | INIAP Breeding program | Not available | Sps |
| S. sessiliflorum | Wild | Lasiocarpa | INIAP ECU-5552 | Morona Santiago-Ecuador | Ss |
| S. stramonifolium | Wild | Lasiocarpa | INIAP Breeding program | Pichincha-Ecuador | St |
| S. betaceum | Cultivated | Puntón Anaranjado | INIAP Breeding program | Pichincha-Ecuador | PA |
| S. betaceum | Cultivated | Gigante Morado | INIAP Breeding program | Pichincha-Ecuador | GM |
| S. betaceum | Cultivated | Gigante Anaranjado | INIAP Breeding program | Pichincha-Ecuador | GA |
| S. betaceum | Cultivated | Puntón Morado | INIAP Breeding program | Pichincha-Ecuador | PM |
| S. betaceum × S. unilobum | Segregant | - | INIAP Breeding program | Pichincha-Ecuador | 12 GR |
| S. betaceum × S. unilobum | Segregant | - | INIAP Breeding program | Pichincha-Ecuador | GT10P8 |
| S. betaceum × S. unilobum | Segregant | - | INIAP Breeding program | Pichincha-Ecuador | GT13P25 |
| S. betaceum × S. unilobum | Segregant | - | INIAP Breeding program | Pichincha-Ecuador | GT33P7 |
| S. betaceum × S. unilobum | Segregant | - | INIAP Breeding program | Pichincha-Ecuador | Cruzam. 5 |
| S. unilobum | Wild | progenitor | INIAP Breeding program | Pichincha-Ecuador | TUP1 |
| S. unilobum | Wild | progenitor | INIAP Breeding program | Pichincha-Ecuador | 25AP1 |
| S. unilobum | Wild | progenitor | INIAP Breeding program | Pichincha-Ecuador | KUYP1 |
| S. unilobum | Wild | progenitor | INIAP Breeding program | Pichincha-Ecuador | M15P1 |
| S. unilobum | Wild | progenitor | INIAP Breeding program | Pichincha-Ecuador | SAN CARLOS |
| SSR Search Results | S. quitoense | S. betaceum |
|---|---|---|
| Total number of sequences examined | 1,400,090 | 1,732,580 |
| Total size of examined sequences (bp) | 274,369,691 | 296,038,400 |
| Total number of identified SSRs | 34,832 | 115,436 |
| Number of SSRs containing sequences | 31,759 | 68,685 |
| Number of sequences containing more than 1 SSR | 2370 | 30,063 |
| Number of SSRs present in compound formation | 2759 | 46,752 |
| Distribution of different repeat-type classes | ||
| Unit size | Number of SSRs | |
| 2 | 18,349 | 16,385 |
| 3 | 13,385 | 91,665 |
| 4 | 1841 | 4348 |
| 5 | 856 | 274 |
| 6 | 401 | 2.764 |
| Primer | S. sessiliflorim | S. stramonifolium | S. hirtum | S. candidum | S. pectinatum | S. pseudolulo | PCR Rate |
|---|---|---|---|---|---|---|---|
| mSq_03 | - | - | - | 236, 239 | - | - | 16.7% |
| mSq_04 | 125 | 125, 170 | 125, 170 | 125, 161 | 125, 146 | 125 | 100.0% |
| mSq_06 | 158 | 182 | 140, 158 | 170 | - | - | 66.7% |
| mSq_08 | - | 173 | 167, 176, 215 | 167 | 158 | 164 | 83.3% |
| mSq_12 | 165 | 165 | 164 | 161 | 164 | 164 | 100.0% |
| mSq_13 | 145, 247 | 145, 247 | 145, 247 | 145 | 142, 145 | 142, 154 | 100.0% |
| mSq_16 | 247 | - | 235, 238, 256 | 247 | 244 | 238 | 83.3% |
| mSq_18 | 128, 137, 140, 143 | 137, 140, 143 | 137, 140, 143 | 122,140, 143 | 140, 143 | - | 83.3% |
| mSq_19 | - | - | - | 224, 227 | 206 | 200, 209 | 66.7% |
| mSq_20 | 195 | - | 195 | 201, 210 | - | - | 50.0% |
| mSq_21 | - | - | 136, 154 | 133 | 136 | 163 | 66.7% |
| mSq_23 | - | - | - | 107, 134 | 140 | - | 33.3% |
| mSq_24 | 104, 158 | 104, 149 | 125, 152 | 131 | 152 | 104, 143, 149 | 100.0% |
| mSq_26 | 105 | 111 | 105, 108 | 108 | 108 | - | 83.3% |
| mSq_27 | 104 | 104 | 98, 104 | 95, 104 | 89, 95, 104 | 98, 104 | 100.0% |
| mSq_28 | 153 | 150 | 153, 168 | 153, 162 | 153, 174 | - | 83.3% |
| mSq_29 | - | - | 145 | - | - | - | 16.7% |
| mSq_31 | - | - | - | 190 | 190 | - | 33.3% |
| mSq_33 | - | - | - | 143 | 134 | 122 | 50.0% |
| mSq_35 | - | 239 | 230, 242, 251 | - | 248 | - | 50.0% |
| mSq_36 | 110, 112 | 121 | 118 | 118, 130 | 112 | 115 | 100.0% |
| mSq_37 | - | 237 | 237, 249 | 234 | 246 | - | 66.7% |
| mSq_38 | 113, 146 | 113, 128 | 113, 128 | 113, 140 | 113, 128 | 113, 128 | 100.0% |
| mSq_40 | 209 | 206, 221 | 206, 221, 230 | 206, 218 | 206 | 206 | 100.0% |
| mSq_43 | - | 133 | - | 133 | - | 133 | 50.0% |
| mSq_44 | 118 | 114 | 126, 134 | 118 | 134 | 130 | 100.0% |
| mSq_46 | - | - | 231 | - | - | 231 | 33.3% |
| mSq_49 | - | 89 | 89 | 89, 91 | 89, 91 | 89 | 83.3% |
| mSq_50 | 160 | 182 | 150 | 150, 172 | 150 | - | 83.3% |
| mSq_51 | - | - | 235 | 247 | - | - | 33.3% |
| mSq_56 | - | 112 | 112 | 130 | - | 112 | 66.7% |
| mSq_57 | - | 158 | 168, 172, 176, 190 | 162 | 172 | - | 66.7% |
| mSq_58 | - | 140 | 144 | 144 | 176 | 160 | 83.3% |
| mSq_59 | - | 183 | 189, 191,193 | 179 | 179 | 179 | 83.3% |
| mSq_63 | - | 147 | 135, 137, 143 | - | - | - | 33.3% |
| mSq_66 | 142 | 156 | 150, 172 | 148 | 154 | 178 | 100.0% |
| mSq_68 | 176 | - | - | 172, 188 | - | - | 33.3% |
| mSq_84 | 82 | 82 | - | - | 88 | 90 | 66.7% |
| mSq_87 | 142 | 142 | - | 130, 142 | 150, 180 | 142, 162 | 83.3% |
| mSq_91 | - | - | 121 | 121, 135, 147 | - | - | 33.3% |
| mSq_93 | - | 129, 159 | 117, 141 | 133, 143 | - | 119, 137 | 66.7% |
| % | 48.8% | 68.3% | 78.0% | 87.8% | 70.7% | 58.5% |
| Primer | Forward Primer | Reverse Primer | Motif | Size (pb) | Alleles |
|---|---|---|---|---|---|
| mSq006 | TTACAGGGGAAGAGGGG | CGTATTTGTGTCTTATGTGGG | (AAT)11 | 170 | 170, 191 |
| mSq012 | TTCAAGTGTCAAGATTCAAG | AATTGTGTCAACTCTTACCC | (TTA)16 | 194 | 164, 194, 203 |
| mSq016 | CCATTATGCCTATCAATTCC | CTCGTCCCAAGAACAAAA | (AAT)12 | 256 | 244, 256 |
| mSq018 | TCTCCAAGATCCATGATT | AGGATGCTTCTTTTGATG | (ATT)13 | 143 | 140, 143, 247 |
| mSq036 | ACCAGCTTCAGAACATCAAA | GATTATTCTAGTAGCCGTCCCT | (AAT)9 | 118 | 118, 124 |
| mSq040 | AGTAAGTCACTCCAGTCTATTCA | CTAGTCCCCAAGCGAA | (ATT)11 | 221 | 209, 221 |
| mSq049 | ACAGGTATTACAAAGTCCACA | TTGGGAGCTTGTTTGTT | (AT)14 | 107 | 89, 107 |
| mSq050 | AATGCGAGGTGTGATAAATG | CATGTTGATGGTTTGGGA | (AT)15 | 172 | 172, 174 |
| mSq058 | AGATAGTCCTTCCCACCT | AAGAAAGTGATTTCGCC | (AT)14 | 162 | 156, 162 |
| mSq059 | TGAAGTCATAGCCACCAAC | CCACAAAGTTCCCTAATAAATC | (TA)15 | 195 | 179, 191, 195 |
| mSq063 | GCTTGAACAAACCAATTTCA | TTGCCACCAACTGAGGA | (TA)14 | 157 | 155, 157 |
| mSq066 | AGTCCCCTTGTATCTGGTG | GGAGAAAGGCAAGTGAGAG | (AT)15 | 162 | 162, 168 |
| mSq068 | TAAAATTAACACGACCCACA | AAGTGGCAAAGACGCA | (TA)17 | 188 | 186, 188 |
| mSq091 | CCGATTATGCAAGAAAGGT | GAGCTAGTTTAGCCTATTTTGGT | (AT)16 | 147 | 147, 165 |
| Primer | Genotypes | Availability Data (Na) | Alleles | Gene Diversity (He) | Heterozygosity (Ho) | PIC | |||
|---|---|---|---|---|---|---|---|---|---|
| mSq006 | 6 | 113 | 6 | 0.65 | 0.019 | 0.06 | 0.019 | 0.61 | 0.019 |
| mSq012 | 7 | 107 | 6 | 0.68 | 0.592 | 0.41 | 0.936 | 0.64 | 0.511 |
| mSq016 | 10 | 112 | 6 | 0.73 | 0.142 | 0.13 | 0.115 | 0.69 | 0.132 |
| mSq018 | 13 | 116 | 6 | 0.71 | 0.492 | 0.66 | 0.875 | 0.66 | 0.371 |
| mSq036 | 9 | 114 | 8 | 0.63 | 0.018 | 0.13 | 0.019 | 0.61 | 0.018 |
| mSq040 | 10 | 113 | 7 | 0.75 | 0.497 | 0.62 | 0.925 | 0.71 | 0.374 |
| mSq049 | 4 | 115 | 4 | 0.55 | 0.499 | 0.46 | 0.964 | 0.49 | 0.375 |
| mSq050 | 9 | 113 | 7 | 0.67 | 0.073 | 0.05 | 0.000 | 0.64 | 0.070 |
| mSq058 | 7 | 106 | 7 | 0.75 | 0.043 | 0.00 | 0.000 | 0.72 | 0.042 |
| mSq059 | 10 | 109 | 7 | 0.75 | 0.203 | 0.08 | 0.061 | 0.71 | 0.189 |
| mSq063 | 6 | 110 | 7 | 0.65 | 0.077 | 0.05 | 0.000 | 0.58 | 0.074 |
| mSq066 | 10 | 108 | 10 | 0.78 | 0.117 | 0.11 | 0.042 | 0.76 | 0.110 |
| mSq068 | 6 | 108 | 5 | 0.71 | 0.499 | 0.01 | 0.000 | 0.66 | 0.375 |
| mSq091 | 6 | 113 | 5 | 0.61 | 0.019 | 0.04 | 0.019 | 0.53 | 0.019 |
| Mean | 8 | 111.2 | 6.4 | 0.69 | 0.23 | 0.19 | 0.28 | 0.64 | 0.178 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Morillo, E.; Buitron, J.; Yanez, D.; Mournet, P.; Vásquez-Castillo, W.; Viteri, P. Genetic Assessment in the Andean Tropical Fruits Solanum quitoense Lam. and S. betaceum Cav.: Efforts Towards a Molecular Breeding Approach. Plants 2025, 14, 874. https://doi.org/10.3390/plants14060874
Morillo E, Buitron J, Yanez D, Mournet P, Vásquez-Castillo W, Viteri P. Genetic Assessment in the Andean Tropical Fruits Solanum quitoense Lam. and S. betaceum Cav.: Efforts Towards a Molecular Breeding Approach. Plants. 2025; 14(6):874. https://doi.org/10.3390/plants14060874
Chicago/Turabian StyleMorillo, Eduardo, Johanna Buitron, Denisse Yanez, Pierre Mournet, Wilson Vásquez-Castillo, and Pablo Viteri. 2025. "Genetic Assessment in the Andean Tropical Fruits Solanum quitoense Lam. and S. betaceum Cav.: Efforts Towards a Molecular Breeding Approach" Plants 14, no. 6: 874. https://doi.org/10.3390/plants14060874
APA StyleMorillo, E., Buitron, J., Yanez, D., Mournet, P., Vásquez-Castillo, W., & Viteri, P. (2025). Genetic Assessment in the Andean Tropical Fruits Solanum quitoense Lam. and S. betaceum Cav.: Efforts Towards a Molecular Breeding Approach. Plants, 14(6), 874. https://doi.org/10.3390/plants14060874







