Identification of the bHLH Transcription Factor Family in Orah Mandarin and the Response of CrbHLH46 to Low-Temperature Stress
Abstract
:1. Introduction
2. Results
2.1. Subsection Identification and Physicochemical Property Analysis of the bHLH TF Family
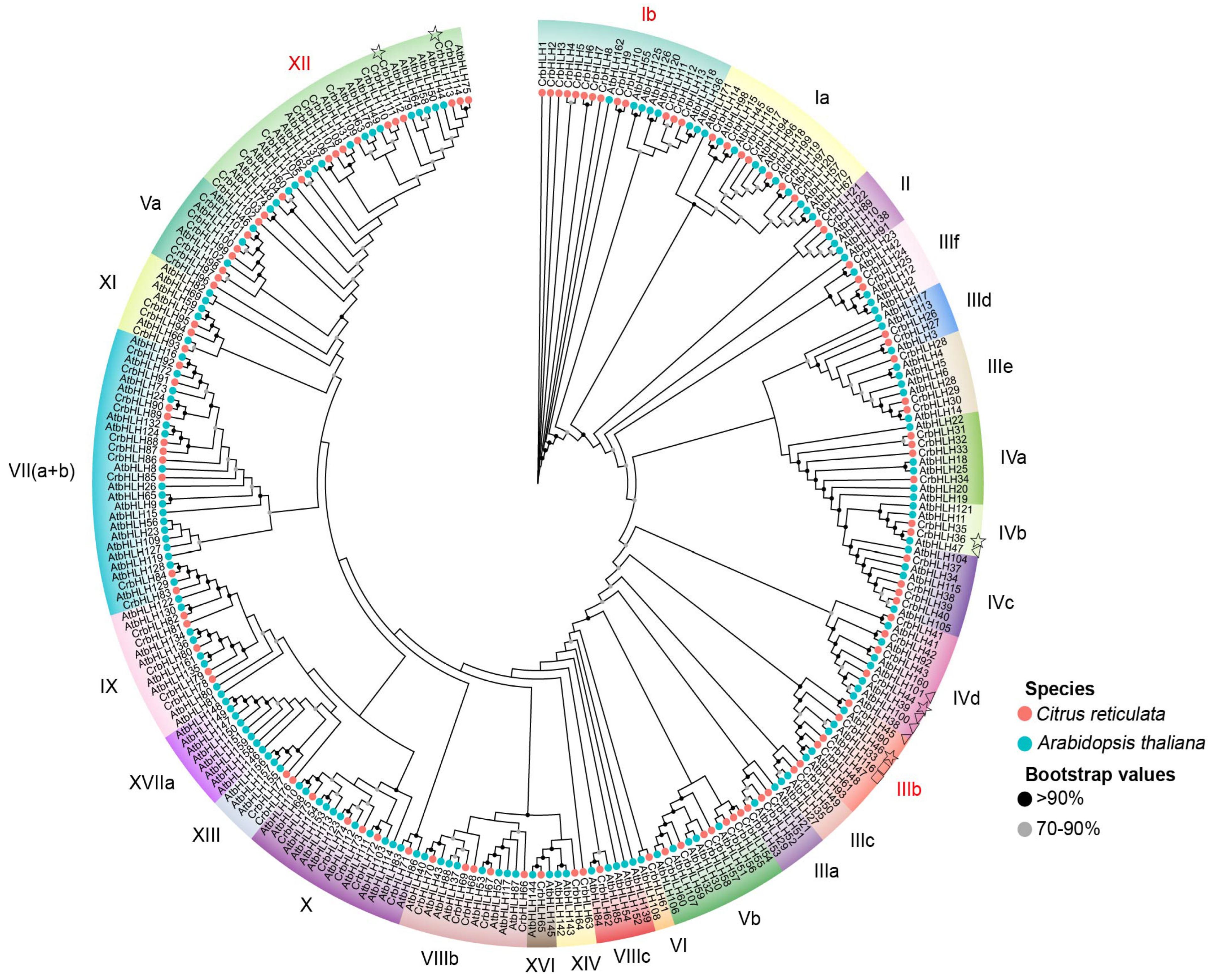
2.2. Evolutionary Analysis
2.3. Analysis of Conserved Motifs and Gene Structure
2.4. Analysis of Cis-Acting Elements
2.5. GO Function Annotation of the CrbHLH TF Family
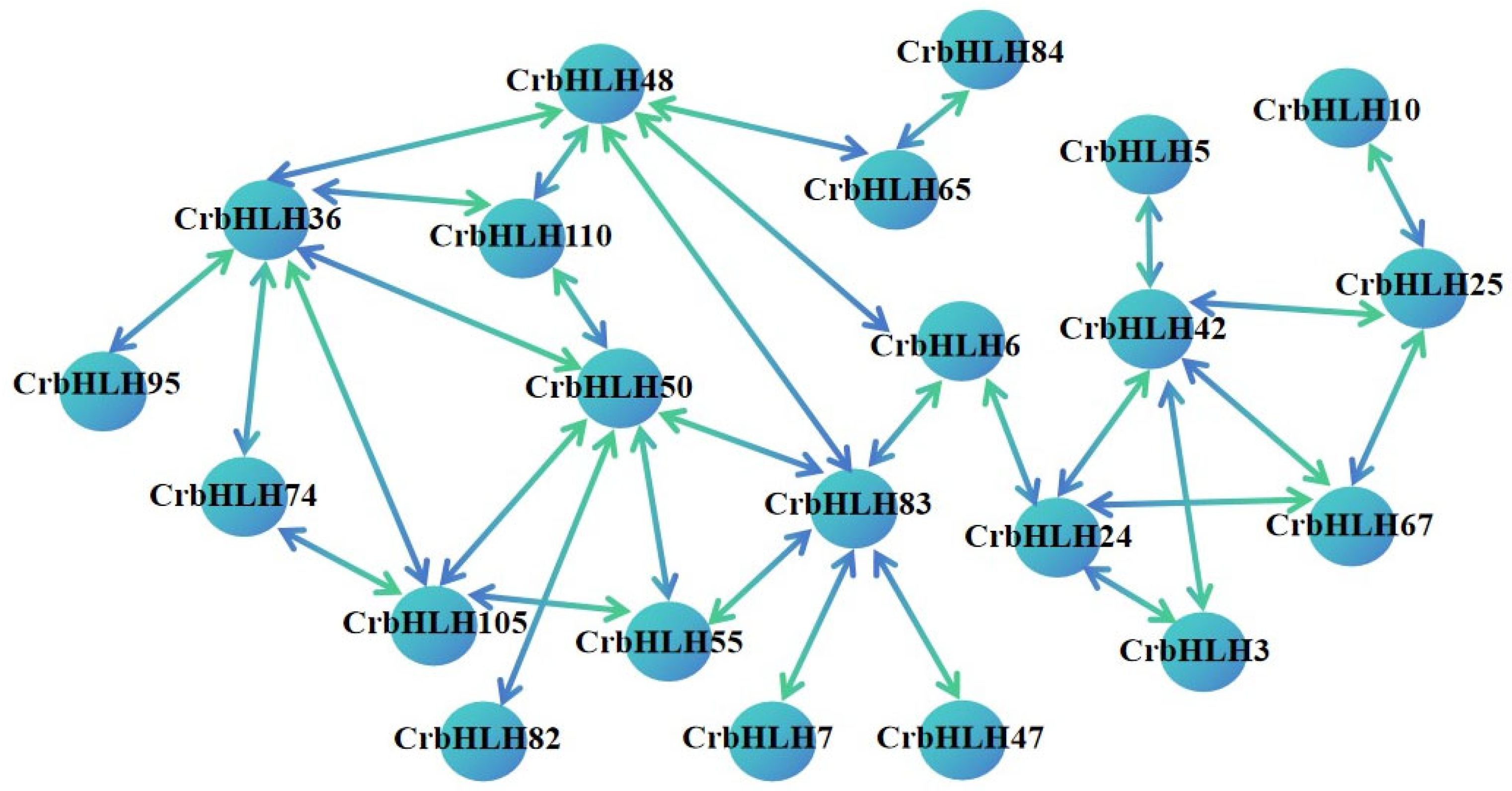
2.6. Analysis of Protein Network Interactions of the bHLH Transcription Factor Family of Orah Mandarin
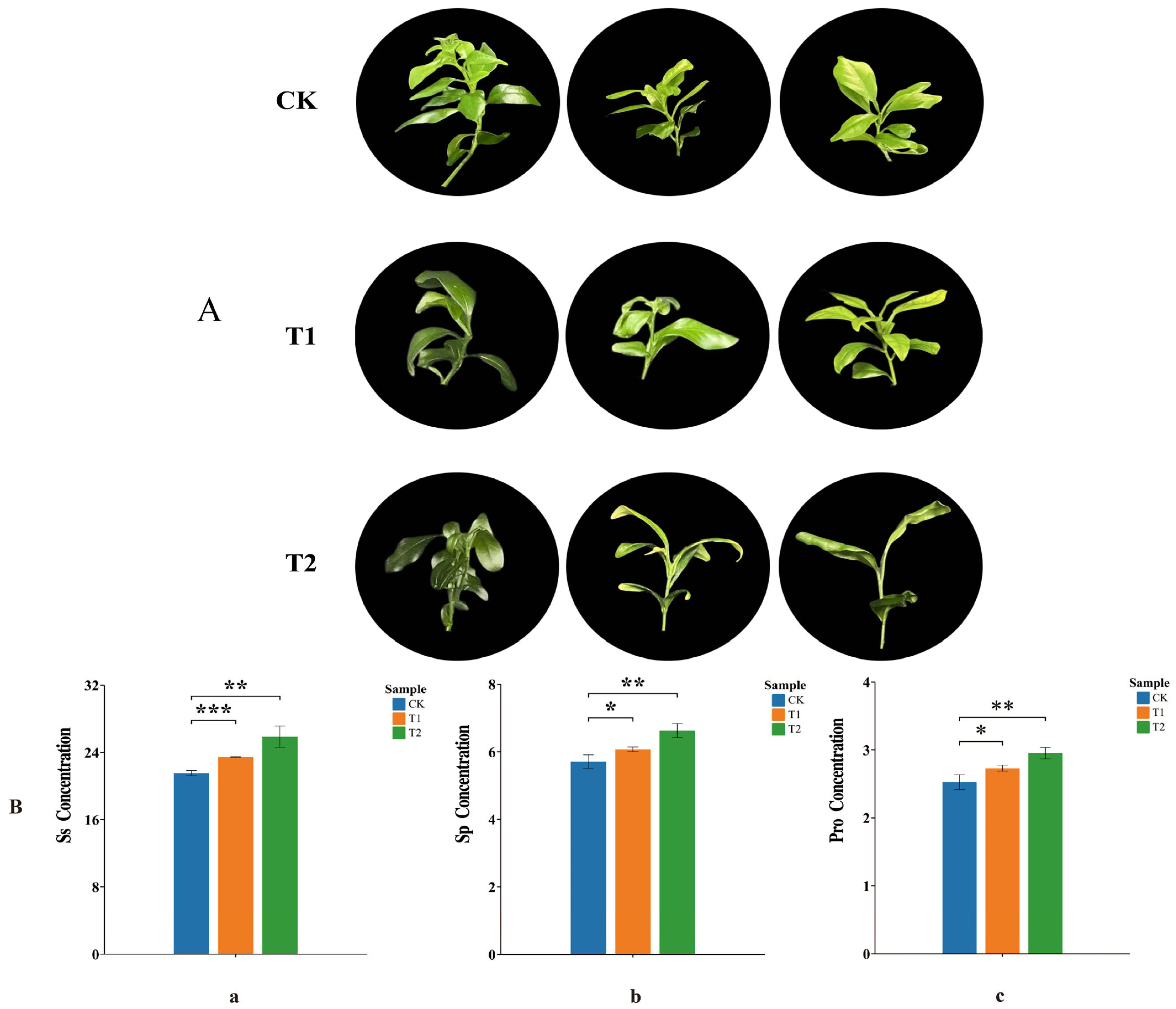
2.7. Analysis of Phenotypic and Osmoregulatory Substance Content Changes in Orah Mandarin Under Low-Temperature Stress
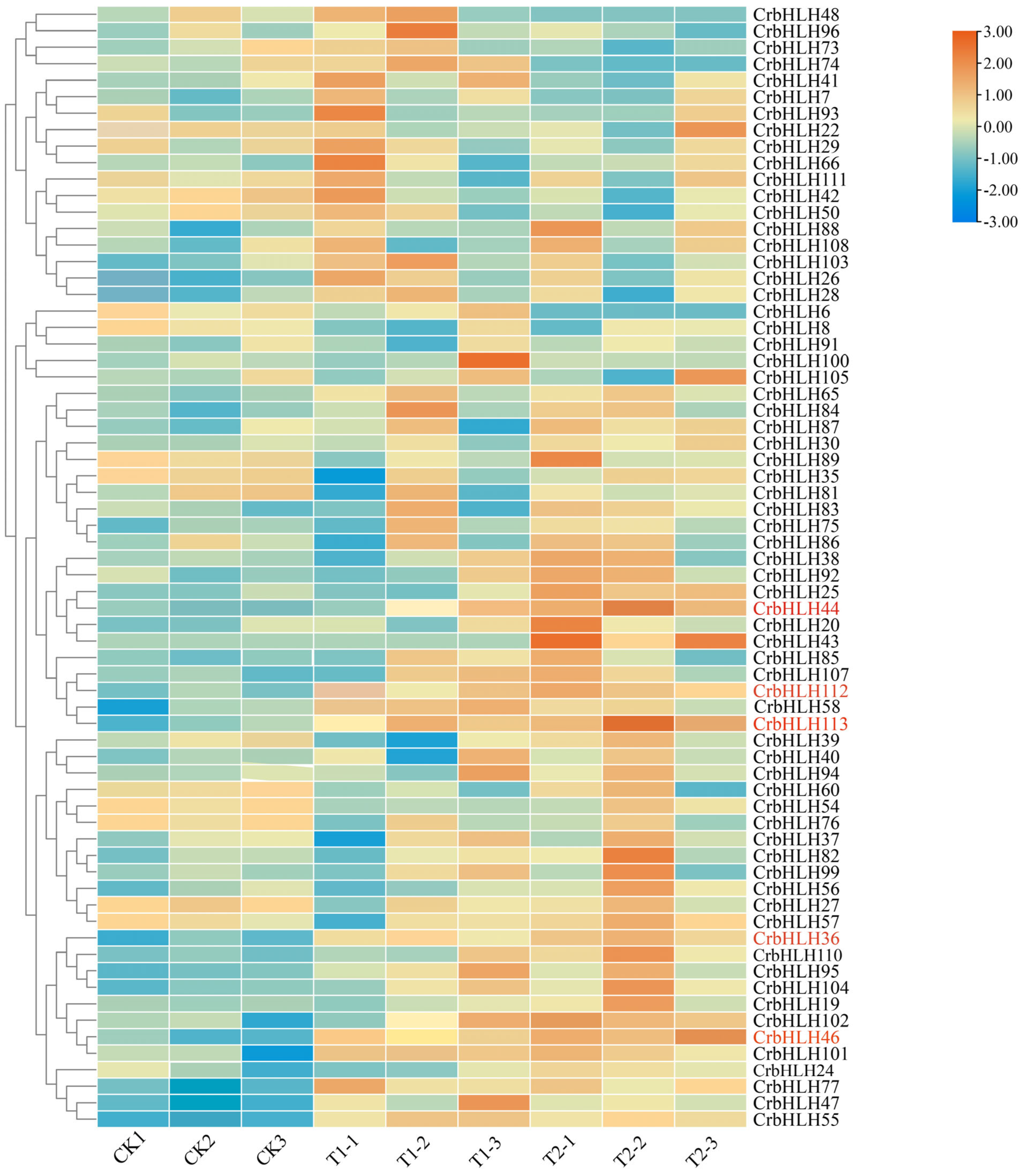
2.8. Analysis of the Expression Patterns of the CrbHLH TF Family in Orah Mandarin Under Low-Temperature Stress
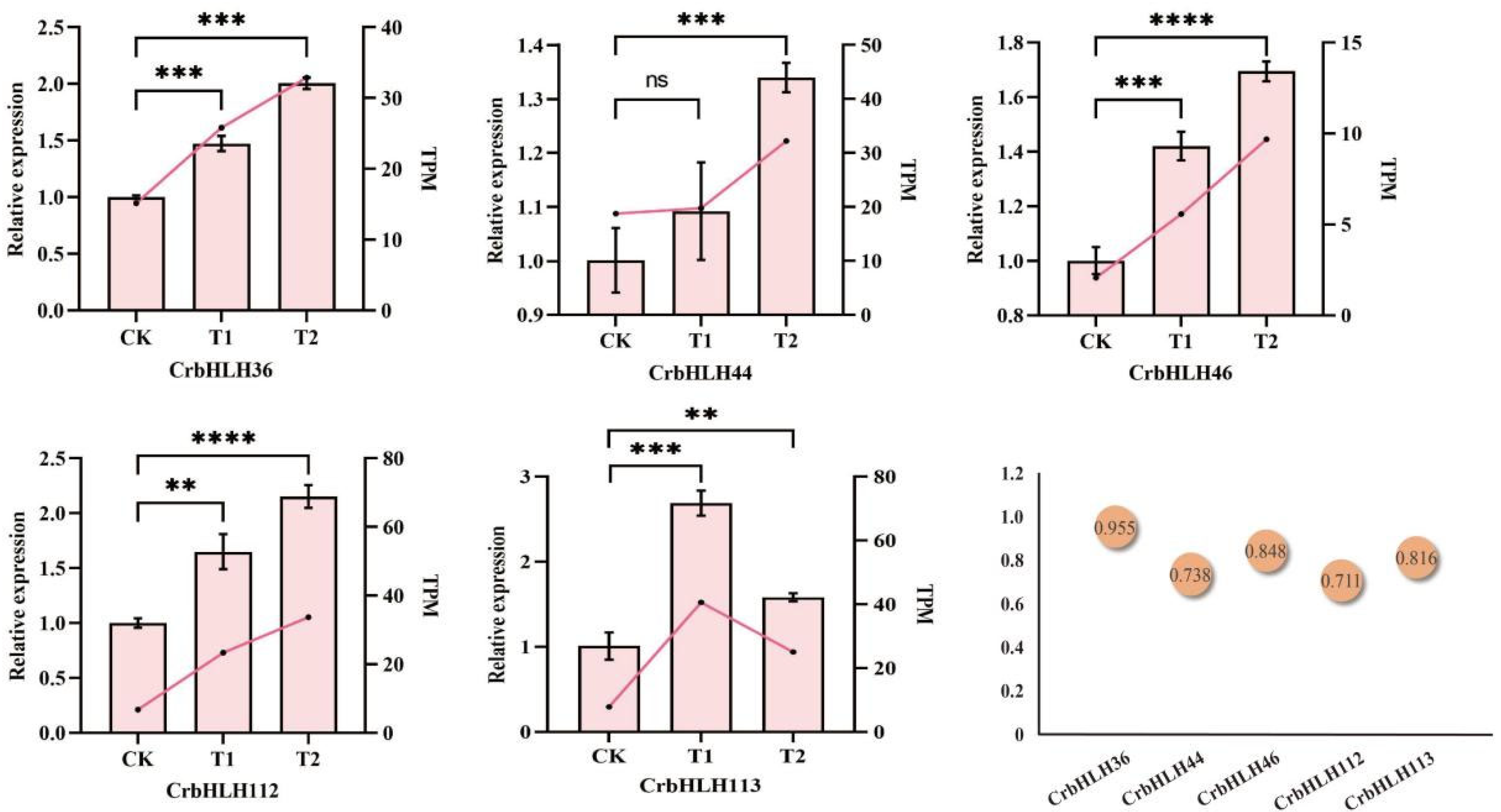
2.9. qRT-PCR
3. Discussion
4. Materials and Methods
4.1. Identification and Physicochemical Properties of the bHLH TF Family
4.2. Evolutionary Analysis
4.3. Analysis of Gene Structure and Conserved Motifs
4.4. Analysis of Cis-Acting Elements
4.5. GO Enrichment Analysis
4.6. Analysis of Protein Network Interactions
4.7. Analysis of CrbHLH Expression Patterns in Orah Mandarin in Response to Low-Temperature Stress
4.8. Determination of Soluble Sugar Content Under Low-Temperature Stress
4.9. Determination of Soluble Protein Content Under Low-Temperature Stress
4.10. Determination of the Proline Content Under Low-Temperature Stress
4.11. Analysis of the Expression Patterns of the CrbHLH TF Family Under Low-Temperature Stress
4.12. Verification via qRT-PCR Under Low-Temperature Stress
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
| Sequence ID | Gene Name | Number of Amino Acids (aa) | Molecular Weight (kDa) | Theoretical pI | Instability Index | Aliphatic Index | Grand Average of Hydropathicity (GRAVY) |
|---|---|---|---|---|---|---|---|
| MSYJ191700.1 | CrbHLH1 | 146 | 16,347.6 | 6.92 | 44.51 | 92.12 | −0.472 |
| MSYJ273420.1 | CrbHLH2 | 164 | 18,394.11 | 6.97 | 48.49 | 93.29 | −0.349 |
| MSYJ273410.1 | CrbHLH3 | 142 | 15,916.98 | 5.95 | 64.53 | 87.25 | −0.436 |
| MSYJ045200.1 | CrbHLH4 | 78 | 6308.10 | 9.18 | 54.75 | 70.35 | −0.726 |
| MSYJ152660.1 | CrbHLH5 | 54 | 6209.97 | 9.40 | 58.85 | 72.41 | −0.785 |
| MSYJ073010.1 | CrbHLH6 | 181 | 20,709.95 | 9.22 | 42.80 | 84.53 | −0.470 |
| MSYJ231530.1 | CrbHLH7 | 181 | 20,682.89 | 9.05 | 43.20 | 90.39 | −0.403 |
| MSYJ185140.7 | CrbHLH8 | 189 | 21,165.74 | 6.09 | 45.38 | 74.87 | −0.629 |
| MSYJ246180.1 | CrbHLH9 | 331 | 36,216.05 | 8.18 | 55.35 | 75.17 | −0.495 |
| MSYJ258050.1 | CrbHLH10 | 265 | 29,635.65 | 6.98 | 55.38 | 74.72 | −0.505 |
| MSYJ210310.1 | CrbHLH11 | 177 | 19,651.56 | 8.35 | 65.55 | 96.21 | −0.326 |
| MSYJ107100.1 | CrbHLH12 | 246 | 28,324.92 | 6.32 | 71.43 | 79.23 | −0.722 |
| MSYJ210300.1 | CrbHLH13 | 245 | 27,704.23 | 6 | 39.57 | 88.65 | −0.559 |
| MSYJ118000.1 | CrbHLH14 | 337 | 36,863.42 | 5.06 | 59.38 | 85.64 | −0.432 |
| MSYJ166830.1 | CrbHLH15 | 183 | 20,360.77 | 6.96 | 43.26 | 116.07 | 0.074 |
| MSYJ064870.1 | CrbHLH16 | 329 | 36,393.74 | 5.58 | 68.81 | 77.14 | −0.469 |
| MSYJ048500.1 | CrbHLH17 | 310 | 34,673.10 | 6.84 | 63.69 | 67.94 | −0.471 |
| MSYJ229290.1 | CrbHLH18 | 300 | 34,083.89 | 5.10 | 62.91 | 79.33 | −0.641 |
| MSYJ104770.2 | CrbHLH19 | 424 | 48,007.90 | 5.35 | 57.36 | 77.26 | −0.654 |
| MSYJ112160.1 | CrbHLH20 | 415 | 47,086.40 | 5.61 | 70.83 | 78.94 | −0.551 |
| MSYJ179400.1 | CrbHLH21 | 355 | 39,839.61 | 5.62 | 49.24 | 78.85 | −0.539 |
| MSYJ107260.3 | CrbHLH22 | 474 | 53,314.32 | 5.65 | 52.49 | 72.41 | −0.686 |
| MSYJ218540.1 | CrbHLH23 | 689 | 76,292.21 | 4.97 | 64.27 | 77.52 | −0.587 |
| MSYJ233170.1 | CrbHLH24 | 613 | 68,767.58 | 5.84 | 45.03 | 83.15 | −0.466 |
| MSYJ110120.1 | CrbHLH25 | 656 | 73,535.07 | 5.18 | 53.80 | 82.21 | −0.434 |
| MSYJ111470.2 | CrbHLH26 | 492 | 54,578.53 | 7.73 | 38.01 | 79.25 | −0.460 |
| MSYJ219740.1 | CrbHLH27 | 502 | 55,604.62 | 6.16 | 46.40 | 73.37 | −0.466 |
| MSYJ000950.1 | CrbHLH28 | 683 | 74,406.52 | 5.50 | 53.34 | 66.54 | −0.613 |
| MSYJ269980.1 | CrbHLH29 | 519 | 58,204.60 | 6.33 | 39.60 | 78.81 | −0.447 |
| MSYJ111900.1 | CrbHLH30 | 512 | 56,810.90 | 5.56 | 60.06 | 78.26 | −0.538 |
| MSYJ143890.1 | CrbHLH31 | 316 | 35,270.32 | 7.75 | 41.30 | 85.16 | −0.345 |
| MSYJ047470.1 | CrbHLH32 | 366 | 40,702.18 | 8.41 | 50.28 | 78.33 | −0.541 |
| MSYJ047460.1 | CrbHLH33 | 364 | 40,665.78 | 6.12 | 58.04 | 80.58 | −0.477 |
| MSYJ242210.1 | CrbHLH34 | 212 | 23,975.01 | 9.66 | 53.17 | 101.08 | −0.320 |
| MSYJ247350.1 | CrbHLH35 | 341 | 37,620.01 | 7.67 | 60.58 | 56.95 | −1.002 |
| MSYJ232700.4 | CrbHLH36 | 241 | 26,523.52 | 5.31 | 59.72 | 82.95 | −0.619 |
| MSYJ239490.2 | CrbHLH37 | 214 | 24,457.47 | 5.52 | 61.03 | 67.10 | −0.946 |
| MSYJ183770.1 | CrbHLH38 | 235 | 25,755.34 | 6.75 | 51.52 | 75.49 | −0.514 |
| MSYJ228520.1 | CrbHLH39 | 236 | 25,950.56 | 8.36 | 57.32 | 74.45 | −0.553 |
| MSYJ237760.1 | CrbHLH40 | 250 | 27,493.21 | 5.75 | 55.86 | 74.2 | −0.506 |
| MSYJ095510.1 | CrbHLH41 | 506 | 56,290.32 | 7.07 | 66.52 | 81.64 | −0.418 |
| MSYJ238820.1 | CrbHLH42 | 234 | 26,616.07 | 6.67 | 35.94 | 76.28 | −0.711 |
| MSYJ000300.1 | CrbHLH43 | 331 | 37,194.71 | 4.57 | 58.16 | 71.36 | −0.802 |
| MSYJ208070.1 | CrbHLH44 | 252 | 28,837.98 | 7.64 | 58.62 | 85.12 | −0.437 |
| MSYJ261990.1 | CrbHLH45 | 459 | 51,646.69 | 6.16 | 44.83 | 85.19 | −0.434 |
| MSYJ280970.1 | CrbHLH46 | 559 | 61,182.96 | 5.27 | 46.09 | 71.18 | −0.606 |
| MSYJ047620.1 | CrbHLH47 | 353 | 39,507.35 | 5.1 | 62.24 | 78.5 | −0.490 |
| MSYJ065830.1 | CrbHLH48 | 273 | 31,273.05 | 4.70 | 55.38 | 69.34 | −0.638 |
| MSYJ143230.1 | CrbHLH49 | 611 | 68,556.92 | 4.89 | 38.49 | 78.63 | −0.543 |
| MSYJ189770.2 | CrbHLH50 | 250 | 28,719.09 | 8.34 | 50.89 | 90.12 | −0.36 |
| MSYJ018900.1 | CrbHLH51 | 367 | 40,965.20 | 5.08 | 33.48 | 81.34 | −0.374 |
| MSYJ018890.1 | CrbHLH52 | 287 | 31,509.16 | 5.25 | 38.47 | 81.57 | −0.497 |
| MSYJ018870.1 | CrbHLH53 | 210 | 23,234.97 | 4.93 | 47.42 | 69.62 | −0.655 |
| MSYJ164470.2 | CrbHLH54 | 269 | 29,985.98 | 5.84 | 54.59 | 92.42 | −0.339 |
| MSYJ186060.1 | CrbHLH55 | 264 | 28,785.67 | 6.86 | 49.62 | 83.94 | −0.423 |
| MSYJ050860.1 | CrbHLH56 | 279 | 31,002.13 | 8.85 | 50.67 | 81.54 | −0.468 |
| MSYJ065270.1 | CrbHLH57 | 584 | 65,471.04 | 8.82 | 52.46 | 74.97 | −0.530 |
| MSYJ209320.2 | CrbHLH58 | 364 | 40,665.61 | 6.44 | 68.19 | 77.23 | −0.711 |
| MSYJ048190.1 | CrbHLH59 | 225 | 25,652.07 | 9.93 | 52.35 | 97.82 | −0.371 |
| MSYJ035990.1 | CrbHLH60 | 238 | 26,927.67 | 7.01 | 61.37 | 91.39 | −0.405 |
| MSYJ134110.1 | CrbHLH61 | 293 | 31,857.65 | 6.70 | 55.86 | 66.89 | −0.472 |
| MSYJ245010.1 | CrbHLH62 | 376 | 40,928.65 | 4.78 | 59.06 | 64.68 | −0.769 |
| MSYJ066560.1 | CrbHLH63 | 366 | 40,651.83 | 5.27 | 49.31 | 65.33 | −0.706 |
| MSYJ076010.1 | CrbHLH64 | 169 | 19,014.44 | 7.07 | 56.25 | 60.65 | −0.615 |
| MSYJ187200.1 | CrbHLH65 | 241 | 26,833.87 | 5.53 | 54.51 | 60.25 | −0.663 |
| MSYJ049300.1 | CrbHLH66 | 417 | 46,170.88 | 5.85 | 46.31 | 64.30 | −0.746 |
| MSYJ131440.1 | CrbHLH67 | 892 | 102,017.78 | 6.58 | 50.21 | 84.69 | −0.306 |
| MSYJ257940.1 | CrbHLH68 | 272 | 29,959.74 | 8.61 | 49.68 | 65.00 | −0.518 |
| MSYJ182490.1 | CrbHLH69 | 267 | 29,843.47 | 9.51 | 59.70 | 71.99 | −0.456 |
| MSYJ082400.1 | CrbHLH70 | 152 | 17,144.66 | 10.13 | 51.79 | 81.58 | −0.513 |
| MSYJ036660.1 | CrbHLH71 | 291 | 32,246.84 | 6.33 | 42.26 | 63.09 | −0.686 |
| MSYJ223830.1 | CrbHLH72 | 247 | 27,979.54 | 5.99 | 55.07 | 86.8 | −0.533 |
| MSYJ183960.1 | CrbHLH73 | 493 | 53,680.28 | 6.14 | 59.63 | 54.08 | −0.640 |
| MSYJ201180.1 | CrbHLH74 | 459 | 499,898.70 | 6.83 | 61.56 | 53.99 | −0.688 |
| MSYJ256210.1 | CrbHLH75 | 431 | 46,477.76 | 6.86 | 65.44 | 69.28 | −0.468 |
| MSYJ258490.2 | CrbHLH76 | 257 | 27,958.09 | 5.49 | 57.57 | 73.58 | −0.585 |
| MSYJ219890.3 | CrbHLH77 | 748 | 82,439.8 | 5.78 | 41.78 | 80.6 | −0.366 |
| MSYJ160330.1 | CrbHLH78 | 93 | 10,447.76 | 7.94 | 93.56 | 101.72 | −0.490 |
| MSYJ232630.1 | CrbHLH79 | 91 | 10,315.60 | 9.09 | 79.13 | 95.38 | −0.652 |
| MSYJ120230.1 | CrbHLH80 | 91 | 10,360.86 | 9.09 | 84.39 | 108.24 | −0.413 |
| MSYJ265660.2 | CrbHLH81 | 404 | 45,419.53 | 6.90 | 60.68 | 59.18 | −0.910 |
| MSYJ116150.1 | CrbHLH82 | 362 | 39,860.15 | 9.08 | 49.78 | 59.50 | −0.744 |
| MSYJ162360.1 | CrbHLH83 | 853 | 93,338.07 | 7.54 | 43.95 | 56.08 | −0.807 |
| MSYJ225570.1 | CrbHLH84 | 371 | 39,339.29 | 8.59 | 62.21 | 58.89 | −0.599 |
| MSYJ281850.1 | CrbHLH85 | 941 | 103,706.16 | 8.34 | 48.03 | 67.89 | −0.493 |
| MSYJ250810.2 | CrbHLH86 | 535 | 58,268.31 | 6.25 | 61.16 | 49.50 | −0.717 |
| MSYJ174150.3 | CrbHLH87 | 744 | 79,818.85 | 5.99 | 54.57 | 60.94 | −0.638 |
| MSYJ112680.1 | CrbHLH88 | 1340 | 149,772.15 | 8.88 | 62.61 | 63.25 | −0.893 |
| MSYJ258060.1 | CrbHLH89 | 293 | 31,542.93 | 5.16 | 60.79 | 69.93 | −0.490 |
| MSYJ182580.1 | CrbHLH90 | 409 | 44,239.29 | 6.33 | 56.81 | 75.75 | −0.466 |
| MSYJ176470.1 | CrbHLH91 | 419 | 46,235.85 | 8.62 | 62.87 | 54.80 | −0.755 |
| MSYJ108410.1 | CrbHLH92 | 464 | 50,717.49 | 8.51 | 59.94 | 55.75 | −0.638 |
| MSYJ074340.1 | CrbHLH93 | 461 | 48,308.26 | 5.61 | 47.01 | 62.08 | −0.590 |
| MSYJ243680.2 | CrbHLH94 | 338 | 35,982.21 | 5.43 | 62.20 | 69.67 | −0.451 |
| MSYJ219480.1 | CrbHLH95 | 294 | 31,303.42 | 5.85 | 55.82 | 79.35 | −0.352 |
| MSYJ122980.1 | CrbHLH96 | 519 | 56,318.99 | 4.96 | 51.52 | 80.42 | −0.462 |
| MSYJ277260.1 | CrbHLH97 | 396 | 44,630.71 | 6.22 | 56.88 | 55.93 | −0.784 |
| MSYJ277250.1 | CrbHLH98 | 279 | 31,322.64 | 8.90 | 50.40 | 73.15 | −0.313 |
| MSYJ265030.2 | CrbHLH99 | 341 | 38,175.63 | 6.35 | 57.82 | 67.24 | −0.862 |
| MSYJ038780.1 | CrbHLH100 | 340 | 37,950.64 | 5.45 | 64.05 | 55.38 | −0.945 |
| MSYJ177500.1 | CrbHLH101 | 565 | 61,509.43 | 9.01 | 51.53 | 58.73 | −0.829 |
| MSYJ280900.2 | CrbHLH102 | 370 | 41,598.09 | 6.14 | 42.93 | 60.68 | −0.801 |
| MSYJ066230.1 | CrbHLH103 | 480 | 52,613.39 | 7.12 | 45.28 | 67.02 | −0.602 |
| MSYJ116110.1 | CrbHLH104 | 756 | 83,682.97 | 5.99 | 45.16 | 74.30 | −0.590 |
| MSYJ090140.1 | CrbHLH105 | 553 | 60,065.44 | 6.06 | 51.20 | 61.81 | −0.724 |
| MSYJ161620.1 | CrbHLH106 | 277 | 31,230.58 | 8.86 | 40.73 | 78.92 | −0.495 |
| MSYJ232170.1 | CrbHLH107 | 276 | 29,821.98 | 5.51 | 53.51 | 60.87 | −0.697 |
| MSYJ287560.1 | CrbHLH108 | 256 | 28,558.69 | 6.37 | 55.97 | 55.62 | −0.927 |
| MSYJ221570.1 | CrbHLH109 | 72 | 8430.11 | 9.46 | 39.93 | 124.17 | 0.266 |
| MSYJ208710.3 | CrbHLH110 | 558 | 59,511.88 | 5.90 | 53.72 | 58.41 | −0.672 |
| MSYJ221550.1 | CrbHLH111 | 481 | 52,707.83 | 7.03 | 49.35 | 65.68 | −0.696 |
| MSYJ174650.1 | CrbHLH112 | 433 | 48,390.10 | 6.29 | 54.59 | 65.57 | −0.718 |
| MSYJ037560.2 | CrbHLH113 | 363 | 41,522.10 | 8.91 | 48.23 | 63.91 | −0.567 |
| MSYJ133760.1 | CrbHLH114 | 242 | 26,942.30 | 6.39 | 48.64 | 65.29 | −0.510 |
References
- Turner, T.; Burri, B. Potential Nutritional Benefits of Current Citrus Consumption. Agriculture 2013, 3, 170–187. [Google Scholar] [CrossRef]
- Sun, Q.; He, Z.; Wei, R.; Zhang, Y.; Ye, J.; Chai, L.; Xie, Z.; Guo, W.; Xu, J.; Cheng, Y.; et al. The Transcriptional Regulatory Module CsHB5-CsbZIP44 Positively Regulates Abscisic Acid-mediated Carotenoid Biosynthesis in Citrus (Citrus spp.). Plant Biotechnol. J. 2024, 22, 722–737. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y. Physiological Mechanism of Seasonal Change of Phosphorus in Leaves and Roots of Different Citrus Cultivars. Master’s Thesis, Southwest University, Chongqing, China, 2024. [Google Scholar]
- Zhu, P. Comparative Study on Fruit Yield and Quality of Citrus in Different Climate Zones. Master’s Thesis, Southwest University, Chongqing, China, 2020. [Google Scholar] [CrossRef]
- Koehler, G.; Wilson, R.C.; Goodpaster, J.V.; Sønsteby, A.; Lai, X.; Witzmann, F.A.; You, J.-S.; Rohloff, J.; Randall, S.K.; Alsheikh, M. Proteomic Study of Low-Temperature Responses in Strawberry Cultivars (Fragaria × ananassa) That Differ in Cold Tolerance. Plant Physiol. 2012, 159, 1787–1805. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Hu, W.; Song, S.; Ye, X.; Ding, Z.; Liu, J.; Wang, Z.; Li, J.; Hou, X.; Xu, B.; et al. MaDREB1F Confers Cold and Drought Stress Resistance through Common Regulation of Hormone Synthesis and Protectant Metabolite Contents in Banana. Hortic. Res. 2023, 10, uhac275. [Google Scholar] [CrossRef]
- Liang, G.; He, H.; Nai, G.; Feng, L.; Li, Y.; Zhou, Q.; Ma, Z.; Yue, Y.; Chen, B.; Mao, J. Genome-Wide Identification of BAM Genes in Grapevine (Vitis vinifera L.) and Ectopic Expression of VvBAM1 Modulating Soluble Sugar Levels to Improve Low-Temperature Tolerance in Tomato. BMC Plant Biol. 2021, 21, 156. [Google Scholar] [CrossRef]
- Han, X.; Yao, F.; Xue, T.; Wang, Z.; Wang, Y.; Cao, X.; Hui, M.; Wu, D.; Li, Y.; Wang, H.; et al. Sprayed Biodegradable Liquid Film Improved the Freezing Tolerance of Cv. Cabernet Sauvignon by up-Regulating Soluble Protein and Carbohydrate Levels and Alleviating Oxidative Damage. Front. Plant Sci. 2022, 13, 1021483. [Google Scholar] [CrossRef]
- Ghasemi Soloklui, A.A.; Ershadi, A.; Fallahi, E. Evaluation of Cold Hardiness in Seven Iranian Commercial Pomegranate (Punica granatum L.) Cultivars. HortScience 2012, 47, 1821–1825. [Google Scholar] [CrossRef]
- Meraj, T.A.; Fu, J.; Raza, M.A.; Zhu, C.; Shen, Q.; Xu, D.; Wang, Q. Transcriptional Factors Regulate Plant Stress Responses Through Mediating Secondary Metabolism. Genes 2020, 11, 346. [Google Scholar] [CrossRef]
- Sun, W.; Ma, Z.; Chen, H.; Liu, M. MYB Gene Family in Potato (Solanum tuberosum L.): Genome-Wide Identification of Hormone-Responsive Reveals Their Potential Functions in Growth and Development. Int. J. Mol. Sci. 2019, 20, 4847. [Google Scholar] [CrossRef]
- Tang, R.; Zhu, Y.; Yang, S.; Wang, F.; Chen, G.; Chen, J.; Zhao, K.; Liu, Z.; Peng, D. Genome-Wide Identification and Analysis of WRKY Gene Family in Melastoma Dodecandrum. Int. J. Mol. Sci. 2023, 24, 14904. [Google Scholar] [CrossRef]
- Hou, S.; Zhang, Q.; Chen, J.; Meng, J.; Wang, C.; Du, J.; Guo, Y. Genome-Wide Identification and Analysis of the GRAS Transcription Factor Gene Family in Theobroma Cacao. Genes 2022, 14, 57. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Chen, W.; Liu, R.; Shi, B.; Shu, Y.; Zhang, H. Genome-Wide Characterization and Expression Analysis of bHLH Gene Family in Physic Nut (Jatropha curcas L.). PeerJ 2022, 10, e13786. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Zhao, P.; Kong, N.; Lu, R.; Pei, Y.; Huang, C.; Ma, H.; Chen, Q. Genome-Wide Identification and Characterization of the Potato bHLH Transcription Factor Family. Genes 2018, 9, 54. [Google Scholar] [CrossRef] [PubMed]
- Atchley, W.R.; Terhalle, W.; Dress, A. Positional Dependence, Cliques, and Predictive Motifs in the bHLH Protein Domain. J. Mol. Evol. 1999, 48, 501–516. [Google Scholar] [CrossRef]
- Li, J.; Chen, S.; Yin, Y.; Shan, Q.; Zheng, C.; Chen, Y. Genome-Wide Analysis of bHLH Family Genes and Identification of Members Associated with Cold/Drought-Induced Photoinhibition in Kandelia Obovata. Int. J. Mol. Sci. 2023, 24, 15942. [Google Scholar] [CrossRef]
- Feller, A.; Machemer, K.; Braun, E.L.; Grotewold, E. Evolutionary and Comparative Analysis of MYB and bHLH Plant Transcription Factors. Plant J. 2011, 66, 94–116. [Google Scholar] [CrossRef]
- Carretero-Paulet, L.; Galstyan, A.; Roig-Villanova, I.; Martínez-García, J.F.; Bilbao-Castro, J.R.; Robertson, D.L. Genome-wide classification and evolutionary analysis of the bHLH family of transcription factors in Arabidopsis, poplar, rice, moss, and algae. Plant Physiol. 2010, 153, 1398–1412. [Google Scholar] [CrossRef]
- Mao, K.; Dong, Q.; Li, C.; Liu, C.; Ma, F. Genome Wide Identification and Characterization of Apple bHLH Transcription Factors and Expression Analysis in Response to Drought and Salt Stress. Front. Plant Sci. 2017, 8, 480. [Google Scholar] [CrossRef]
- Sun, H.; Fan, H.-J.; Ling, H.-Q. Genome-Wide Identification and Characterization of the bHLH Gene Family in Tomato. BMC Genom. 2015, 16, 9. [Google Scholar] [CrossRef]
- Dong, Y.; Wang, C.; Han, X.; Tang, S.; Liu, S.; Xia, X.; Yin, W. A Novel bHLH Transcription Factor PebHLH35 from Populus Euphratica Confers Drought Tolerance through Regulating Stomatal Development, Photosynthesis and Growth in Arabidopsis. Biochem. Biophys. Res. Commun. 2014, 450, 453–458. [Google Scholar] [CrossRef]
- Ito, S.; Song, Y.H.; Josephson-Day, A.R.; Miller, R.J.; Breton, G.; Olmstead, R.G.; Imaizumi, T. FLOWERING BHLH Transcriptional Activators Control Expression of the Photoperiodic Flowering Regulator CONSTANS in Arabidopsis. Proc. Natl. Acad. Sci. USA 2012, 109, 3582–3587. [Google Scholar] [CrossRef] [PubMed]
- Xie, X.; Li, S.; Zhang, R.; Zhao, J.; Chen, Y.; Zhao, Q.; Yao, Y.; You, C.; Zhang, X.; Hao, Y. The bHLH Transcription Factor MdbHLH3 Promotes Anthocyanin Accumulation and Fruit Colouration in Response to Low Temperature in Apples. Plant Cell Environ. 2012, 35, 1884–1897. [Google Scholar] [CrossRef] [PubMed]
- Yao, P.; Sun, Z.; Li, C.; Zhao, X.; Li, M.; Deng, R.; Huang, Y.; Zhao, H.; Chen, H.; Wu, Q. Overexpression of Fagopyrum Tataricum FtbHLH2 Enhances Tolerance to Cold Stress in Transgenic Arabidopsis. Plant Physiol. Biochem. 2018, 125, 85–94. [Google Scholar] [CrossRef]
- Yang, T.; Yao, S.; Hao, L.; Zhao, Y.; Lu, W.; Xiao, K. Wheat bHLH-Type Transcription Factor Gene TabHLH1 Is Crucial in Mediating Osmotic Stresses Tolerance through Modulating Largely the ABA-Associated Pathway. Plant Cell Rep. 2016, 35, 2309–2323. [Google Scholar] [CrossRef] [PubMed]
- Zhao, M.; Li, J.; Zhu, L.; Chang, P.; Li, L.; Zhang, L. Identification and Characterization of MYB-bHLH-WD40 Regulatory Complex Members Controlling Anthocyanidin Biosynthesis in Blueberry Fruits Development. Genes 2019, 10, 496. [Google Scholar] [CrossRef]
- Zhao, L.; Gong, X.; Gao, J.; Dong, H.; Zhang, S.; Tao, S.; Huang, X. Transcriptomic and Evolutionary Analyses of White Pear (Pyrus bretschneideri) β-Amylase Genes Reveal Their Importance for Cold and Drought Stress Responses. Gene 2019, 689, 102–113. [Google Scholar] [CrossRef]
- Xi, D.; Yin, T.; Han, P.; Yang, X.; Zhang, M.; Du, C.; Zhang, H.; Liu, X. Genome-Wide Identification of Sweet Orange WRKY Transcription Factors and Analysis of Their Expression in Response to Infection by Penicillium digitatum. Curr. Issues Mol. Biol. 2023, 45, 1250–1271. [Google Scholar] [CrossRef]
- Seo, M.-H.; Kim, P.M. The Present and the Future of Motif-Mediated Protein-Protein Interactions. Curr. Opin. Struct. Biol. 2018, 50, 162–170. [Google Scholar] [CrossRef]
- Zhu, L.; Yin, T.; Zhang, M.; Yang, X.; Wu, J.; Cai, H.; Yang, N.; Li, X.; Wen, K.; Chen, D.; et al. Genome-Wide Identification and Expression Pattern Analysis of the Kiwifruit GRAS Transcription Factor Family in Response to Salt Stress. BMC Genom. 2024, 25, 12. [Google Scholar] [CrossRef]
- Zuo, Z.-F.; Sun, H.-J.; Lee, H.-Y.; Kang, H.-G. Identification of bHLH Genes through Genome-Wide Association Study and Antisense Expression of ZjbHLH076/ZjICE1 Influence Tolerance to Low Temperature and Salinity in Zoysia japonica. Plant Sci. 2021, 313, 111088. [Google Scholar] [CrossRef]
- Noshi, M.; Tanabe, N.; Okamoto, Y.; Mori, D.; Ohme-Takagi, M.; Tamoi, M.; Shigeoka, S. Clade Ib Basic Helix-Loop-Helix Transcription Factor, bHLH101, Acts as a Regulatory Component in Photo-Oxidative Stress Responses. Plant Sci. 2018, 274, 101–108. [Google Scholar] [CrossRef] [PubMed]
- Yu, D.; Wei, W.; Fan, Z.; Chen, J.; You, Y.; Huang, W.; Zhan, J. VabHLH137 Promotes Proanthocyanidin and Anthocyanin Biosynthesis and Enhances Resistance to Colletotrichum gloeosporioides in Grapevine. Hortic. Res. 2022, 10, uhac261. [Google Scholar] [CrossRef] [PubMed]
- Wu, T.-Y.; Goh, H.; Azodi, C.B.; Krishnamoorthi, S.; Liu, M.-J.; Urano, D. Evolutionarily Conserved Hierarchical Gene Regulatory Networks for Plant Salt Stress Response. Nat. Plants 2021, 7, 787–799. [Google Scholar] [CrossRef] [PubMed]
- Zeng, M.; Zhu, P.; Tang, Y.; Lin, Y.; He, T.; Rong, J.; Zheng, Y.; Chen, L. Genome-Wide Identification and Role of the bHLH Gene Family in Dendrocalamus latiflorus Flowering Regulation. Int. J. Mol. Sci. 2024, 25, 10837. [Google Scholar] [CrossRef]
- Wang, P.; Su, L.; Gao, H.; Jiang, X.; Wu, X.; Li, Y.; Zhang, Q.; Wang, Y.; Ren, F. Genome-Wide Characterization of bHLH Genes in Grape and Analysis of Their Potential Relevance to Abiotic Stress Tolerance and Secondary Metabolite Biosynthesis. Front. Plant Sci. 2018, 9, 64. [Google Scholar] [CrossRef]
- Lim, S.-H.; Kim, D.-H.; Jung, J.-A.; Lee, J.-Y. Alternative Splicing of the Basic Helix–Loop–Helix Transcription Factor Gene CmbHLH2 Affects Anthocyanin Biosynthesis in Ray Florets of Chrysanthemum (Chrysanthemum morifolium). Front. Plant Sci. 2021, 12, 669315. [Google Scholar] [CrossRef]
- Zhou, X.; Liao, Y.; Kim, S.-U.; Chen, Z.; Nie, G.; Cheng, S.; Ye, J.; Xu, F. Genome-Wide Identification and Characterization of bHLH Family Genes from Ginkgo biloba. Sci. Rep. 2020, 10, 13723. [Google Scholar] [CrossRef]
- Liu, R.; Song, J.; Liu, S.; Chen, C.; Zhang, S.; Wang, J.; Xiao, Y.; Cao, B.; Lei, J.; Zhu, Z. Genome-Wide Identification of the Capsicum bHLH Transcription Factor Family: Discovery of a Candidate Regulator Involved in the Regulation of Species-Specific Bioactive Metabolites. BMC Plant Biol. 2021, 21, 262. [Google Scholar] [CrossRef]
- Li, J.; Wang, T.; Han, J.; Ren, Z. Genome-Wide Identification and Characterization of Cucumber bHLH Family Genes and the Functional Characterization of CsbHLH041 in NaCl and ABA Tolerance in Arabidopsis and Cucumber. BMC Plant Biol. 2020, 20, 272. [Google Scholar] [CrossRef]
- Liang, J.; Fang, Y.; An, C.; Yao, Y.; Wang, X.; Zhang, W.; Liu, R.; Wang, L.; Aslam, M.; Cheng, Y.; et al. Genome-Wide Identification and Expression Analysis of the bHLH Gene Family in Passion Fruit (Passiflora edulis) and Its Response to Abiotic Stress. Int. J. Biol. Macromol. 2023, 225, 389–403. [Google Scholar] [CrossRef]
- Dong, J.; Wu, Y.; Dong, Y.; Pu, R.; Li, X.; Lyu, Y.; Bai, T.; Zhang, J. Genome-Wide Identification of the bHLH Gene Family in Rhododendron delavayi and Its Expression Analysis in Different Floral Tissues. Genes 2024, 15, 1256. [Google Scholar] [CrossRef] [PubMed]
- Hudson, K.A.; Hudson, M.E. The Basic Helix-Loop-Helix Transcription Factor Family in the Sacred Lotus, Nelumbo nucifera. Trop. Plant Biol. 2014, 7, 65–70. [Google Scholar] [CrossRef]
- Duek, P.D.; Fankhauser, C. bHLH Class Transcription Factors Take Centre Stage in Phytochrome Signalling. Trends Plant Sci. 2005, 10, 51–54. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Lv, W.; Zhang, H.; Ma, L.; Li, P.; Ge, L.; Li, G. Genome-Wide Analysis of the Basic Helix-Loop-Helix (bHLH) Transcription Factor Family in Maize. BMC Plant Biol. 2018, 18, 235. [Google Scholar] [CrossRef]
- Kamran, H.M.; Fu, X.; Wang, H.; Yang, N.; Chen, L. Genome-Wide Identification and Expression Analysis of the bHLH Transcription Factor Family in Wintersweet (Chimonanthus praecox). Int. J. Mol. Sci. 2023, 24, 13462. [Google Scholar] [CrossRef]
- Zhan, H.; Liu, H.; Ai, W.; Han, X.; Wang, Y.; Lu, X. Genome-Wide Identification and Expression Analysis of the bHLH Transcription Factor Family and its Response to Abiotic Stress in Mongolian Oak (Quercus mongolica). Curr. Issues Mol. Biol. 2023, 45, 1127–1148. [Google Scholar] [CrossRef]
- Ke, Q.; Tao, W.; Li, T.; Pan, W.; Chen, X.; Wu, X.; Nie, X.; Cui, L. Genome-Wide Identification, Evolution and Expression Analysis of Basic Helix-Loop-Helix (bHLH) Gene Family in Barley (Hordeum vulgare L.). Curr. Genom. 2020, 21, 624–644. [Google Scholar] [CrossRef]
- Xu, G.; Guo, C.; Shan, H.; Kong, H. Divergence of Duplicate Genes in Exon–Intron Structure. Proc. Natl. Acad. Sci. USA 2012, 109, 1187–1192. [Google Scholar] [CrossRef]
- De Wit, M.; Galvão, V.C.; Fankhauser, C. Light-Mediated Hormonal Regulation of Plant Growth and Development. Annu. Rev. Plant Biol. 2016, 67, 513–537. [Google Scholar] [CrossRef]
- Cheng, Y.-Z.; He, G.-Q.; Yang, S.-D.; Ma, S.-H.; Ma, J.-P.; Shang, F.-H.-Z.; Li, X.-F.; Jin, H.-Y.; Guo, D.-L. Genome-Wide Identification and Expression Analysis of JmjC Domain–Containing Genes in Grape under MTA Treatment. Funct. Integr. Genom. 2022, 22, 783–795. [Google Scholar] [CrossRef]
- Feng, Y.; Cui, R.; Wang, S.; He, M.; Hua, Y.; Shi, L.; Ye, X.; Xu, F. Transcription Factor BnaA9.WRKY47 Contributes to the Adaptation of Brassica napus to Low Boron Stress by Up-regulating the Boric Acid Channel Gene BnaA3.NIP5;1. Plant Biotechnol. J. 2020, 18, 1241–1254. [Google Scholar] [CrossRef] [PubMed]
- Radani, Y.; Li, R.; Korboe, H.M.; Ma, H.; Yang, L. Transcriptional and Post-Translational Regulation of Plant bHLH Transcription Factors during the Response to Environmental Stresses. Plants 2023, 12, 2113. [Google Scholar] [CrossRef] [PubMed]
- Goulao, L.F.; Vieira-Silva, S.; Jackson, P.A. Association of Hemicellulose- and Pectin-Modifying Gene Expression with Eucalyptus Globulus Secondary Growth. Plant Physiol. Biochem. 2011, 49, 873–881. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, D.; Gorka, M.; Erban, A.; Graf, A.; Kopka, J.; Zuther, E.; Hincha, D.K. Both Cold and Sub-Zero Acclimation Induce Cell Wall Modification and Changes in the Extracellular Proteome in Arabidopsis Thaliana. Sci. Rep. 2019, 9, 2289. [Google Scholar] [CrossRef]
- Pieslinger, A.M.; Hoepflinger, M.C.; Tenhaken, R. Cloning of Glucuronokinase from Arabidopsis Thaliana, the Last Missing Enzyme of the Myo-Inositol Oxygenase Pathway to Nucleotide Sugars. J. Biol. Chem. 2010, 285, 2902–2910. [Google Scholar] [CrossRef]
- Tenhaken, R.; Thulke, O. Cloning of an Enzyme That Synthesizes a Key Nucleotide-Sugar Precursor of Hemicellulose Biosynthesis from Soybean:UDP-Glucose Dehydrogenase. Plant Physiol. 1996, 112, 1127–1134. [Google Scholar] [CrossRef]
- Reboul, R.; Geserick, C.; Pabst, M.; Frey, B.; Wittmann, D.; Lütz-Meindl, U.; Léonard, R.; Tenhaken, R. Down-Regulation of UDP-Glucuronic Acid Biosynthesis Leads to Swollen Plant Cell Walls and Severe Developmental Defects Associated with Changes in Pectic Polysaccharides. J. Biol. Chem. 2011, 286, 39982–39992. [Google Scholar] [CrossRef]
- Thakur, N.; Flowerika; Chaturvedi, S.; Tiwari, S. Wheat Derived Glucuronokinase as a Potential Target for Regulating Ascorbic Acid and Phytic Acid Content with Increased Root Length under Drought and ABA Stresses in Arabidopsis thaliana. Plant Sci. 2023, 331, 111671. [Google Scholar] [CrossRef]
- Duan, J.; Zhang, M.; Zhang, H.; Xiong, H.; Liu, P.; Ali, J.; Li, J.; Li, Z. OsMIOX, a Myo-Inositol Oxygenase Gene, Improves Drought Tolerance through Scavenging of Reactive Oxygen Species in Rice (Oryza sativa L.). Plant Sci. 2012, 196, 143–151. [Google Scholar] [CrossRef]
- Feng, Y.J.; Zhao, W.; Li, Y.L.; Shen, Y.J.; Sun, Y.C.; Meng, X.Y.; Li, J.; Wu, W.; Zhang, G.X.; Liu, M.Y.; et al. Genome-Wide Identification of Wheat USP Gene Family and Functional Dissection of TaUSP85 Involved in Heat Tolerance. Plant Physiol. Biochem. 2025, 219, 109359. [Google Scholar] [CrossRef]
- Wen, K.; Li, X.; Yin, T.; Chen, C.; Zhu, L.; Zi, Y.; Zhao, K.; Liu, X.; Zhang, H. Genome-Wide Identification of Carotenoid Cleavage Oxygenase Genes in Orah Mandarin and the Mechanism by Which CrCCD4b1 Affects Peel Color. Sci. Hortic. 2024, 338, 113652. [Google Scholar] [CrossRef]
- Madeira, F.; Park, Y.; Lee, J.; Buso, N.; Gur, T.; Madhusoodanan, N.; Basutkar, P.; Tivey, A.R.N.; Potter, S.C.; Finn, R.D.; et al. The EMBL-EBI Search and Sequence Analysis Tools APIs in 2019. Nucleic Acids Res. 2019, 47, W636–W641. [Google Scholar] [CrossRef] [PubMed]
- Yin, T.; Han, P.; Xi, D.; Yu, W.; Zhu, L.; Du, C.; Yang, N.; Liu, X.; Zhang, H. Genome-Wide Identification, Characterization, and Expression Profile ofNBS-LRRgene Family in Sweet Orange (Citrussinensis). Gene 2023, 854, 147117. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Lu, S.; Wang, J.; Chitsaz, F.; Derbyshire, M.K.; Geer, R.C.; Gonzales, N.R.; Gwadz, M.; Hurwitz, D.I.; Marchler, G.H.; Song, J.S.; et al. CDD/SPARCLE: The Conserved Domain Database in 2020. Nucleic Acids Res. 2020, 48, D265–D268. [Google Scholar] [CrossRef]
- Zi, Y.; Zhang, M.; Yang, X.; Zhao, K.; Yin, T.; Wen, K.; Li, X.; Liu, X.; Zhang, H. Identification of the Sweet Orange (Citrus sinensis) bHLH Gene Family and the Role of CsbHLH55 and CsbHLH87 in Regulating Salt Stress. Plant Genome 2024, 17, e20502. [Google Scholar] [CrossRef]
- Katoh, K.; Rozewicki, J.; Yamada, K.D. MAFFT Online Service: Multiple Sequence Alignment, Interactive Sequence Choice and Visualization. Brief. Bioinform. 2019, 20, 1160–1166. [Google Scholar] [CrossRef]
- Nguyen, L.-T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A Fast and Effective Stochastic Algorithm for Estimating Maximum-Likelihood Phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef]
- Li, X.; Wen, K.; Zhu, L.; Chen, C.; Yin, T.; Yang, X.; Zhao, K.; Zi, Y.; Zhang, H.; Luo, X.; et al. Genome-Wide Identification and Expression Analysis of the Eriobotrya japonica TIFY Gene Family Reveals its Functional Diversity Under Abiotic Stress Conditions. BMC Genom. 2024, 25, 468. [Google Scholar] [CrossRef]
- Brown, P.; Baxter, L.; Hickman, R.; Beynon, J.; Moore, J.D.; Ott, S. MEME-LaB: Motif Analysis in Clusters. Bioinformatics 2013, 29, 1696–1697. [Google Scholar] [CrossRef]
- Lescot, M. PlantCARE, a Database of Plant Cis-Acting Regulatory Elements and a Portal to Tools for in Silico Analysis of Promoter Sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef] [PubMed]
- Wimalanathan, K.; Lawrence-Dill, C.J. Gene Ontology Meta Annotator for Plants (GOMAP). Plant Methods 2021, 17, 54. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A Fast Spliced Aligner with Low Memory Requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Shi, W. FeatureCounts: An Efficient General Purpose Program for Assigning Sequence Reads to Genomic Features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated Estimation of Fold Change and Dispersion for RNA-Seq Data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Li, J.L.; Li, H.; Zhao, J.J.; Yang, P.; Xiang, X.; Wei, S.Y.; Wang, T.; Shi, Y.J.; Huang, J.; He, F. Genome-Wide Identification and Characterization of the RZFP Gene Family and Analysis of Its Expression Pattern under Stress in Populus trichocarpa. Int. J. Biol. Macromol. 2024, 255, 128108. [Google Scholar] [CrossRef]
- Li, L.; Zhang, H.; Liu, J.; Huang, T.; Zhang, X.; Xie, H.; Guo, Y.; Wang, Q.; Zhang, P.; Qin, P. Grain Color Formation and Analysis of Correlated Genes by Metabolome and Transcriptome in Different Wheat Lines at Maturity. Front. Nutr. 2023, 10, 1112497. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Duricki, D.A.; Soleman, S.; Moon, L.D.F. Analysis of Longitudinal Data from Animals with Missing Values Using SPSS. Nat. Protoc. 2016, 11, 1112–1129. [Google Scholar] [CrossRef]




| Gene | Sequence (5′–3′) | |
|---|---|---|
| MSYJ123420.2 (Actin) | F | ACTGAGCACCACATTCCCATACA |
| R | GCAAGTCATAACTATTGGAGCCG | |
| MSYJ232700.4 | F | GAGGAGACGCAGGAAGAAGCTC |
| R | GTCAATGCTGAACCAGGAGGG | |
| MSYJ208070.1 | F | TTCCAAGCCTTGGGTGGC |
| R | GCTCGCTGGCATTGTGATAGA | |
| MSYJ280900.2 | F | GCAAGGAGGGGTCAAGCAAC |
| R | CTTGTCACAGCCAGGAACAAGC | |
| MSYJ174650.1 | F | CAACAAGTGAAGCCCGACCC |
| R | ATTCAAAGCAGCCACTGACACC | |
| MSYJ037560.2 | F | TGCAGAGCTAAAGCCGAGCAT |
| R | CTTACGGTACGAACACGGGGTA | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, C.; Li, X.; Wen, K.; Yin, T.; Tian, P.; Zhao, K.; Zhang, L.; Zhou, X.; Liu, X.; Zhang, H. Identification of the bHLH Transcription Factor Family in Orah Mandarin and the Response of CrbHLH46 to Low-Temperature Stress. Plants 2025, 14, 882. https://doi.org/10.3390/plants14060882
Chen C, Li X, Wen K, Yin T, Tian P, Zhao K, Zhang L, Zhou X, Liu X, Zhang H. Identification of the bHLH Transcription Factor Family in Orah Mandarin and the Response of CrbHLH46 to Low-Temperature Stress. Plants. 2025; 14(6):882. https://doi.org/10.3390/plants14060882
Chicago/Turabian StyleChen, Chaoying, Xulin Li, Ke Wen, Tuo Yin, Ping Tian, Ke Zhao, Li Zhang, Xianyan Zhou, Xiaozhen Liu, and Hanyao Zhang. 2025. "Identification of the bHLH Transcription Factor Family in Orah Mandarin and the Response of CrbHLH46 to Low-Temperature Stress" Plants 14, no. 6: 882. https://doi.org/10.3390/plants14060882
APA StyleChen, C., Li, X., Wen, K., Yin, T., Tian, P., Zhao, K., Zhang, L., Zhou, X., Liu, X., & Zhang, H. (2025). Identification of the bHLH Transcription Factor Family in Orah Mandarin and the Response of CrbHLH46 to Low-Temperature Stress. Plants, 14(6), 882. https://doi.org/10.3390/plants14060882






