MIKC-Type MADS-Box Gene Analysis Reveals the Role of PlSOC1 in Bud Dormancy Transition in Herbaceous Peony
Abstract
:1. Introduction
2. Results
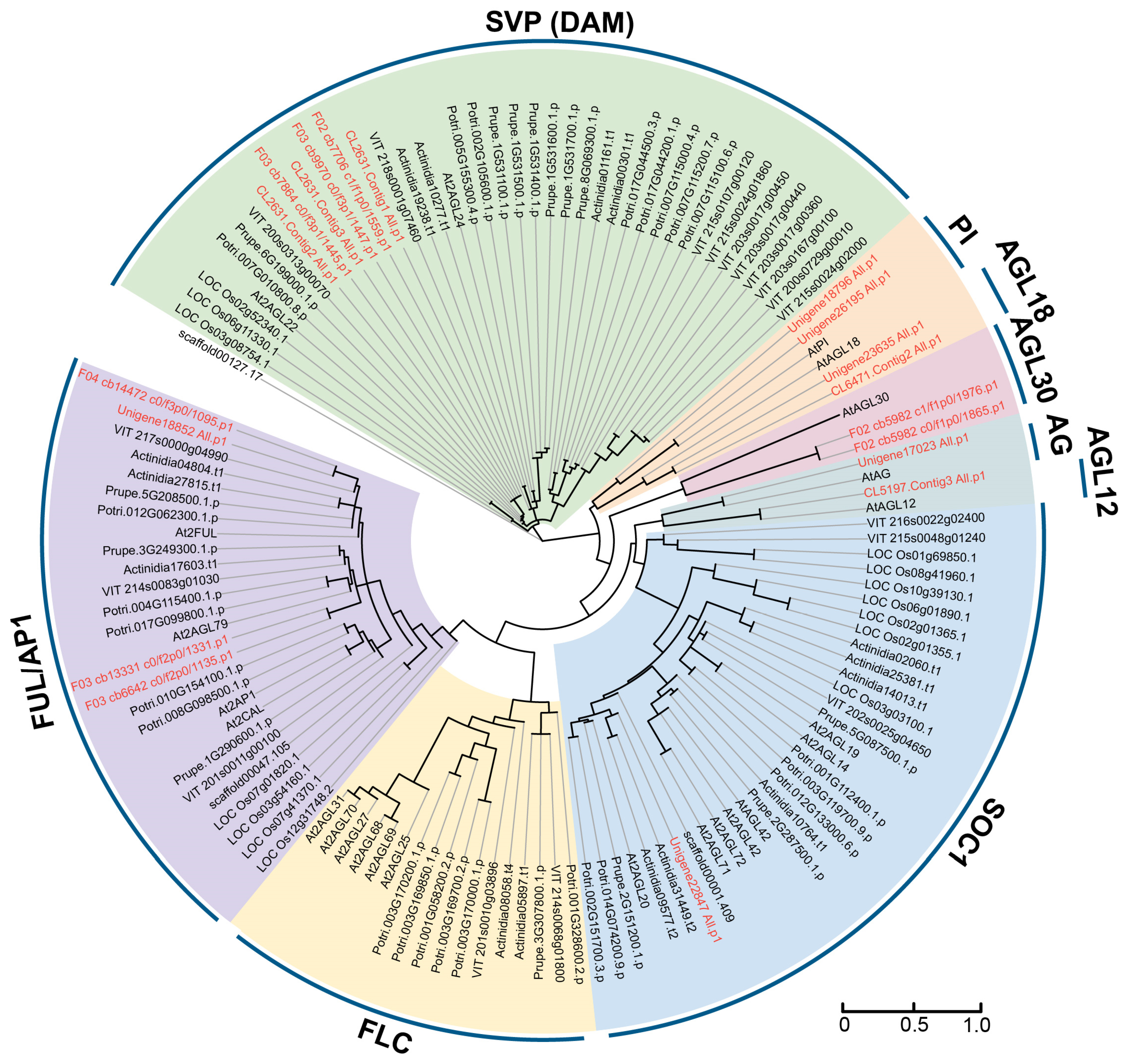
2.1. Identification and Phylogenetic Analysis of MIKC-Type Genes in Herbaceous Peony
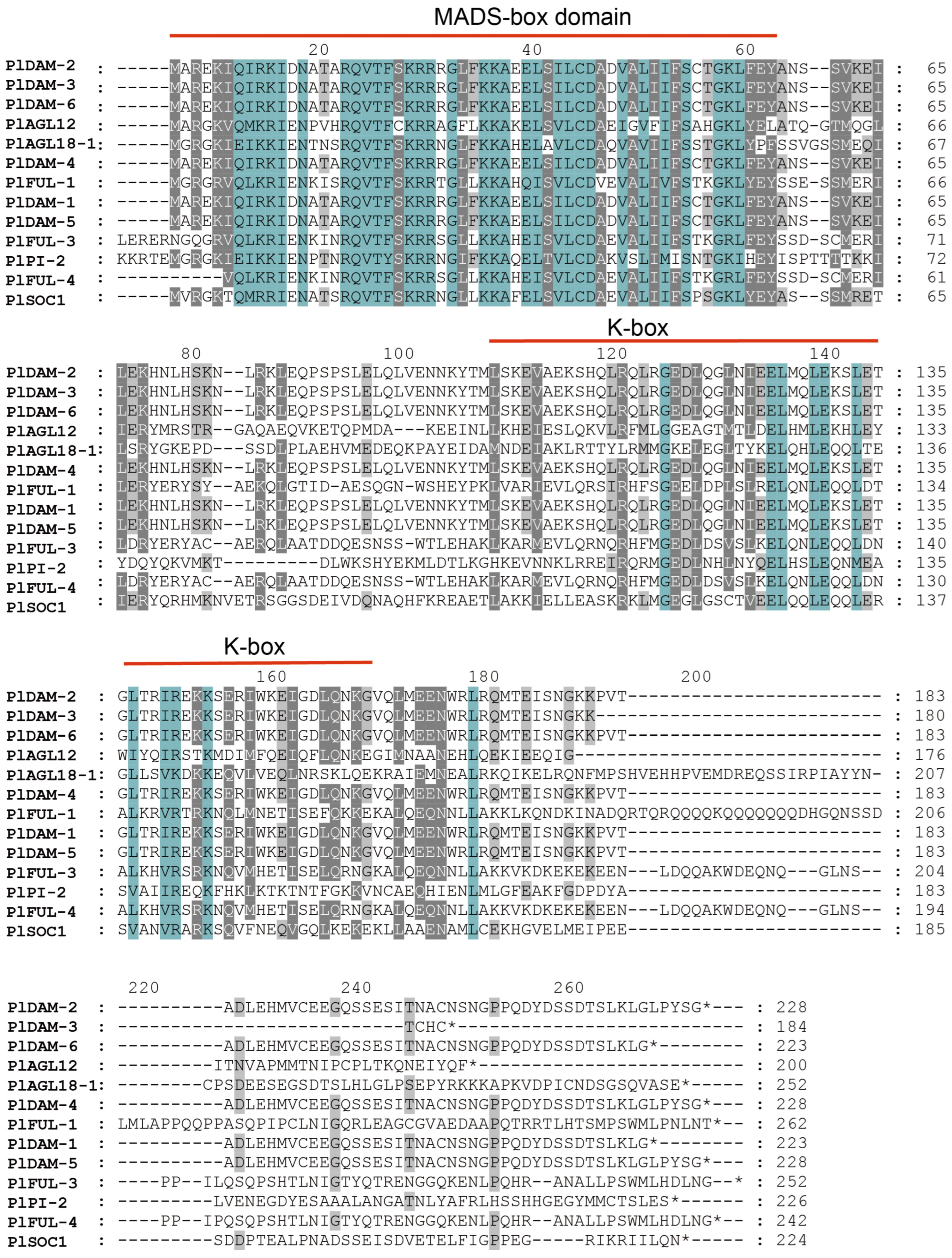
2.2. Multiple-Sequence Alignment of PlMIKCs
2.3. Protein Structure and Conserved Motif Distribution Analysis
2.4. Low-Temperature Triggers BDT in Herbaceous Peony
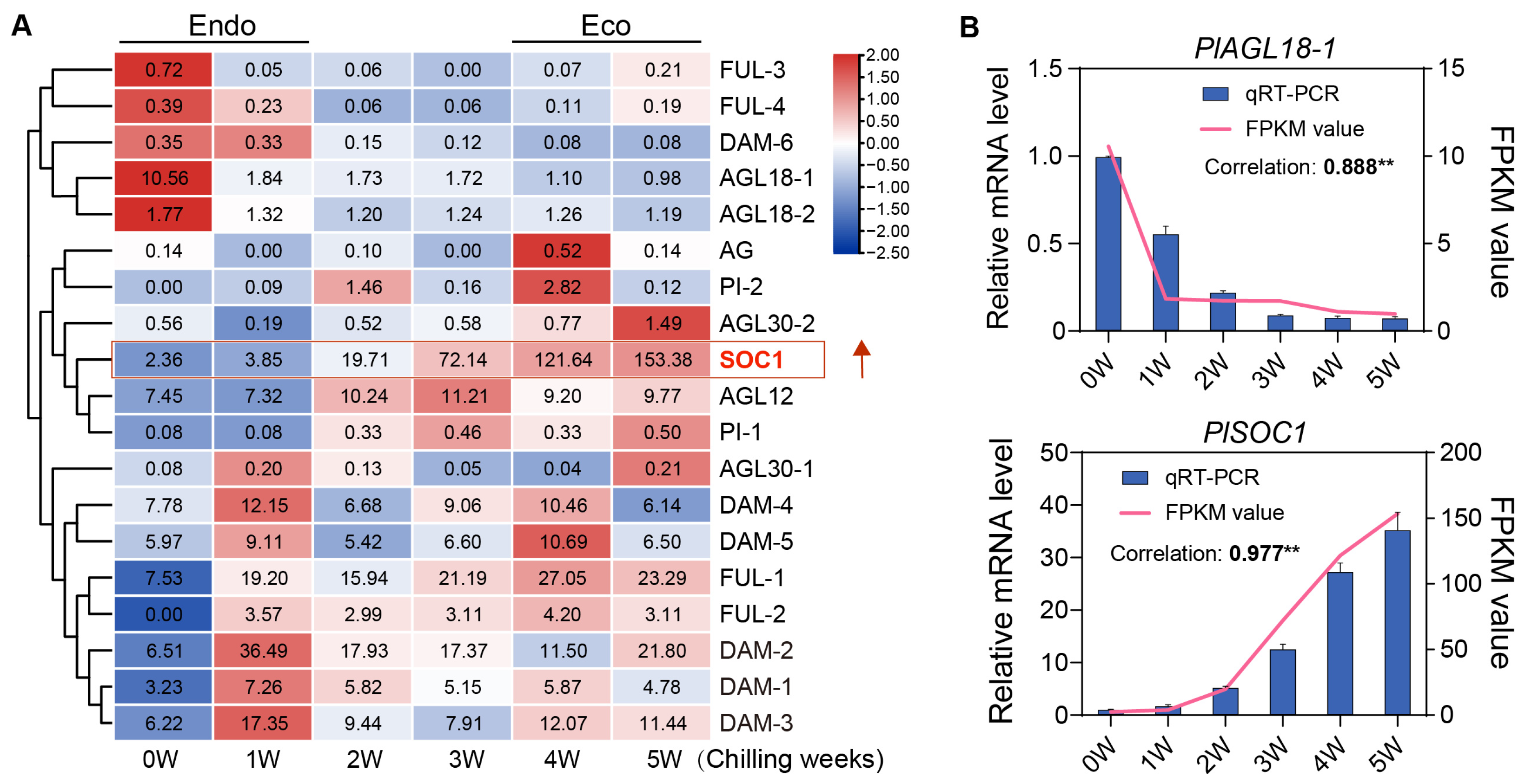
2.5. Expression Pattern of the PlMIKC Genes During BDT
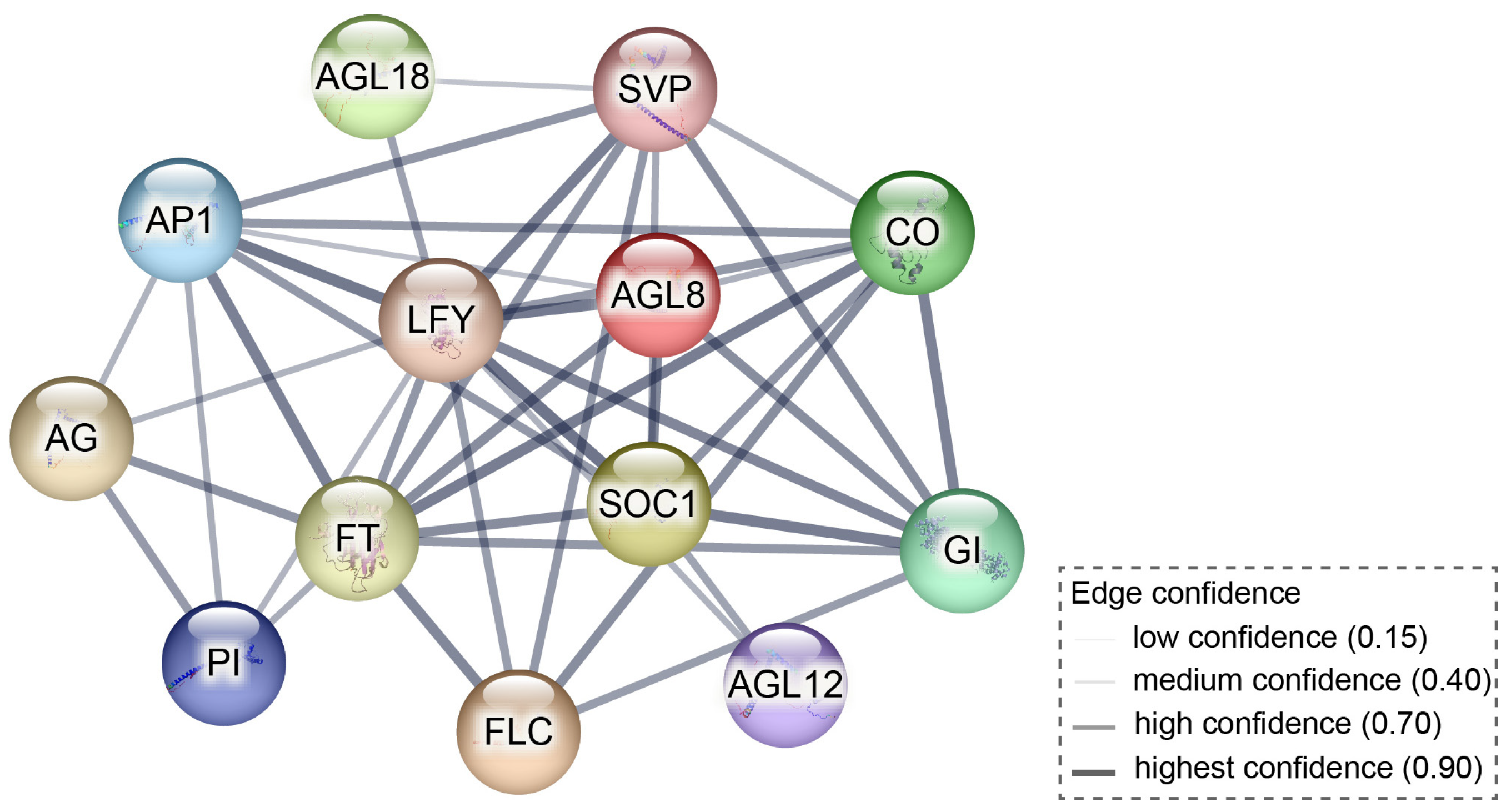
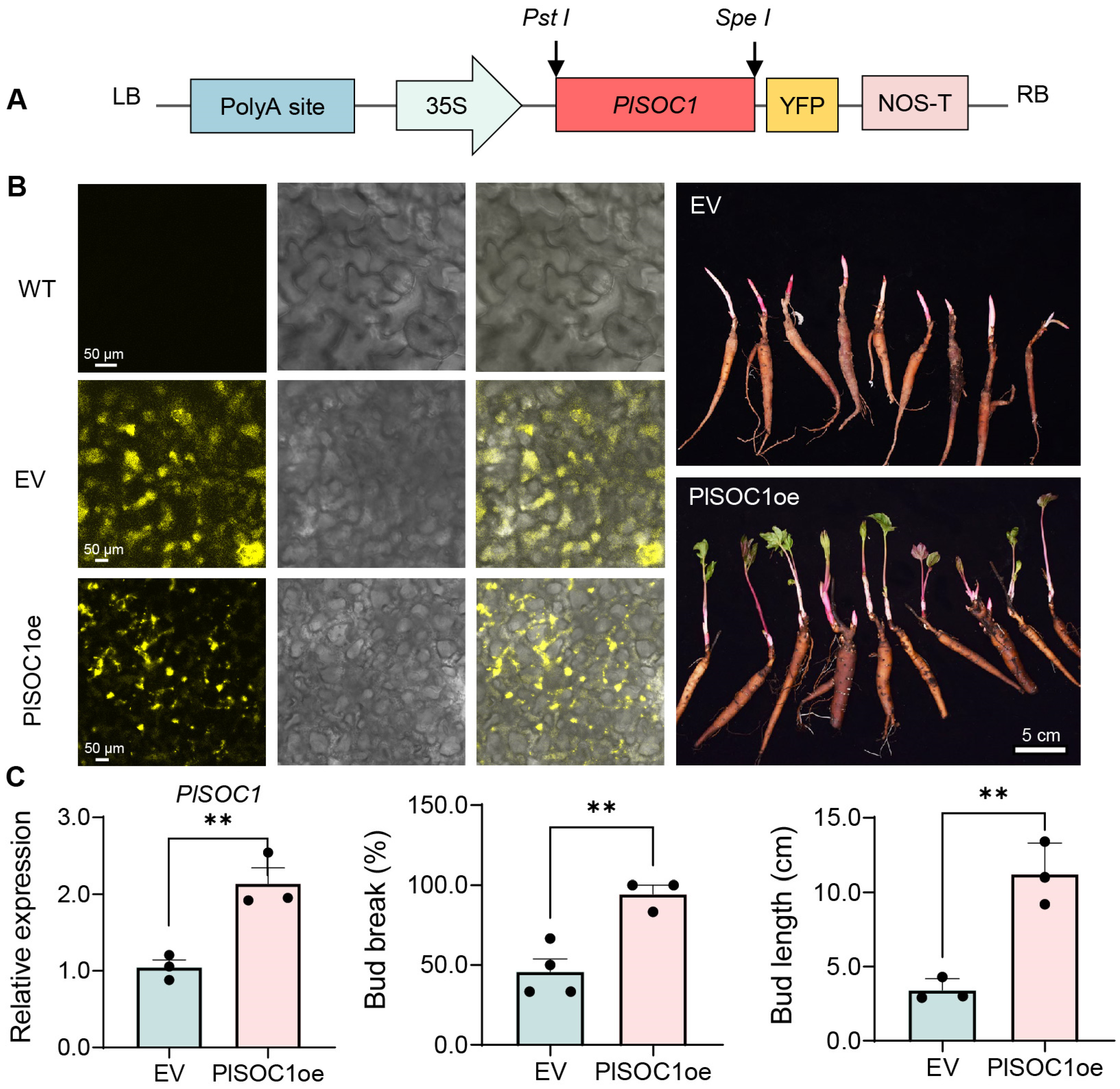
2.6. PlSOC1 Promotes BDT in Herbaceous Peony
3. Discussion
3.1. MIKC-Type Genes in Peony
3.2. Functional Analysis of PlMIKC Genes Based on Gene Expression Pattern
3.3. SOC1: Traditionally Known for Promoting Flowering, but Also Facilitates Dormancy Transition
4. Materials and Methods
4.1. Plant Materials
4.2. Low-Temperature Treatment and Transcriptome Analysis
4.3. Identification and Phylogenetic Analysis of MIKC Genes
4.4. Physicochemical Characteristics and Subcellular Localization Prediction
4.5. Protein Structure and Conserved Motif Analysis
4.6. Expression Profiles of PlMIKC Genes During Dormancy Transition
4.7. Transient Overexpression of PlSOC1 in Herbaceous Peony
4.8. Quantitative Real-Time PCR (qRT–PCR) Analysis
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Anderson, J.V.; Gesch, R.W.; Jia, Y.; Chao, W.S.; Horvath, D.P. Seasonal shifts in dormancy status, carbohydrate metabolism, and related gene expression in crown buds of leafy spurge. Plant Cell Environ. 2005, 28, 1567–1578. [Google Scholar] [CrossRef]
- Rohde, A.; Bhalerao, R.P. Plant dormancy in the perennial context. Trends Plant Sci. 2007, 12, 217–223. [Google Scholar] [CrossRef]
- Lang, G.A.; Early, J.D.; Martin, G.C.; Darnell, R.L. Endo-, Para-, and Ecodormancy: Physiological Terminology and Classification for Dormancy Research. HortScience 1987, 22, 371–377. [Google Scholar] [CrossRef]
- Takahashi, H.; Nishihara, M.; Yoshida, C.; Itoh, K. Gentian FLOWERING LOCUS T orthologs regulate phase transitions: Floral induction and endodormancy release. Plant Physiol. 2022, 188, 1887–1899. [Google Scholar] [CrossRef]
- Johnson, N.C.; Xie, S.P.; Kosaka, Y.; Li, X. Increasing occurrence of cold and warm extremes during the recent global warming slowdown. Nat. Commun. 2018, 9, 1724. [Google Scholar] [CrossRef] [PubMed]
- Ochieng, J.; Kirimi, L.; Mathenge, M. Effects of climate variability and change on agricultural production: The case of small scale farmers in Kenya. NJAS-Wagening. J. Life Sci. 2021, 77, 71–78. [Google Scholar] [CrossRef]
- Sieber, J. Impacts of, and adaptation options to, extreme weather events and climate change concerning thermal power plants. Clim. Change 2013, 121, 55–66. [Google Scholar] [CrossRef]
- Jewaria, P.K.; Hanninen, H.; Li, X.; Bhalerao, R.P.; Zhang, R. A hundred years after: Endodormancy and the chilling requirement in subtropical trees. New Phytol. 2021, 231, 565–570. [Google Scholar] [CrossRef]
- Grimplet, J.; Martínez-Zapater, J.M.; Carmona, M.J. Structural and functional annotation of the MADS-box transcription factor family in grapevine. BMC Genom. 2016, 17, 80. [Google Scholar] [CrossRef]
- Becker, A.; Theissen, G. The major clades of MADS-box genes and their role in the development and evolution of flowering plants. Mol. Phylogenetics Evol. 2003, 29, 464–489. [Google Scholar] [CrossRef]
- Ma, M.M.; Zhang, H.F.; Tian, Q.; Wang, H.C.; Zhang, F.Y.; Tian, X.; Zeng, R.F.; Huang, X.M. MIKC type MADS-box transcription factor LcSVP2 is involved in dormancy regulation of the terminal buds in evergreen perennial litchi (Litchi chinensis Sonn.). Hortic. Res. 2024, 11, uhae150. [Google Scholar] [CrossRef] [PubMed]
- Henschel, K.; Kofuji, R.; Hasebe, M.; Saedler, H.; Münster, T.; Theißen, G. Two Ancient Classes of MIKC-type MADS-box Genes are Present in the Moss Physcomitrella patens. Mol. Biol. Evol. 2002, 19, 801–814. [Google Scholar] [CrossRef]
- Pařenicová, L.; de Folter, S.; Kieffer, M.; Horner, D.S.; Favalli, C.; Busscher, J.; Cook, H.E.; Ingram, R.M.; Kater, M.M.; Davies, B.; et al. Molecular and Phylogenetic Analyses of the Complete MADS-Box Transcription Factor Family in Arabidopsis: New Openings to the MADS World. Plant Cell 2003, 15, 1538–1551. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Fu, X.; Dong, Y.; Lu, J.; Ren, M.; Zhou, N.; Wang, C. MIKC(C)-type MADS-box genes in Rosa chinensis: The remarkable expansion of ABCDE model genes and their roles in floral organogenesis. Hortic. Res. 2018, 5, 25. [Google Scholar] [CrossRef]
- Horvath, D.P.; Chao, W.S.; Suttle, J.C.; Thimmapuram, J.; Anderson, J.V. Transcriptome analysis identifies novel responses and potential regulatory genes involved in seasonal dormancy transitions of leafy spurge (Euphorbia esula L.). BMC Genom. 2008, 9, 536. [Google Scholar] [CrossRef]
- Li, D.; Shao, L.; Zhang, J.; Wang, X.; Zhang, D.; Horvath, D.P.; Zhang, L.; Zhang, J.; Xia, Y. MADS-box transcription factors determine the duration of temporary winter dormancy in closely related evergreen and deciduous Iris spp. J. Exp. Bot. 2022, 73, 1429–1449. [Google Scholar] [CrossRef]
- Bielenberg, D.G.; Wang, Y.; Li, Z.; Zhebentyayeva, T.; Fan, S.; Reighard, G.L.; Scorza, R.; Abbott, A.G. Sequencing and annotation of the evergrowing locus in peach [Prunus persica (L.) Batsch] reveals a cluster of six MADS-box transcription factors as candidate genes for regulation of terminal bud formation. Tree Genet. Genomes 2008, 4, 495–507. [Google Scholar] [CrossRef]
- Zhao, Y.L.; Li, Y.; Cao, K.; Yao, J.L.; Bie, H.L.; Khan, I.A.; Fang, W.C.; Chen, C.W.; Wang, X.W.; Wu, J.L.; et al. MADS-box protein PpDAM6 regulates chilling requirement-mediated dormancy and bud break in peach. Plant Physiol. 2023, 193, 448–465. [Google Scholar] [CrossRef]
- Horvath, D.P.; Sung, S.; Kim, D.; Chao, W.; Anderson, J. Characterization, expression and function of DORMANCY ASSOCIATED MADS-BOX genes from leafy spurge. Plant Mol. Biol. 2010, 73, 169–179. [Google Scholar] [CrossRef]
- Yang, Q.; Yang, B.; Li, J.; Wang, Y.; Tao, R.; Yang, F.; Wu, X.; Yan, X.; Ahmad, M.; Shen, J.; et al. ABA-responsive ABRE-BINDING FACTOR3 activates DAM3 expression to promote bud dormancy in Asian pear. Plant Cell Environ. 2020, 43, 1360–1375. [Google Scholar] [CrossRef]
- Sasaki, R.; Yamane, H.; Ooka, T.; Jotatsu, H.; Kitamura, Y.; Akagi, T.; Tao, R. Functional and expressional analyses of PmDAM genes associated with endodormancy in Japanese apricot. Plant Physiol. 2011, 157, 485–497. [Google Scholar] [CrossRef]
- Zhang, Y.; Niu, D.; Yuan, Y.; Liu, F.; Wang, Z.; Gao, L.; Liu, C.; Zhou, G.; Gai, S. PsSOC1 is involved in the gibberellin pathway to trigger cell proliferation and budburst during endodormancy release in tree peony. New Phytol. 2024, 243, 1017–1033. [Google Scholar] [CrossRef]
- Voogd, C.; Wang, T.; Varkonyi-Gasic, E. Functional and expression analyses of kiwifruit SOC1-like genes suggest that they may not have a role in the transition to flowering but may affect the duration of dormancy. J. Exp. Bot. 2015, 66, 4699–4710. [Google Scholar] [CrossRef] [PubMed]
- Azeez, A.; Miskolczi, P.; Tylewicz, S.; Bhalerao, R.P. A tree ortholog of APETALA1 mediates photoperiodic control of seasonal growth. Curr. Biol. 2014, 24, 717–724. [Google Scholar] [CrossRef]
- Pandey, S.K.; Maurya, J.P.; Aryal, B.; Drynda, K.; Nair, A.; Miskolczi, P.; Singh, R.K.; Wang, X.; Ma, Y.; de Souza Moraes, T.; et al. A regulatory module mediating temperature control of cell-cell communication facilitates tree bud dormancy release. Embo J. 2024, 43, 5793–5812. [Google Scholar] [CrossRef] [PubMed]
- Zhao, P.-X.; Miao, Z.-Q.; Zhang, J.; Chen, S.-Y.; Liu, Q.-Q.; Xiang, C.-B. Arabidopsis MADS-box factor AGL16 negatively regulates drought resistance via stomatal density and stomatal movement. J. Exp. Bot. 2020, 71, 6092–6106. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.H.; Wu, J.; Zhang, Z.S.; Miao, Z.Q.; Zhao, P.X.; Wang, Z.; Xiang, C.B. Arabidopsis MADS-Box Transcription Factor AGL21 Acts as Environmental Surveillance of Seed Germination by Regulating ABI5 Expression. Mol. Plant 2017, 10, 834–845. [Google Scholar] [CrossRef]
- Daminato, M.; Masiero, S.; Resentini, F.; Lovisetto, A.; Casadoro, G. Characterization of TM8, a MADS-box gene expressed in tomato flowers. BMC Plant Biol. 2014, 14, 319. [Google Scholar] [CrossRef]
- Immink, R.G.; Pose, D.; Ferrario, S.; Ott, F.; Kaufmann, K.; Valentim, F.L.; de Folter, S.; van der Wal, F.; van Dijk, A.D.; Schmid, M.; et al. Characterization of SOC1’s central role in flowering by the identification of its upstream and downstream regulators. Plant Physiol. 2012, 160, 433–449. [Google Scholar] [CrossRef]
- Moon, J.; Lee, H.; Kim, M.; Lee, I. Analysis of flowering pathway integrators in Arabidopsis. Plant Cell Physiol. 2005, 46, 292–299. [Google Scholar] [CrossRef]
- Tapia-López, R.; García-Ponce, B.; Dubrovsky, J.G.; Garay-Arroyo, A.; Pérez-Ruíz, R.V.; Kim, S.-H.; Acevedo, F.; Pelaz, S.; Alvarez-Buylla, E.R. An AGAMOUS-Related MADS-Box. Gene, XAL1 (AGL12), Regulates Root Meristem Cell Proliferation and Flowering Transition in Arabidopsis. Plant Physiol. 2008, 146, 1182–1192. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Chen, H.; Er, H.L.; Soo, H.M.; Kumar, P.P.; Han, J.H.; Liou, Y.C.; Yu, H. Direct interaction of AGL24 and SOC1 integrates flowering signals in Arabidopsis. Development 2008, 135, 1481–1491. [Google Scholar] [CrossRef] [PubMed]
- Jang, S.; Torti, S.; Coupland, G. Genetic and spatial interactions between FT, TSF and SVP during the early stages of floral induction in Arabidopsis. Plant J. 2009, 60, 614–625. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Shi, X.; Zhang, R.; Zhang, K.; Shao, L.; Xu, T.; Li, D.; Zhang, D.; Zhang, J.; Xia, Y. Impact of summer heat stress inducing physiological and biochemical responses in herbaceous peony cultivars (Paeonia lactiflora Pall.) from different latitudes. Ind. Crop Prod. 2022, 184, 115000. [Google Scholar] [CrossRef]
- Zhang, R.; Wang, X.; Shi, X.; Shao, L.; Xu, T.; Xia, Y.; Li, D.; Zhang, J. Chilling Requirement Validation and Physiological and Molecular Responses of the Bud Endodormancy Release in Paeonia lactiflora ‘Meiju’. Int. J. Mol. Sci. 2021, 22, 8382. [Google Scholar] [CrossRef]
- Wang, X.; Zhang, R.; Huang, Q.; Shi, X.; Li, D.; Shao, L.; Xu, T.; Horvath, D.P.; Xia, Y.; Zhang, J. Comparative Study on Physiological Responses and Gene Expression of Bud Endodormancy Release Between Two Herbaceous Peony Cultivars (Paeonia lactiflora Pall.) with Contrasting Chilling Requirements. Front. Plant Sci. 2021, 12, 772285. [Google Scholar] [CrossRef]
- Wang, X.; Li, D.; Zhang, D.; Shi, X.; Wu, Y.; Qi, Z.; Ding, H.; Zhu, K.; Xia, Y.; Zhang, J. Improving crucial details and selecting the optimal model for evaluating the chilling requirement of Paeonia lactiflora Pall. at low latitudes during four winters. Sci. Hortic. 2020, 265, 109175. [Google Scholar] [CrossRef]
- Wang, X.; Zhang, R.; Zhang, K.; Shao, L.; Xu, T.; Shi, X.; Li, D.; Zhang, J.; Xia, Y. Development of a Multi-Criteria Decision-Making Approach for Evaluating the Comprehensive Application of Herbaceous Peony at Low Latitudes. Int. J. Mol. Sci. 2022, 23, 14342. [Google Scholar] [CrossRef]
- Zhao, Y.; Liu, C.; Sui, J.; Liang, J.; Ge, J.; Li, J.; Pan, W.; Yi, M.; Du, Y.; Wu, J. A wake-up call: Signaling in regulating ornamental geophytes dormancy. Ornam. Plant Res. 2022, 2, 8. [Google Scholar] [CrossRef]
- Zhang, J.; Wu, Y.; Li, D.; Wang, G.; Li, X.; Xia, Y. Transcriptomic analysis of the underground renewal buds during dormancy transition and release in ‘Hangbaishao’ peony (Paeonia lactiflora). PLoS ONE 2015, 10, e0119118. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Won, S.Y.; Jung, J.A.; Kim, J.S. Genome-wide analysis of the MADS-Box. gene family in Chrysanthemum. Comput. Biol. Chem. 2021, 90, 107424. [Google Scholar] [CrossRef] [PubMed]
- Arora, R.; Agarwal, P.; Ray, S.; Singh, A.K.; Singh, V.P.; Tyagi, A.K.; Kapoor, S. MADS-box gene family in rice: Genome-wide identification, organization and expression profiling during reproductive development and stress. BMC Genom. 2007, 8, 242. [Google Scholar] [CrossRef]
- Hou, H.; Tian, M.; Liu, N.; Huo, J.; Sui, S.; Li, Z. Genome-wide analysis of MIKCC-type MADS-box genes and roles of CpFUL/SEP/AGL6 superclade in dormancy breaking and bud formation of Chimonanthus praecox. Plant Physiol. Biochem. 2023, 196, 893–902. [Google Scholar] [CrossRef] [PubMed]
- Gramzow, L.; Theißen, G. Phylogenomics reveals surprising sets of essential and dispensable clades of MIKC(c)-group MADS-box genes in flowering plants. J. Exp. Zool. B Mol. Dev. Evol. 2015, 324, 353–362. [Google Scholar] [CrossRef]
- Yuan, J.; Jiang, S.; Jian, J.; Liu, M.; Yue, Z.; Xu, J.; Li, J.; Xu, C.; Lin, L.; Jing, Y.; et al. Genomic basis of the giga-chromosomes and giga-genome of tree peony Paeonia ostii. Nat. Commun. 2022, 13, 7328. [Google Scholar] [CrossRef]
- Yang, X.; Guo, Q.; Liu, S.; Wang, D.; Zuo, D.; Niu, T.; Wei, D.; Guo, L.; Hou, X. Genome-wide characterization of the MADS-box gene family in Paeonia ostii and expression analysis of genes related to floral organ development. BMC Genom. 2025, 26, 49. [Google Scholar] [CrossRef]
- Li, Y.; Lu, J.; Chang, Y.; Tang, W.; Yang, Q. Comparative analysis of tree peony petal development by transcriptome sequencing. Acta Physiol. Plant 2017, 39, 216. [Google Scholar] [CrossRef]
- Wenlong, T.; Pengfei, Z.; Yonghua, L.I.; Dan, H.E.; Jiuxing, L.U. Cloning and Expression Analysis of Transcription Factor Gene PsAGL6 in Tree Peony (Paeonia suffruticosa L.). Acta Bot. Boreali-Occident. Sin. 2018, 38, 439–444. [Google Scholar]
- Lu, J.; Zheng, Y.; Wang, H.; Wang, Z.; Li, Y.; Gao, G.; Li, Y. Cloning and Expression Analysis of PsPI Gene in Tree Peony (Paeonia suffruticosa). J. Plant Physiol. 2019. [Google Scholar] [CrossRef]
- Melzer, S.; Lens, F.; Gennen, J.; Vanneste, S.; Rohde, A.; Beeckman, T. Flowering-time genes modulate meristem determinacy and growth form in Arabidopsis thaliana. Nat. Genet. 2008, 40, 1489–1492. [Google Scholar] [CrossRef] [PubMed]
- Adamczyk, B.J.; Fernandez, D.E. MIKC* MADS Domain Heterodimers Are Required for Pollen Maturation and Tube Growth in Arabidopsis. Plant Physiol. 2009, 149, 1713–1723. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, Y.; Zhang, Y.; Guan, S.; Liu, C.; Zheng, G.; Gai, S. Isolation and Characterization of a SOC1-Like Gene from Tree Peony (Paeonia suffruticosa). Plant Mol. Biol. Rep. 2014, 33, 855–866. [Google Scholar] [CrossRef]
- Hou, D.; Li, L.; Ma, T.; Pei, J.; Zhao, Z.; Lu, M.; Wu, A.; Lin, X. The SOC1-like gene BoMADS50 is associated with the flowering of Bambusa oldhamii. Hortic. Res. 2021, 8, 133. [Google Scholar] [CrossRef]
- Kitamura, Y.; Takeuchi, T.; Yamane, H.; Tao, R. Simultaneous down-regulation of DORMANCY-ASSOCIATED MADS-box6 and SOC1 during dormancy release in Japanese apricot (Prunus mume) flower buds. J. Hortic. Sci. Biotech. 2016, 91, 476–482. [Google Scholar] [CrossRef]
- Gómez-Soto, D.; Ramos-Sánchez, J.M.; Alique, D.; Conde, D.; Triozzi, P.M.; Perales, M.; Allona, I. Overexpression of a SOC1-Related Gene Promotes Bud Break in Ecodormant Poplars. Front. Plant Sci. 2021, 12, 670497. [Google Scholar] [CrossRef] [PubMed]
- Trainin, T.; Bar-Ya’akov, I.; Holland, D. ParSOC1, a MADS-box gene closely related to Arabidopsis AGL20/SOC1, is expressed in apricot leaves in a diurnal manner and is linked with chilling requirements for dormancy break. Tree Genet. Genomes 2013, 9, 753–766. [Google Scholar] [CrossRef]
- Zhang, J.; Li, D.; Shi, X.; Zhang, D.; Qiu, S.; Wei, J.; Zhang, J.; Zhou, J.; Zhu, K.; Xia, Y. Mining and expression analysis of candidate genes involved in regulating the chilling requirement fulfillment of Paeonia lactiflora ‘Hang Baishao’. BMC Plant Biol. 2017, 17, 262. [Google Scholar] [CrossRef]
- Zhang, K.; Wang, X.; Chen, X.; Zhang, R.; Guo, J.; Wang, Q.; Li, D.; Shao, L.; Shi, X.; Han, J.; et al. Establishment of a Homologous Silencing System with Intact-Plant Infiltration and Minimized Operation for Studying Gene Function in Herbaceous Peonies. Int. J. Mol. Sci. 2024, 25, 4412. [Google Scholar] [CrossRef]
- Finn, R.D.; Clements, J.; Eddy, S.R. HMMER web server: Interactive sequence similarity searching. Nucleic Acids Res. 2011, 39, W29–W37. [Google Scholar] [CrossRef]
- Falavigna, V.D.S.; Guitton, B.; Costes, E.; Andres, F. I Want to (Bud) Break Free: The Potential Role of DAM and SVP-Like Genes in Regulating Dormancy Cycle in Temperate Fruit Trees. Front. Plant Sci. 2018, 9, 1990. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Standley, D.M. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree: Computing large minimum evolution trees with profiles instead of a distance matrix. Mol. Biol. Evol. 2009, 26, 1641–1650. [Google Scholar] [CrossRef] [PubMed]
- Nicholas, K.B.; Nicholas, H.B. GeneDoc: A Tool for Editing and Annotating Multiple Sequence Alignments. 1997. Available online: https://api.semanticscholar.org/CorpusID:81058551 (accessed on 3 January 2025).
- Chou, K.C.; Shen, H.B. Plant-mPLoc: A top-down strategy to augment the power for predicting plant protein subcellular localization. PLoS ONE 2010, 5, e11335. [Google Scholar] [CrossRef]
- Wang, J.; Chitsaz, F.; Derbyshire, M.K.; Gonzales, N.R.; Gwadz, M.; Lu, S.; Marchler, G.H.; Song, J.S.; Thanki, N.; Yamashita, R.A.; et al. The conserved domain database in 2023. Nucleic Acids Res. 2023, 51, D384–D388. [Google Scholar] [CrossRef]
- Bailey, T.L.; Johnson, J.; Grant, C.E.; Noble, W.S. The MEME Suite. Nucleic Acids Res. 2015, 43, W39–W49. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Wu, Y.; Li, J.; Wang, X.; Zeng, Z.; Xu, J.; Liu, Y.; Feng, J.; Chen, H.; He, Y.; et al. TBtools-II: A “one for all, all for one” bioinformatics platform for biological big-data mining. Mol. Plant 2023, 16, 1733–1742. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Kirsch, R.; Koutrouli, M.; Nastou, K.; Mehryary, F.; Hachilif, R.; Gable, A.L.; Fang, T.; Doncheva, N.T.; Pyysalo, S.; et al. The STRING database in 2023: Protein-protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res 2023, 51, D638–D646. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−∆∆C(T) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]



| Gene Name | Gene ID | Amino Acid | Molecular Weight | PI | Instability Index | GRAVY | Subcellular Location |
|---|---|---|---|---|---|---|---|
| PlDAM-2 | CL2631.Contig1_All | 228 | 26,050.66 | 6.92 | 59.02 | −0.734 | Nucleus |
| PlDAM-3 | CL2631.Contig2_All | 184 | 21,467.82 | 9.26 | 59.32 | −0.723 | Nucleus |
| PlDAM-6 | CL2631.Contig3_All | 223 | 25,533.08 | 6.92 | 59.25 | −0.749 | Nucleus |
| PlAGL12 | CL5197.Contig3_All | 200 | 23,194.02 | 7.02 | 47.74 | −0.408 | Nucleus |
| PlAGL18 | CL6471.Contig2_All | 252 | 28,738.64 | 6.04 | 54.01 | −0.762 | Nucleus |
| PlAGL30-1 | F02_cb5982_c0/f1p0/1865 | 211 | 24,086.83 | 9.82 | 38.24 | −0.559 | Nucleus |
| PlAGL30-2 | F02_cb5982_c1/f1p0/1976 | 351 | 39,547.65 | 6.12 | 49.89 | −0.663 | Nucleus |
| PlDAM-4 | F02_cb7706_c1/f1p0/1559 | 228 | 26,050.66 | 6.92 | 59.02 | −0.734 | Nucleus |
| PlFUL-1 | F03_cb13331_c0/f2p0/1331 | 262 | 30,262.34 | 9.55 | 58.59 | −0.929 | Nucleus |
| PlFUL-2 | F03_cb6642_c0/f2p0/1135 | 127 | 14,302.88 | 10.01 | 40.17 | 0.035 | Nucleus |
| PlDAM-1 | F03_cb7864_c0/f3p1/1445 | 223 | 25,533.08 | 6.92 | 59.25 | −0.749 | Nucleus |
| PlDAM-5 | F03_cb9970_c0/f3p1/1447 | 228 | 26,050.66 | 6.92 | 59.02 | −0.734 | Nucleus |
| PlFUL-3 | F04_cb14472_c0/f3p0/1095 | 252 | 29,265.89 | 8.68 | 56.03 | −1.028 | Nucleus |
| PlAG | Unigene17023_All | 168 | 19,064.71 | 10.17 | 47.30 | −0.702 | Nucleus |
| PlPI-2 | Unigene18796_All | 265 | 30,655.26 | 9.35 | 37.83 | −0.609 | Nucleus |
| PlFUL-4 | Unigene18852_All | 242 | 28,053.55 | 8.38 | 56.58 | −0.992 | Nucleus |
| PlSOC1 | Unigene22847_All | 224 | 25,382.85 | 5.65 | 54.89 | −0.637 | Nucleus |
| PlAGL18-2 | Unigene23635_All | 188 | 21,515.76 | 5.86 | 58.56 | −0.422 | Nucleus |
| PlPI-1 | Unigene26195_All | 185 | 21,385.22 | 6.10 | 55.92 | −0.764 | Nucleus |
| Primer Names | Primer Sequences (5′-3′) |
|---|---|
| p35S::PlSOC1_F | CTTGGATCCTCGAGCTGCAGGAAGCTCAATCAGTTTCATCCCAA |
| p35S::PlSOC1_R | CCCTTGCTCACCATACTAGTGTTCTGTAATATAATGCGTTTAATTCTGC |
| Primer Names | Primer Sequences (5′-3′) |
|---|---|
| PlATUBA_F | GACGTGGGCTCGATCTGAAT |
| PlATUBA_R | ATCCCTTGCCTGAGCATCAC |
| PlSOC1_F | ATGCGCGAGACAATTGAACG |
| PlSOC1_R | TCAACGGTGCATGATCCCAA |
| PlAGL18-1_F | CGCTGCGCAAGCAGATTAAG |
| PlAGL18-1_R | TAAGGCTCAGAAGGCAACCC |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, Q.; Chen, X.; Zhong, S.; Wu, S.; Guo, J.; Wang, Q.; Li, J.; Li, D.; Xia, Y.; Zhang, J.; et al. MIKC-Type MADS-Box Gene Analysis Reveals the Role of PlSOC1 in Bud Dormancy Transition in Herbaceous Peony. Plants 2025, 14, 928. https://doi.org/10.3390/plants14060928
Huang Q, Chen X, Zhong S, Wu S, Guo J, Wang Q, Li J, Li D, Xia Y, Zhang J, et al. MIKC-Type MADS-Box Gene Analysis Reveals the Role of PlSOC1 in Bud Dormancy Transition in Herbaceous Peony. Plants. 2025; 14(6):928. https://doi.org/10.3390/plants14060928
Chicago/Turabian StyleHuang, Qiaoyu, Xiaoxuan Chen, Shuyun Zhong, Shuangzhe Wu, Junhong Guo, Qiyao Wang, Jiahe Li, Danqing Li, Yiping Xia, Jiaping Zhang, and et al. 2025. "MIKC-Type MADS-Box Gene Analysis Reveals the Role of PlSOC1 in Bud Dormancy Transition in Herbaceous Peony" Plants 14, no. 6: 928. https://doi.org/10.3390/plants14060928
APA StyleHuang, Q., Chen, X., Zhong, S., Wu, S., Guo, J., Wang, Q., Li, J., Li, D., Xia, Y., Zhang, J., & Wang, X. (2025). MIKC-Type MADS-Box Gene Analysis Reveals the Role of PlSOC1 in Bud Dormancy Transition in Herbaceous Peony. Plants, 14(6), 928. https://doi.org/10.3390/plants14060928








