Abstract
The importance of fruit shape studies extends beyond fundamental plant biology, as it holds significant implications for breeding. Understanding the genetic and hormonal regulation of fruit morphology can facilitate targeted breeding strategies to enhance yield, quality, and stress resistance, ultimately contributing to sustainable farming and nutrition security. The diversity in fruit shapes is the result of complex hormone regulation and molecular pathways that affect key traits, including carpel number, fruit length, and weight. Fruit shape is a quality attribute that directly influences consumer preference, marketability and the ease of post-harvest processing. This article focuses on investigations carried out on molecular, genetic and hormonal regulation mechanisms of fruit shape, color, maturation in fruit plants and key genetic pathways such as CLV-WUS and OVATE, as well as their roles in shaping non-climacteric fruits such as strawberries, grapes and raspberries. Plant hormones, especially abscisic acid (ABA) and indole-3-acetic acid (IAA), play a crucial role in enhancing desirable traits such as color and taste, while regulating anthocyanin synthesis and growth time. In addition, the dynamic interactions between auxin, gibberellin, and ethylene are crucial for the ripening process. Jasmonate enhances stress response, brassinosteroids promote ripening and cytokinins promote early fruit development. In addition, this review also studied the fruit morphology of species such as tomatoes and cucumbers, emphasizing the importance of the CLV-WUS pathway, which regulates the number of carpels through genes such as WUSCHEL (WUS), FRUITFULL1 (FUL1), and auxin response factor 14 (ARF14). The weight of fresh fruit is affected by microRNAs such as miRNA156, which emphasizes the importance of post transcriptional regulation. The involvement of transcription factors such as SISHN1, CaOvate, and CISUN25-26-27a further emphasizes the complexity of hormone regulation. Understanding these regulatory mechanisms can enhance our understanding of fruit development and have a profound impact on agricultural practices and crop improvement strategies aimed at meeting the growing global demand for high-quality agricultural products.
1. Introduction
The diversity of plant fruit shapes represents a significant evolutionary adaptation that enhances seed dispersal and survival in dynamic environments. Fruits are generally classified into two main categories: fleshy and dry. Fleshy fruits develop through both cell division and expansion, undergoing substantial changes in texture, color, and flavor during ripening, regulated by complex metabolic and hormonal processes [1,2]. In contrast, dry fruits do not exhibit such pronounced expansion or compositional transformations, and their developmental regulatory networks are comparatively well understood [2,3]. Fleshy fruits evolved from ancestral siliques through a conserved genetic network, developing diverse edible structures for seed dispersal. Examples include drupes (mango, cherry), berries (Schisandra, tomato, grape), pomes (apple, pear), hesperidiums (orange, lemon), and pepos (watermelon, pumpkin). In contrast, dry fruits lack a fleshy pericarp and are classified as dehiscent (legumes, capsules) or indehiscent (nuts, achenes, grains), relying on wind, water, or animals for dispersal [4,5].
Crop improvement and domestication have enhanced fruit diversity by selecting desirable traits, including shape and color. Squash varieties exhibit scallop and long crookneck forms, while tomatoes vary in shape, including round, ellipsoid, rectangular, heart, oxheart, and obovoid. Cucumbers are typically long cylindrical or round, and peaches are generally flat or round (Table 1). These fruit shapes serve as models for studying shape variation in Cucurbitaceae, Solanaceae, and Rosaceae fruit crops [6,7]. Fruit development in tomatoes unfolds through distinct stages: floral organ formation, cell division, cell expansion, and maturation. The initial stage lasts for 14 to 21 days, laying the foundation for the identity and shape of the flower. Subsequently, there is a two-week period of cell division leading to fertilization, followed by significant cell expansion, which can increase in size by more than 20 times. Maturity stabilizes size and shape, while triggering rapid biochemical changes such as changes in color and aroma [8]. The maturation process is subject to complex regulations and differs between climacteric fruits such as bananas and tomatoes, which mature through the production of ethylene after harvest, and non-climacteric fruits, such as grapes and strawberries, which ripen on the plant without undergoing significant post-harvest changes [9,10]. The balance of sugar and acid has a crucial impact on flavor, and indicators such as titratable acidity play a key role in determining maturity [11]. This review explores the genetic, hormonal, and environmental interactions that regulate fruit shape diversity, providing insights into the molecular mechanisms underlying fruit development. It examines the genetic control and hormonal regulation of fruit shape, color, taste, and ripening processes.

Table 1.
Variation in fruit shapes among different fruit crops.
Table 1.
Variation in fruit shapes among different fruit crops.
| Fruit Shape | Examples | Fruit Family | Reference |
|---|---|---|---|
| Spherical | Malus domestica, Citrus sinensis, Solanum lycopersicum | Rosaceae, Rutaceae, Solanaceae | [12] |
| Oval | Musa paradisiaca Linn., Vitis vinifera L., Carica papaya | Musaceae, Vitaceae, Caricaceae | [13] |
| Elongated | Cucumis sativus Solanum melongena Capsicum | Cucurbitaceae, Solanaceae | [14] |
| Irregular | Fragaria x ananassa, Ficus carica, Actinidia deliciosa | Rosaceae, Moraceae, Actinidiaceae | [15] |
| Oblong | Mangifera indica, Persea americana, Prunus domestica | Anacardiaceae, Lauraceae, Rosaceae | [16] |
| Globular | Vaccinium koreanum, Vitis vinifera L., Prunus avium | Ericaceae, Vitaceae, Rosaceae | [17] |
| Pear-shaped | Pyrus communis, Carica papaya Guava | Rosaceae, Caricaceae, Myrtaceae | [18] |
| Spherical | Malus pumila, Citrus sinensis, Solanum lycopersicum | Rosaceae, Rutaceae, Solanaceae | [12] |
2. Molecular and Genetic Regulation Mechanisms of Fruit Shape
In recent years, with the introduction of innovative tools such as the tomato analyzer developed by [19], there has been an increasing trend in determining the diversity of fruit shapes. It categorizes tomato fruit shapes into eight different types, such as long, bovine heart, heart-shaped, flat, rectangular, circular, inverted spherical, and elliptical [20]. In contrast, gourd family crops such as cucumber, melon, and watermelon typically exhibit more uniform shapes, mainly cylindrical or elliptical [21]. The morphological occurrence and development of fruit shape are influenced by various internal factors, including protein interactions, regulatory genes, and external environmental conditions. This study classified various fruit shapes and delved into the regulatory mechanisms that drive their formation.
2.1. Carpel Number
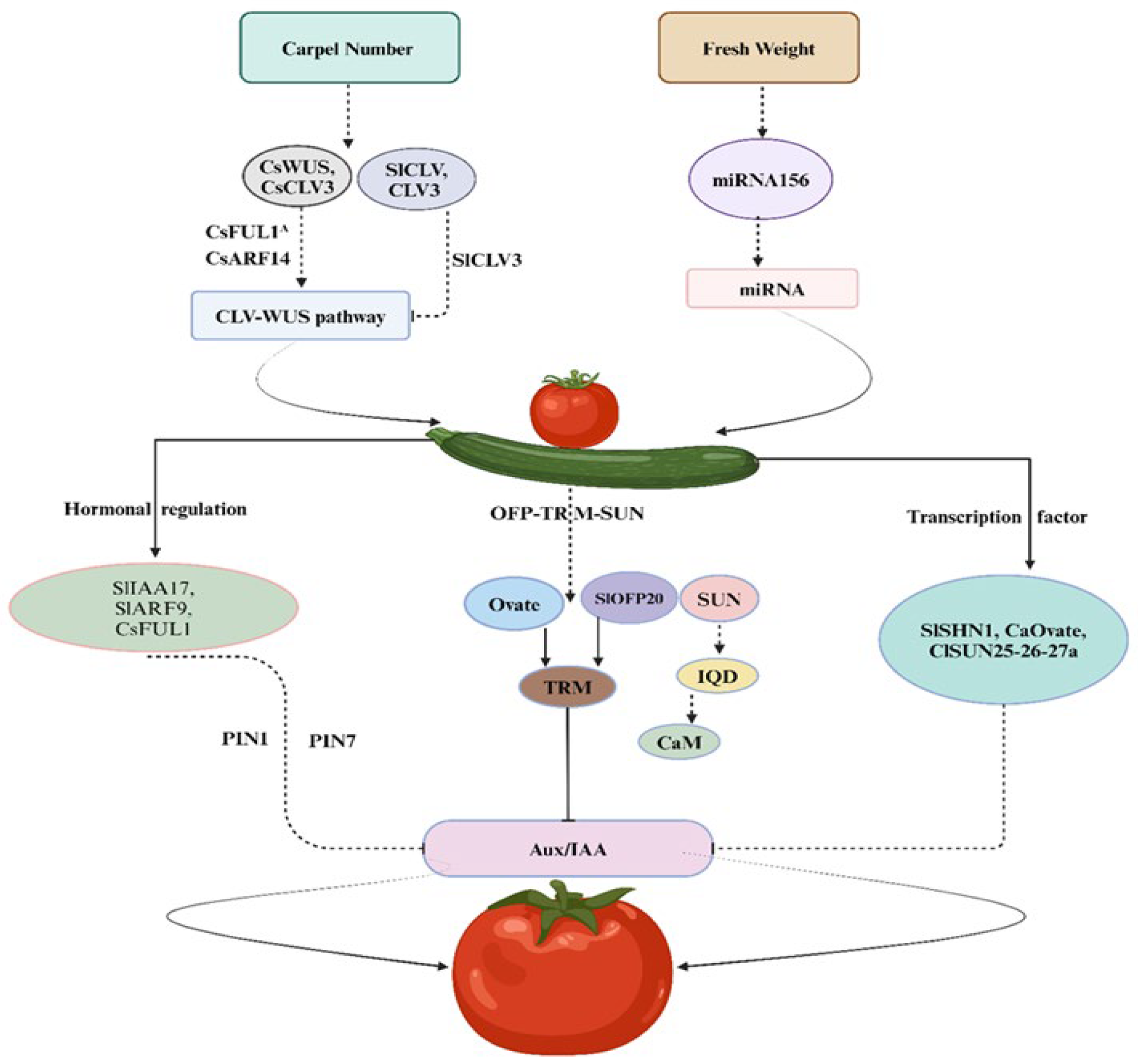
The carpel number (CN) stands as a pivotal fruit trait among vegetables that effects both shape and size of the fruits. In the natural course, typically fruits produce two or more than two carpels, with variations attributed to domestication and mutations. In Arabidopsis, research shows that shared regulatory factors are important to determine CN, WUSCHEL (WUS) and CLAVATA (CLV) involved in negative feedback mechanisms that regulate the floral organs’ number and meristem size (Figure 1). Genetic mutation belonging to the CLV family leads to an increased number of undifferentiated cells mostly present in the central area, leading to expansion of the meristem cells and eventually resulting in shaping the fruit [22]. Larger floral meristems correspond to increased CN and fruit width [23]. Significantly, in higher plant CLV-WUS pathways, function is conserved [24], regulating the fruit CN in plants belonging to Brassicaceae Cucurbitaceae and Solanaceae families [25,26,27]. In cucumbers, transcription factors (TFs), CsARF14, and CsFUL1A genes (FRUITFULL-like MADS-box) regulate the CsWUS-CsCLV3 pathway, playing an important role in the cucumber CN-regulation mechanism [25]. It is observed in tomato plants that a loss of function mutation in SlCLV3 genes leads to an increase in the size of the fruit due to an increased number of locules [26,28]. Moreover, the upregulation of gene (WOX1 TF), belonging to the SlWUS family, affects fruit shape and carpel number [29]. The locule number (LC) in tomatoes is regulated by the WUS gene, while the FAS gene encodes a regulator that influences CLV3 expression, collectively controlling fruit locule development, mutations in LC or FAS, resulting in a high yield of locule count [30]. Besides, the transcriptional regulation of non-coding RNAs (long or small) shows a critical role in fruit shape development. Locule number in tomato can be altered by overexpression of MIR156 (156 MicroRNA) [31]. In rapeseed, CLV1 disruption may result in a trilocular phenotype [27] (Table 2). Furthermore, a plant-specific TF belonging to the YABBY family includes a number of members capable of increasing the locule number and influencing the flat fruit shape in tomatoes [20,32].
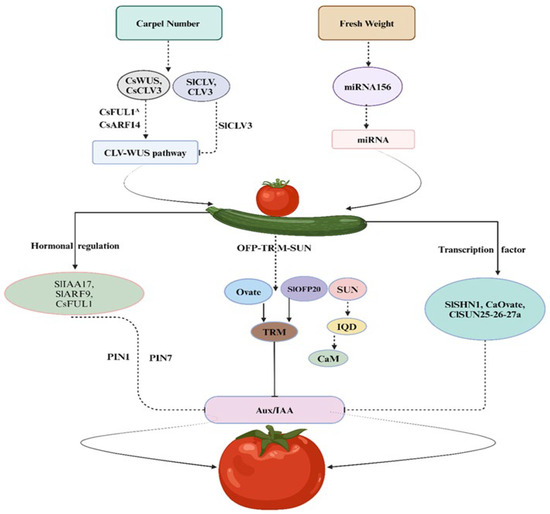
Figure 1.
CLV-WUS and OVATE-based model pathway and role of associated genes in fruit traits.

Table 2.
Genes associated with fruit shape development.
Table 2.
Genes associated with fruit shape development.
| Gene | Function | Fruit Species | Reference |
|---|---|---|---|
| FASCIATED (FAS) | Cell division regulation | Tomato, Capsicum | [33] |
| OVATE (O) | Fruit shape determination | Tomato | [34] |
| SUN | Cell expansion control | Tomato, Pepper | [35] |
| SGR | Cell wall metabolism | Tomato | [36] |
| LCY-B | Carotenoid biosynthesis | Tomato, Pepper | [37] |
| CsFUL1A, CsARF14 | Regulates carpel number | Cucumber | [25] |
| SlCLV3 | Increases locule number and fruit size | Tomato | [26,28] |
| SlWUS (WOX1 TF) | Affects fruit shape and carpel number | Tomato | [29] |
| CLV1 | Disruption induces a trilocular phenotype | Rapeseed | [27] |
| YABBY TFs | Enhances locule number and promotes flat fruit shape | Tomato | [20,32] |
2.2. Fruit Length
The phenotypic trait of fruit length (FL) is significantly influenced by diverse factors, treated as a quantitative trait, and has been the subject of investigation through quantitative trait loci (QTL) mapping, leading to the identification of various relevant loci. A multitude of candidate genes linked to FL, including fl3.2, mfl3, fl7.1, fl4.1, FS3.2, FS3.3, and fs10.1, have been discerned [38,39,40]. Moreover, an abundance of genes and regulatory pathways affecting FL have been unveiled. In cucumbers, genes like short fruit (sf1) and CsFUL1 have been reported for FL regulators [41,42]. The sf3 mutation in cucumbers is associated with the candidate gene CsKTN1, a homolog of KTN1, which was first identified in A. thaliana and is involved in the regulation of nuclear transport; this gene also encodes a katanin p60 subunit and plays a role in influencing FL regulation [43]. In tomatoes, regulatory factors such as Tonneau1 recruitment motif (TRM), SUN and Ovate family proteins (OFP) have individual or collective effects on FL. The identification of the regulatory gene Ovate in pear-shaped tomato fruit by Liu et al. marked a significant discovery [44]. Extensive research on plant-specific TFs OVATE and SlOFP20 has shown their impact on tomato fruit size and shape. OVATE induces pear-shaped fruit, while SlOFP20 regulates floral organ and pollen tube development by modulating brassinosteroid (BL) and gibberellin (GA) signaling pathways [45].
Research involving the overexpression of AtOFP1 in Arabidopsis, like the overexpression of OVATE and SlOFP20 in tomatoes, suggests a shortened length of floral organs [44]. Tomato fruit morphology is controlled by a complex regulatory network of hormonal signaling and genetic interactions. The fruit shape and size are modulated by auxins, gibberellins (GAs), ethylene, and brassinosteroids (BRs) through key regulatory pathways (Figure 2). In tomatoes, egg-shaped mutations induce changes in fruit cell-division patterns, affecting the number of cells in both proximal and distal directions [33]. In addition, the locus inhibitors of OVATE 1 (SOV1) enhance the impact of OVATE mutations [46]. Furthermore, due to the overexpression of homologous OFP genes (AtOFP15, AtOFP16, AtOFP13, and AtOFP18) in Arabidopsis, the reduction in siliqua length was exaggerated [47]. These genes are also responsible for regulating fruit shape, indicating that this gene family may also affect organ shape in other plant species [48].
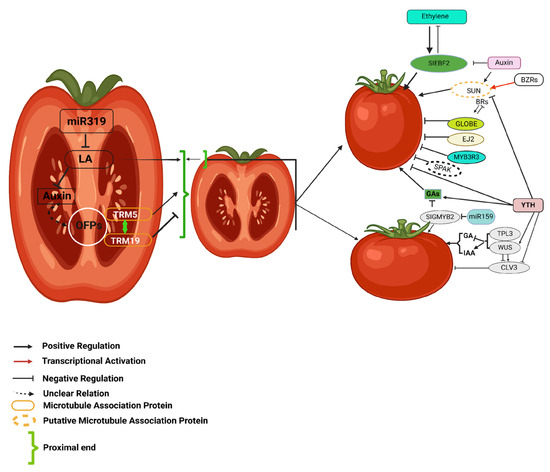
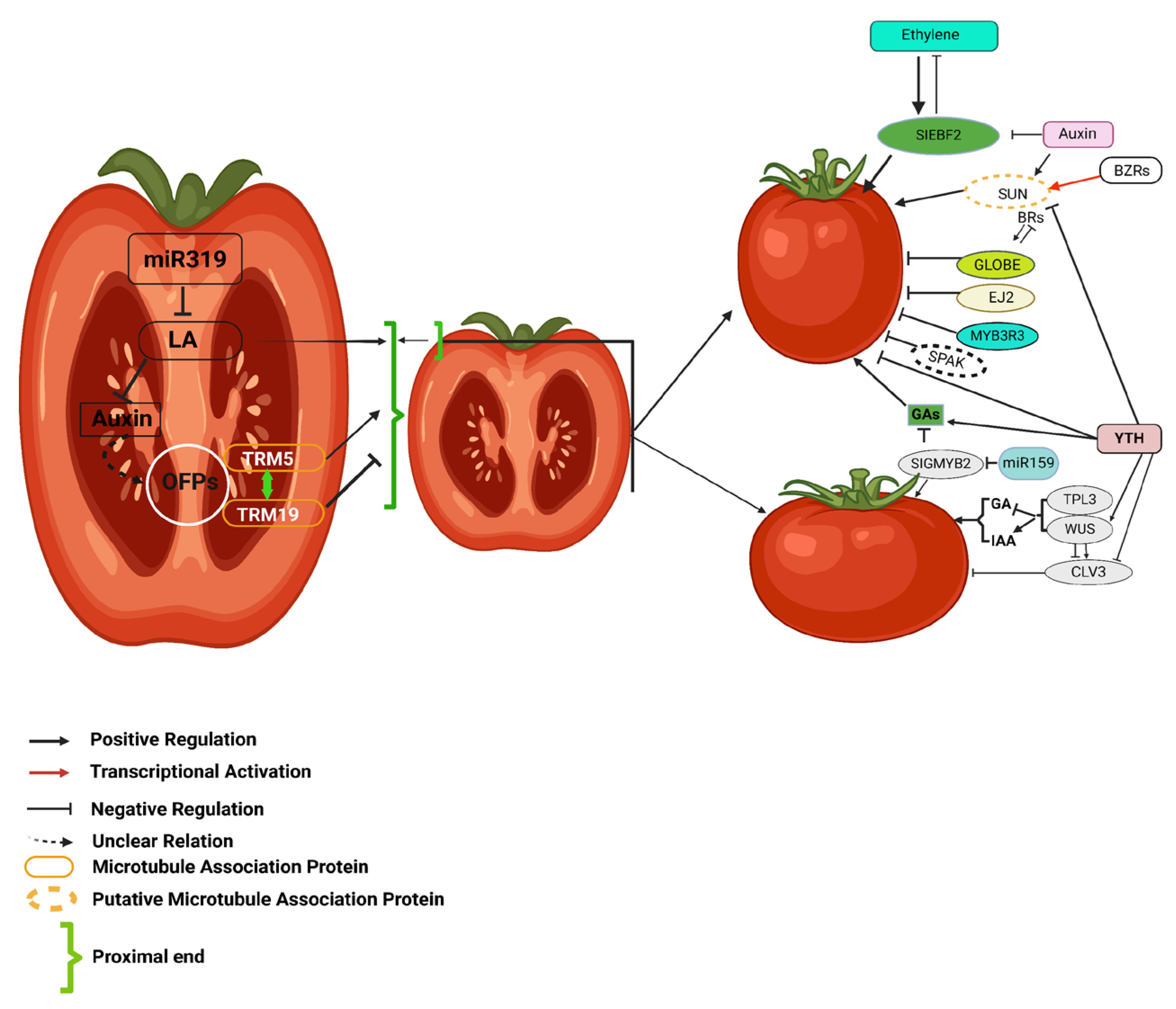
Figure 2.
This figure illustrates the regulatory network of hormonal and genetic interactions influencing tomato fruit morphology. Auxins, gibberellins (GAs), ethylene, and brassinosteroids (BRs) coordinate fruit shape and size by modulating key genetic regulators. CLV3, controlled by WUS, TPL3, and hormonal signals, determines locule number. The interplay between SUN, GLOBE, EJ2, and microtubule-associated proteins, regulated by BRs and ethylene, further refines fruit morphology. Additionally, miR159 and miR319 modulate transcription factors affecting auxin and GA pathways. This intricate network highlights the coordinated influence of hormonal and genetic factors in shaping tomato fruit development. The figure is adapted with slight modifications from Li et al. (2023) [49].
Although the molecular mechanism of OVATE is not clear yet, it has been shown that it works together with some microtubule-associated proteins, such as TRM [50]. SlOFP20 and OVATE engage with the TRM M8 motif by the OFP domain. When co-expressed OVATE or SlOFP20 with SlTRM3/4 induce the relocation of TRMs and OFPs from microtubules toward cytoplasm, this indicates that the OFP–TRM protein complex is important for organ growth and cell division by maintaining a dynamic equilibrium among micro tubular and cytoplasmic localization [34,51]. The CaOvate gene suppression in peppers resulted in an elongated fruit shape, indicating the role of OFP families in regulating fruit size [52]. SUN is a member of the family (IQ 67 domain (IQD)) that encodes a protein involved in Calmodulin (CaM) binding [46,53]. SUN in melons, cucumbers and tomatoes is known as a fruit length (FL) and shape regulator [33,53,54,55]. The IQD, a conserved region of 67 amino acids consisting of three regularly spaced IQ motifs that facilitate binding of CaM in the occurrence of Ca2+ [56,57], triggers changes in fruit shape through SUN during the cell division stage, 7–10 days post-pollination [58]. This change involves increased cell division and elongation beside the proximo-distal axis, allowing the regulation of fruit structure by affecting microtubule dynamics [59]. Lazzaro et al. (2018) proposed a model illustrating the interaction between SUN, OFP, and TRM in tomato fruit-shape regulation, particularly in connection with microtubule development [52]. Additionally, plant-specific Rho GTPases (ROPs) play an important role in organizing microtubules and influencing the cytoskeleton to determining the absolute shape of a cell [60].
Several IQD proteins have been identified as key facilitators of ROP domains’ formation, contributing to the regulation of cytoskeletal structure at the plasma membrane. Within this interaction network, the upregulation of TRMs and SUNs, or the downregulation of OFPs, can lead to an elongation of tomato fruit [52]. Research shows that SUN/IQD TRMs and OFPs collectively influence the microtubule activity, consequently impacting the shape of the fruit. SUN family proteins (Csa1G575000, CmSUN-14, CsGy1G026840.1, and Cla011257) are known as FL regulators in cucurbitaceous crops [21,52,54,61]. Further investigation shows that the Cla011257 gene impacts the ovary before anthesis to influence FL during the development period [62]. MADS-box transcription factors (TFs) are closely linked with development of a plant, significantly contributing to fruit formation [63]. FUL is a vital TF member of the MADS-box family that plays an important role in regulating FL in various plants. For instance, MBP7/FUL2 in tomatoes regulate the shape of fruits by modulating cell division and enlargement. Fruit fails to elongate after fertilization as a result of knocking out FUL genes and encoding MADS-box proteins in different species, with a significant reduction in valve size [52,64]. In cucumbers, CsFUL1A acts as a negative regulator by inhibiting auxin transport and cell division, thereby affecting FL. CsFUL1A, another MADS-box TF, binds to the CArG-box, regulating cell division and expansion by repressing the expression of auxin transporters PIN1 and PIN7, which reduces auxin accumulation and controls FL [42].
2.3. Fruit Weight
The fruit weight (FW) is intricately linked to the size of the fruit. During the breeding of vegetables, multiple loci governing FW have been identified (Table 1). FW, being a quantitative characteristic, is under the control of numerous loci [65]. In Solanaceae vegetables and especially in tomatoes, several genes, including fw1.1, fw11.2, fw3.3, FW3.2, and the Cell Size Regulator (CSR), involved in regulating FW have been successfully cloned [66]. Notably, FAS and LC homologous to YABBY2 and WUS, respectively, have been identified as genes influencing the locule number (CN) to enhance FW [23,30]. In the Cucurbitaceae family, a number of loci are associated with FW. Cucumbers, for instance, have three QTLs described such as fw6.1, fw2.1 and fw4.1 [67]. In melons, FW has been regulated by genomic regions FWQM11 and FWQM8 [68]. A CYP78A gene with the subfamily KLUH primarily controls the size of organs in Arabidopsis. Various members of subfamily CYP78A are from vegetables, such as GmCYP78A72 (soybean), BnaA9.CYP78A9 (rapeseed), CaKLUH (pepper), and SlKLUH (tomato). GmCYP78A10 have been said to regulate FW, and a recent research also indicated that the upregulation of the transcription factor gene SHINE1 (SlSHN1) can lead to a decline in the FW of a tomato [69,70,71].
2.4. Regulation of Fruit Shapes by Hormone
Phytohormones, responsive to both external and endogenous stimuli during a plant’s development, play a crucial role in various fruit development stages, influencing growth and eventually determining the fruit shape and size [72,73]. Among the numerous plant hormones, comprising cytokinin (CK), ethylene, gibberellin (GA), abscisic Acid (ABA) and auxin, several have been identified for their impact on fruit shape (Table 3). Auxin, in particular, plays a pivotal function in the fleshy fruit development [7,74]. In the study of different cucumber inbred lines by Liu et al. (2020), endogenous hormone content, especially indole-3-acetic acid (IAA), was positively correlated with fruit size and cell growth at different developmental stages [7]. Processing the LA1589 near the isogenic line of eggplant, which is characterized by slender pear-shaped fruits and ovaries, early flowering with auxin (2,4-D) can lead to an increase in cell size and quantity near the fruit or ovary [75]. The gene CsYUC10b, which synthesizes auxin, is associated with fruit arch, and its upregulation induces the development of straight fruit [76]. In addition, SlIAA17 is a transcriptional repressor of auxin/indole-3-acetic acid (Aux/IAA) and is associated with an increase in tomato fruit size [77].
Members of the auxin response factors (ARFs) family, such as SlARF9 in tomatoes and BnaA9.ARF in Brassica rapa, have been identified as regulators of fruit size [69]. Apart from auxin, additional hormones collaborate with specific regulatory factors to influence fruit shape [78]. Cytokinins (CKs), primarily responsible for regulating cell division in plants, exhibit a positive relationship with fruit cell division activity [54,79,80]. The gene responsible for CK biosynthetic (CYP735A) has the capacity to alter cell size and biomass accumulation, thereby impacting size of the fruit [81]. In cucumbers, initial stages of fruit development are dependent on trans-Zeatin riboside (tZR), particularly in the early ovary development phase, influencing cell division. Zeatin (ZT) content, on the other hand, increases in the early stages after flowering, facilitating the horizontal expansion of cells [7]. Gibberellin (GA) plays an important role to stimulate fruit and seed development and regulate flowering [54]. The GA application induces cell increase and can result in parthenocarpy [82]. In cucumbers, GA promotes cell expansion during fruit development [83], and 9–12 days after anthesis, at the mid-to-early stage, counteracts indole-3-acetic acid (IAA), potentially impeding fruit enlargement by delaying cell division. The accumulation of GA during fruit development in tomatoes aligns with the direction of cell division in the early stages [84], while ethylene is predominantly associated with fruit ripening in tomatoes [85], and recent reports suggest its regulatory role in cucumber fruit length.
The ethylene-based content also plays an important role in influencing plant development. The enzyme 1-aminocyclopropane-1-carboxylate synthase 2 (ACS2) is important for catalyzing ethylene biosynthesis [84], and the cucumber mutant with decreased ethylene, acs2, exhibits a reduction in fruit length. Conversely, in sf1 mutants, an excess of ethylene can result in the same phenotype [41]. Hence, an imbalanced ethylene concentration, whether excessive or insufficient, can impact fruit length. The homologous ACS gene, CmACS7, has been linked to the round shape of melon fruits [86]. These findings indicate that the function of dosage-dependent ethylene in Cucurbitaceae is comparatively conserved. In a distinct research based on tomatoes, a number of Aux/IAA-like genes such as DR1, DR3, DR4, and DR8, associated with indole-3-acetic acid (IAA), were controlled by ethylene [87]. Abscisic acid (ABA), known as plant growth inhibitory hormone [88], is noted to have an impact on fruit size and cell shape in tomatoes, as evidenced by the small sized fruit and altered shape of cells in the ABA-deficient mutant [88,89]. The research highlights the synergistic effects or potential antagonistic effect between plant hormones, collectively governing development and ultimately effecting the shape of fruits.

Table 3.
Hormones involved in fruit development.
Table 3.
Hormones involved in fruit development.
| Hormone | Function | Effects on Fruit Shape | Reference |
|---|---|---|---|
| Auxins | Cell elongation, fruit growth | Promotes elongation, determines fruit shape | [90] |
| Cytokinins | Cell division, cell differentiation | Regulates fruit size, influences shape development | [91] |
| Gibberellins | Cell elongation, seed germination | Stimulates elongation, affects fruit size and shape | [92] |
| Abscisic Acid | Seed dormancy, stress response | Regulates fruit maturation, influences shape and ripening | [93] |
| Ethylene | Fruit ripening, senescence | Controls fruit ripening, affects texture and shape | [94] |
3. Role of Hormones in Fruit Ripening
Traditionally, comparative genomic analyses in hot peppers (Capsicum spp.) and tomatoes as models for fruits, respectively, reveal commonalities in gene expression. Specifically, genes encoding transcription factors like tomato AGAMOUS-like 1 (TAGLI), non-ripening (NOR), ripening inhibitor (RIN), and components related to the ethylene signaling pathway are identified as shared features in both fruit types [95]. The presence of MADS-box genes in both categories further suggests overlapping molecular regulatory processes in the maturing of climacteric and also in non-climacteric fruits [96]. Plant hormones play a pivotal role in regulating fruit ripening [97,98]. The combined function of hormones such as cytokinin, gibberellins and auxin helps in normal growth for fruit and the shape of the fruit, even if they have no fertilization; this process is known as parthenocarpy. The subsequent discussion provides an overview of plant hormones involved in the ripening of non-climacteric fruits, specifically in grape, strawberry and raspberry, along with a discussion on potential hormone crosstalk [99]. The subsequent section also discusses various hormones and their associated genes, highlighting their roles in fruit development, as summarized in Table 4.

Table 4.
Key plant hormones, their roles, and associated genes in fruit development.
Table 4.
Key plant hormones, their roles, and associated genes in fruit development.
| Hormones | Role in Fruit Development | Associated Genes | Reference |
|---|---|---|---|
| Auxins (IAA) | Regulates fruit initiation, cell expansion, and ripening | ARF14, ARF7, ARF8, TIR1, FUL1 | [77] |
| Gibberellins (GA) | Promotes fruit growth, elongation, and seed development | GA20ox, GA3ox, RGA, GID1 | [100] |
| Cytokinins (CKs) | Stimulates early fruit development and delays senescence | IPT, CKX, ARR-B, LOG | [101] |
| Abscisic Acid (ABA) | Controls ripening, color development, and anthocyanin synthesis | NCED, PYR/PYL, SnRK2, ABF | [102] |
| Ethylene | Induces ripening, softening, and aroma production | ACS, ACO, EIN2, ETR1 | [103] |
| Jasmonates (JA) | Enhances stress responses and influences ripening | JAZ, MYC2, COI1 | [104] |
| Brassinosteroids (BRs) | Modulates fruit size, weight, and ripening | BRI1, BZR1, DWF4 | [105] |
| Salicylic Acid (SA) | Regulates defense mechanisms and fruit quality traits | NPR1, TGA, PR1 | [106] |
3.1. Abscisic Acid (ABA)
One of the most important phytohormones is abscisic acid (ABA), which significantly influences various plant processes, particularly stress responses, seed dormancy and fruit development [107]. In strawberries and grapes, ABA acts as an important part in the ripening process, especially in the absence of ethylene spikes characteristic of climacteric fruits [93]. Historically recognized as being responsible for grape berry ripening, ABA impacts sugar accumulation, coloration, and softening, essential traits for desirable fruit quality [108]. ABA level during early stages of fruit development is typically low but increases significantly as ripening progresses. This interplay highlights how enhanced flavonoid levels contribute to the overall quality and appeal of the fruit, illustrating ABA’s multifaceted role in fruit ripening and quality enhancement.
3.2. Regulation of Fruit Quality by Exogenous ABA
The influence of ABA goes beyond simple maturation; it also increases the anthocyanin content in strawberries and grapes by upregulating enzymes such as anthocyanin synthase (ANS) and glucosyltransferase (GT) [109]. This relationship emphasizes the regulatory role of ABA not only in the ripening process, but also in fruit color development and nutritional value. In strawberries, ABA treatment accelerates softening, enhances coloration, and increases ethylene production while activating phenylalanine ammonia-lyase (PAL), a key enzyme in the phenylpropanoid pathway that synthesizes flavonoids and other metabolites [109]. Additionally, ABA treatment elevates l-ascorbic acid levels, highlighting its essential role in improving fruit quality. Interestingly, sucrose-induced maturation is associated with an increase in ABA biosynthesis, indicating a synergistic relationship between sugar levels and ABA activity [110]. In “flame seedless” grapes, the application of ABA can increase anthocyanin content, improve color, and accelerate softening—a key factor in consumer acceptance and marketability [111]. These findings emphasize the regulatory role of ABA in regulating the essential metabolites that define mature fruits, creating a feedback loop where these metabolites can affect ABA biosynthesis itself, further illustrating the complexity of fruit ripening mechanisms.
3.3. The Effect of Inhibitors on ABA Biosynthesis and Fruit Characteristics
Exploring the dynamics of ABA during fruit ripening and revealing the effect of inhibitors on its synthesis, a type of ABA synthesis inhibitor treated with fluoroketone leads to a decrease in ABA levels and helps maintain fruit texture under storage conditions [112]. In strawberries, the use of dehydroguaiacolic acid (NDGA), an NCED enzyme inhibitor, not only reduces ABA content but also prevents red coloration in fruit containers, indicating that the enzyme plays a critical role in fruit ripening [113]. This observation reinforces the importance of ABA in forming ideal fruit characteristics. By studying the expression of the key gene FaNCED1 in ABA synthesis, the relationship between ABA and its biosynthesis was further elucidated. In fruit tissue sucrose culture experiments, FaNCED1 expression and subsequent ABA accumulation indicate that this gene is controlled by the accumulation of metabolites such as sucrose during strawberry ripening [114]. This complex interaction illustrates how metabolic signals regulate hormone levels and establish a complex regulatory network during fruit development.
3.3.1. Elucidation of ABA Signaling Pathway
Recent advances in understanding the ABA signaling pathway, particularly in Arabidopsis, have identified two key pathways: the “ABA-ABAR-WRKY40-ABI5” ABA and PYR/PYL/RCAR-PP2C-SnRK2 pathways [115,116,117]. In the first pathway, the binding of ABA to the PYR1 receptor activates sucrose non-fermentation-associated kinase 2 (SnRK2) through inactivation of phosphatase 2C (PP2C) protein. This signaling cascade subsequently activates ABA response element-binding transcription factors (AREB/ABF), leading to the expression of various downstream genes, including genes related to NADPH oxidase and ion channels [88,116]. In contrast, another pathway begins at the ABAR receptor, which interacts with the WRKY40 transcription factor. This interaction occurs under elevated ABA conditions and promotes upregulation of ABA responsive gene expression, which is associated with key transcription factors such as ABI4 and ABI5 [118,119,120]. Characterizing these pathways in crops such as grapes and strawberries can provide a deeper understanding of how ABA signaling affects fruit ripening.
3.3.2. The Agronomic Benefits of ABA on Fruit Quality
In grape varieties such as ’Kyoho’, the expression of a PYR1-like gene (VlPYL1) increases during fruit development, which is associated with improved ABA sensitivity and fruit quality, particularly in terms of anthocyanin content [121]. The overexpression of this gene enhances the transcription of ABA responsive genes, creating an environment conducive to high-quality fruit development. Similarly, in strawberries, downregulation of FaABAR hinders ripening, highlighting the importance of this receptor in fruit ripening [122]. Ultimately, understanding the regulatory role of ABA in fruit ripening is of great significance for agriculture. By utilizing the influence of ABA on basic fruit quality attributes such as color, hardness, and nutritional composition, growers can optimize ripening conditions to improve harvest quality. Innovative strategies have been proposed to improve fruit quality, extend shelf life and enhance consumer satisfaction in response to the ABA signaling pathway [48]. A deeper understanding of the complex relationship between ABA and fruit development can pave the way for advances in horticultural science and sustainable agriculture, ensuring that the growing demand for high-quality agricultural products is met while improving the economic feasibility of farmers [123].
In short, the regulatory function of ABA goes beyond academic interest. It has transformative potential for agricultural practices, promoting healthier diets and sustainable food systems. The in-depth understanding of the complex interactions between ABA and metabolic pathways emphasizes the necessity of continuous research. Understanding the role of ABA in fruit ripening can lead to innovative agricultural technologies that improve fruit quality and sustainability. With the increasing demand for high-quality agricultural products, strategic ABA applications may provide solutions that benefit consumers and producers, thereby fostering a resilient agricultural sector. Utilizing these insights can ensure a sustained supply of nutritious and attractive fruits for a growing population.
3.4. Indole-3-Acetic Acid (IAA)
Auxins, especially indole-3-acetic acid (IAA), are key plant hormones that can coordinate various developmental processes, including fruit development [124]. Its role in climacteric and non-climacteric fruits has been extensively studied, highlighting its dual function as a growth promoter and regulator of development time. The involvement of auxin in fruit development is complex, involving interactions with other hormones, transport mechanisms, and gene expression regulation [125].
3.4.1. The Role of Auxin in Fruit Growth Kinetics
Auxin is mainly synthesized through a two-step process, involving tryptophan as a precursor. In Arabidopsis, this biosynthetic pathway is promoted by the YUCCA (YUC) family, containing flavin monooxygenases, and the Arabidopsis tryptophan aminotransferase 1 (TAA1) family. Indole-3-pyruvic acid is formed through tryptophan conversion with the help of the TAA1 protein family, followed by conversion to YUC protein and IAA. Studies on grapes have shown that this biosynthetic pathway is active throughout the entire berry development process, with specific TAR and YUCCA genes showing high levels of expression during early fruit development and the onset of ripening [126]. This indicates that auxin synthesis is crucial during critical developmental stages, demonstrating its role in establishing the fruit setting and subsequent growth. In non-climacteric fruits such as strawberries, auxin plays a crucial role in the development of achenes and flower beds. The main source of auxin in strawberries is achene, which promotes the growth of flower beds. Removing achenes from immature containers can hinder growth and expansion, leading to upregulation of maturation-related genes [127,128]. This indicates that auxin is not only crucial for fruit size, but also for ripening time and overall fruit quality.
As is well known, auxin can stimulate fruit growth, so its impact on ripening is more subtle. In grapes, the high concentration of IAA in young fruits decreases with the maturation process, indicating that auxin may inhibit sugar accumulation and anthocyanin production, leading to a complex relationship of delayed maturation [129]. Throughout the entire maturation process, the regulation of IAA levels highlights the dual role of this hormone as a growth promoter and maturation kinetics regulator.
In strawberries, transcriptional analysis of fruit treated with auxin showed downregulation of genes involved in flavonoid biosynthesis, aroma production, and cell wall modification. For example, genes encoding chalcone synthase, alcohol acyltransferase and pectin lyase were significantly affected by auxin treatment, indicating that it not only affects growth but also directly affects metabolic pathways associated with maturation [10].
3.4.2. Auxin Transport and Internal Balance
The polar transport of auxin is promoted by specific transport proteins, including AUX/LAX and PIN proteins, which play a crucial role in establishing the auxin gradient required for directed growth and development. In grapevines, radiolabeled IAA studies revealed a basal distribution pattern in the skin cells, attributed to the activity of the speculated VvPIN protein. It is worth noting that during the process of young fruit abscission, the expression of several VvPIN genes decreases, indicating that auxin homeostasis is crucial for fruit setting [130].
In strawberries, auxin transporters exhibit developmentally specific transcription patterns, indicating their role in fruit growth and maturation. The identification of 10 FvPIN genes and 4 FvAUX/LAX genes in the genome of forest strawberries emphasizes the genetic basis of auxin transport in this species [131]. In addition, the expression of these genes at different stages of fruit development indicates that auxin transport dynamics are essential for the development of both achenes and flower beds.
3.4.3. Auxin Binding and Metabolism
The homeostasis of auxin is also regulated through binding processes, which activate or deactivate IAA. The IAA amide synthase (GH3) family plays a crucial role in this process, promoting the binding of IAA to amino acids, thereby affecting its availability and activity in plants. For example, in peaches, a potential IAA amide hydrolase was observed to remain upregulated during fruit ripening, similar to IAA-Leucine RESISTANT 1 (ILR1) in Arabidopsis, indicating the existence of a mechanism regulating auxin levels at this critical stage [132]. Under the stimulation of exogenous auxin, the expression of VvGH3.1 in grapes is associated with berry ripening, highlighting the importance of auxin binding in regulating fruit ripening [97]. The balance between IAA binding and hydrolysis seems crucial for maintaining auxin levels that are beneficial for fruit ripening and overall quality.
The interaction between auxin and various other plant hormones, particularly abscisic acid (ABA), ethylene, and gibberellin (GA), is crucial for fruit development and maturation. For example, in climacteric fruits such as tomatoes, auxin interacts with ethylene to regulate the ripening process [133]. The RIN (RIPENING INHIBITOR) gene is a MADS-box transcription factor that responds to auxin and plays a central role in this interaction. Recent studies have shown that RIN is optional in the early stages of fruit ripening and suggest that auxin signaling may be compensated by other MADS-box proteins during this stage [134].
In strawberries, the interaction between auxin, ethylene, and ABA also affects ripening kinetics. As is well known, ethylene increases the expression of auxin responsive genes, making the regulatory network that controls fruit ripening more complex. The balance between these hormones may determine the timing and quality of strawberry ripening and have an impact on commercial fruit production.
3.4.4. Genetic Regulation of Auxin Signaling Pathway
Transcription factors involved in auxin signaling, such as auxin response factor (ARF) and F-box protein, are key mediators of a plant’s response to auxin. In cucumbers, two F-box auxin receptors, CsTIR1 and CsAFB2, were identified, and their expression was highest in young fruit tissues [27]. In transgenic tomato lines overexpressing these genes, monosexual fruit development was observed, emphasizing the potential for manipulating auxin signaling to promote fruit setting and development. In strawberries, the differential expression of ARF throughout the entire maturation stage indicates a complex regulatory framework, in which not all ARF genes have a consistent response to auxin levels. For example, the comparative expression patterns of ARF1 and ARF4 during ripening indicate that specific ARF genes may promote or inhibit fruit development, depending on the developmental environment [135].
Although substantial progress has been made in understanding the role of auxin in fruit development, there are still many issues that need to be addressed. Further clarification is needed on the genetic and molecular mechanisms of auxin biosynthesis, transport, and signaling in non-climacteric fruits. Specifically, exploring the interactions between auxin and other hormones during fruit development and ripening remains a promising field for future research. The transcription factors that mediate auxin response, such as auxin response factor (ARF) and MADS-box proteins, particularly their interactions with other hormone pathways, deserve further investigation. The use of advanced molecular technologies such as CRISPR-based gene editing and RNA interference can provide new insights into the regulatory networks that affect fruit development, ultimately improving fruit quality and supporting sustainable agricultural practices.
3.5. Gibberellins
Gibberellin (GA) is a cyclic diterpenoid compound that is important in many growth processes, such as cell division, seed germination, flower induction, fruit growth, and elongation [126]. There are multiple GAs present in plants, with only a few having biological activity [136,137]. The balance of bioactive gibberellins is maintained through synthesis and inactivation processes, mainly mediated by enzymes such as gibberellin 2-oxidase (GA2ox) and gibberellin 3-oxidase (GA3ox) [138]. Thompson (1969) documented preliminary observations on the effects of exogenous GA application on strawberry receptacle development, and subsequent studies linked GA to fruit ripening, particularly in strawberries [139]. During the development of strawberry fruit, it has been found that the levels of bioactive GA increase, especially in stages 1, 3, and 4, with GA4 concentration reaching its peak during the white development stage.
3.5.1. The Mechanism of Action of Gibberellin in Fruit Growth
The mechanism by which GAs affect fruit growth involves complex genetic and molecular pathways. Key genes related to the GA pathway, such as DELLA, FaGID1c, and FaGID1 (GIBBERELLIN-INSENSITIVE DWARF1b), as well as proteins such as FaRGA (GA REPRESSOR), are upregulated in strawberry receptacle tissue during development [136]. It is worth noting that FaGID1c exhibits GA binding, interacts with FaRGA in vitro, and enhances GA response when expressed ectopically in Arabidopsis. When gibberellin stimulates overexpression of the transcription 2 (FaGAST2) gene in transgenic strawberries, it reduces fruit size, indicating that FaGAST2 is associated with cell elongation and fruit size regulation [140]. In contrast, silencing FaGAST2 resulted in the increased expression of FaGAST1 without altering cell size, indicating complex interactions between these transcription factors in determining fruit cell size and development.
3.5.2. Gibberellins in Viticulture
The application of exogenous GAs has garnered significant attention in viticulture, particularly regarding grape development. Pre-bloom GA3 application has been shown to foster seedlessness and increase the size of berries in seedless grapevines [141]. Transcriptome sequencing studies have indicated a potential role for grapevine miRNAs in berry development and responses to environmental conditions [142]. Furthermore, GA application in the ’Kyoho’ grape stimulated flower opening, facilitated fruit coloring, and led to seed abortion [143]. In Rubus genus, the role of GAs in fruit development is not well studied; some studies indicate that GA application can induce asexual fruit growth in cloudberry (Rubus chamaemorus L.) [144] and affect flower numbers in raspberries [145]. Future research should focus on elucidating the complex regulatory networks governing GA action in fruit development, including advanced molecular techniques like CRISPR and RNA interference, to enhance our understanding of GAs’ influence on fruit growth and ripening.
3.6. Ethylene
In recent years, the involvement of ethylene fruit ripening (especially non-climacteric fruits) has attracted widespread attention. The results of various studies indicate that it plays an important role in the ripening process of different grape varieties, with the peak of ethylene occurring before grape ripening [146]. The use of ethylene receptor inhibitor 1-methylcyclopropene (1-MCP) leads to a reduction in ripening-related factors and berry size in Cabernet Sauvignon grapes, such as anthocyanin accumulation [147]. On the other hand, ethylene treatment increased berry size and was associated with high expression levels of xyloglucan endoglucanase (XTHs), polygalacturonase, aquaporin, elastin, and cellulase [148]. On the contrary, the application of 1-methylcyclopropene prior to its presence reduced ABA levels in Muscat Hamburg grapes, indicating a possible interaction between ABA and ethylene during maturation [149]. In Fragaria x ananassa (strawberry), the concentration of ethylene is relatively low, and its yield varies depending on the stage of fruit development. The ethylene content is moderate in green fruits, lower in white fruits, and significantly increases during the red stage of fruit ripening. The increase in ethylene production during the red fruit stage is consistent with the increase in respiratory rate, like the maturation pattern observed in climacteric fruits [10].
3.6.1. Production Modes of Ethylene in Different Fruits
Raspberry fruit exhibits a unique ethylene production mode. Unlike fruits such as strawberries and grapes, strawberries and grapes reach their peak ethylene activity in the early stages of fruit development [150], while raspberry’s ethylene production increases continuously during ripening, which is in stark contrast to the typical patterns observed in strawberries and wine [10,151]. The ethylene production of raspberry fruits is negatively correlated with hardness loss, and containers have been identified as the main source of ethylene [152,153]. In addition, experiments conducted on raspberry fruits in the white stage showed that the loss of fruit hardness was delayed during 1-MCP in vitro treatment when stored at 10 °C. These findings suggest that ethylene may play a partial role in regulating the softening process during raspberry ripening. In strawberries, the segregation of four FaACS genes and three FaACO genes has been recorded, with each gene exhibiting different expression patterns throughout the entire maturation process [154]. Similarly, various grape varieties such as Cabernet Sauvignon, Hamburg Muscat, and Thompson Seedless have shown that the presence of the VvACO gene is associated with an increase in ethylene, which occurs prior to its presence [23]. It is worth noting that during the development of strawberry fruit, the expression dynamics of two FaACO genes and the ethylene response sensor (FaErs1) have been observed, indicating a correlation between the expression of these genes and ethylene production.
3.6.2. Ethylene Signal Transduction and Its Genetic Regulation
Many studies have emphasized changes in gene expression related to the ethylene signaling pathway during the ripening process of climacteric and non-climacteric fruits [155,156]. These genes include genes encoding ethylene receptor (ETR), ethylene response sensor (ERS), ethylene insensitive protein (EIN), and constitutive triple reactive protein (CTR1). CTR1 is located on the endoplasmic reticulum (ER) membrane and serves as an intermediary between ETRs and EIN2s, playing a critical role in ethylene signaling transduction [155]. The ETR family, composed of transmembrane proteins in the ER, forms stable dimers with two disulfide bonds at the N-terminus upon ethylene binding. These receptors act as negative regulators of the ethylene pathway, blocking downstream signaling when ethylene is absent [155,156]. In transgenic tomatoes, early ripening was triggered by downregulating the SlETR4 gene [157].
In grape development, VvETR2 expression increased at the start of ripening, while VvETR1 remained consistently expressed. Similarly, VvERS1 and VvEIN4 exhibited higher expression levels shortly after anthesis. In strawberries, the expression of ethylene receptor genes FaEtr1, FaErs1, and FaEtr2 coincided with increased ethylene production, with FaEtr2 being predominantly expressed in ripe fruit. This suggests that even the low levels of ethylene produced during strawberry ripening are sufficient to activate ripening processes [158].
Further analysis revealed that the ethylene biosynthesis gene FaSAMS1 and the signaling gene FaCTR1 were transcriptionally induced in parallel with ethylene production during the fruit’s color change. The downregulation of these genes through the tobacco rattle virus-induced gene silencing (VIGS) system affects the production of red color, hardness, and ethylene. In addition, the application of ethephon (a synthetic ethylene-releasing agent) promotes the natural softening and color development of strawberries, partially restoring the biosynthesis of anthocyanins, although the effect on hardness is relatively small. This suggests that FaCTR1 may play a role in regulating strawberry ripening, although it is unclear whether ethylene is involved in the early ripening stage of this non-climacteric fruit [159].
3.7. Jasmonates
Jasmonic acid (JA) and its bioactive isoleucine conjugate (JA Ile) are key signals in various plant stress responses, affecting root growth, seed germination, stamen development, and senescence [160].
3.7.1. The Endogenous Effect of JA
A recent study suggests that endogenous JA levels (including JA Ile) and the expression of their biosynthetic genes decrease synchronously from the flowering to maturity stage of strawberry fruits [161]. During the early development of grape berries, elevated levels of JA and JA Ile were detected, followed by a sharp decline as the fruit matured [162]. This suggests that JA Ile may be related to the early fruit development of strawberries and grape berries.
It is worth noting that the accumulation pattern of anthocyanins (PA) during the development of strawberries and grape berries reflects the accumulation pattern of JA Ile, reaching its peak in the early stages of fruit development and decreasing with fruit ripening [163]. Observations have shown that the application of chemical inhibitors targeting JAR1 increases PA content. JAR1 is a key enzyme in JA Ile biosynthesis, indicating a potential link between the JA pathway and PA biosynthesis in strawberry fruit [164].
In Fragaria chiloensis fruit, exogenous methyl jasmonate (MeJA) significantly altered the expression of maturation-related genes, including genes involved in the biosynthesis of ethylene and jasmonic acid (JA) [165]. MeJA treatment also promoted anthocyanin accumulation by upregulating key genes in the anthocyanin biosynthesis pathway, such as chalcone synthase (FcCHS), chalcone flavonoid isomerase (FcMHI), flavanone 3-hydroxylase (FcF3H), dihydroflavonol 4-reductase (FcDFR), anthocyanin synthase (Fc ANS), and anthocyanin 3-O-glucosyltransferase (Fc0FGT). The increase in anthocyanin levels is also related to the upregulation of JA biosynthesis genes, including 13 lipoxygenase (FcLOX), propadiene oxide synthase (FCAO), and 12 oxo plant diesterase 3 (FcOPR3) [165]. Similarly, the application of MeJA in Fragaria x ananassa resulted in a significant increase in anthocyanin content and elevated levels of JA, JA Ile, and MeJA. JA has been shown to increase anthocyanin production in grape cell suspension, and the application of MeJA has increased the content of proanthocyanidins (PA) in two wine grape varieties [166]. In addition, research on raspberries emphasizes the role of jasmonate in enhancing phenylalanine ammonia lyase (PAL) activity, resulting in higher levels of polyphenolic compounds such as tannic acid, quercetin, and myricetin [167].
3.7.2. Jasmonic Acid and Isoleucine Signaling Pathways
The biological processes regulated by JA Ile involve the activation of the jasmonic acid (JA) signaling pathway, in which the F-box protein CORONATINE INSENSITIVE1 (COI1) forms a co-receptor with the jasmonic acid ZIM-DOMAIN (JAZ) protein. In the absence of sufficient JA Ile, JAZ repressors bind to transcription factors such as MYC2, inhibiting the expression of early JA responsive genes. When JA Ile levels increase, COI1 interacts with JAZ protein, leading to its ubiquitination and subsequent degradation by the 26S proteasome. This degradation releases MYC2 and other transcription factors, thereby activating JA responsive genes. Eleven JAZ members have been identified in grapevines, which respond to various stresses, hormones, and abiotic treatments [168]. Recent studies have shown that 12 potential JAZ proteins and two MYC transcription factor genes in strawberries are highly expressed during the flower and early fruit stages, corresponding to the downregulation of JA Ile observed during fruit development [161].
3.8. Brassinosteroids
Brassinosteroids (BR) are essential steroid plant hormones that regulate various plant processes, including cell division, elongation, vascular differentiation, flowering, pollen development, and photomorphogenesis [169]. They also play an important role in the fruit development and maturation of crops such as tomatoes, cucumbers, grapes, and strawberries [65,170]. Notably, BR has been shown to enhance anthocyanin biosynthesis, which contributes to the color development in non-climacteric fruits like strawberries and grapes. Additionally, BR influences ripeness by modulating key transcription factors that regulate fruit pigmentation and skin quality in these species [171].
3.8.1. The Impact of BR on Maturity
In grape berries, the use of brassinolide (BR), especially brassinolide, has been found to enhance berry color and accelerate ripening. On the contrary, the use of BR biosynthesis inhibitor brasinazole (BZ) has the opposite effect. The enzyme BR 6-oxidase is responsible for converting 6-deoxytestosterone into active BR testosterone [172]. Overexpression of the grape BR 6-oxidase gene (VvBR6OX1) has been shown to restore the normal height of dwarf tomato plants lacking functional dwarf genes, enabling them to reach the same height as wild-type plants [172]. In strawberries, BL application has been observed to promote maturation and increase the expression of FaBRI1 receptors. In addition, temporary inhibition of the FaBRI1 gene leads to delayed maturation, resulting in the fruit clusters remaining white [170,173]. These findings highlight the critical role of BR signaling in the ripening of non-climacteric fruits.
3.8.2. The Regulatory Role of BR and ABA
Brassinolide (BR) is a crucial plant hormone that regulates various growth processes, such as cell elongation and differentiation, by modulating gene expression related to cell wall loosening. Its ability to enhance cell division and expansion is vital for fruit size and shape development. BR is considered an initial signal of grape fruit ripening and may affect ethylene levels [129]. In terms of BR response genes, the late embryogenesis enriched (LEA) domain protein 1 (LDP1) gene is expressed in the early developmental stages of F. chilonensis and F. vesca, particularly in containers [174]. The promoter region of the LDP1 gene contains several motifs that respond to both BR and abscisic acid (ABA). Research has shown that the transient expression of FcLDP promoter GFP fusion is regulated by BR and ABA, highlighting the regulatory effects of these two hormones on FcLDP expression during fruit development in F. chilonensis [174].
3.9. Cytokinins
The study of cytokinins (CKs) in the development and maturation of non-climacteric fruits is an area that requires further exploration. Bombarely et al. (2016) identified two genes related to the CK signaling pathway in various F. x ananassa varieties from a fruit cDNA library, particularly the histidine phosphotransferase protein (AHP) and nuclear reaction regulatory factor (ARR) genes [175]. Afterwards, Kang et al. (2013) examined the transcriptome of F. vesca during the pre-fertilization and post-fertilization stages of fruit development and identified 17 differentially expressed genes (DEGs) associated with CK biosynthesis, signal transduction, and various fruit tissue degradation. The report indicates that CKs play a crucial role in the early stages of strawberry fruit development [176], just as they do in the early development of climacteric fruits such as tomatoes [177]. In grapes, the synthetic cytokinin forchlorfenuron (N-(2-chloro-4-pyridyl)-N’-phenylurea), known as CPPU, has been associated with increased berry weight, although it also led to reductions in sugar and anthocyanin content [178,179]. Notably, the only documented role of CKs in the Rubus genus suggests potential synergies between gibberellins (GAs) and CKs during flower induction in raspberries [180].
4. Conclusions
In conclusion, the molecular investigation of non-climacteric fruit models, such as grape, strawberry, and lesser-studied species like raspberry, has revealed that various phytohormones (including ABA, auxin, ethylene, and others) interact or regulate one another to influence multiple molecular and biochemical processes that determine fruit quality during the onset of ripening. Common genes such as FW2.2, OVATE, SUN, and CLV-WUS are frequently identified as crucial regulators of fruit shape across various fruit plants. These genes are involved in key processes like cell division and expansion, impacting overall fruit morphology. The growing body of research identifying and characterizing key genes associated with the signaling and perception of these hormones in grapes and strawberries could enhance our understanding of the ripening processes in other non-climacteric fruits, including under-researched species like Rubus. This knowledge may also guide strategies to improve post-harvest quality and food security.
Author Contributions
Conceptualization, M.R. and M.G.; methodology, Z.X.; software, A.S.; validation, S.F., K.C. and H.X.; formal analysis, J.Y.; investigation, C.C.; resources, C.C.; data curation, M.R.; writing—original draft preparation, Z.X.; writing—review and editing, M.G.; visualization, C.C.; supervision, C.C.; project administration, M.R.; funding acquisition, C.C. All authors have read and agreed to the published version of the manuscript.
Funding
The authors gratefully acknowledge financial support from Jiangxi Province Double Thousand Talent-Leader of Natural Science Project (JXSQ2023101038), Jiangxi Province Urgently Overseas Talent Project (2022BCJ25027), and The Key Research Projects in Jiangxi Province (20223BBH8007 and 20232BBG70014). This work was also funded by the key research program of Jiujiang City (S2023ZDYFN796, S2024ZDSYS037, S2024KXJJ0001).
Data Availability Statement
The authors declare no data to share.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Giovannoni, J.J. Genetic regulation of fruit development and ripening. Plant Cell 2004, 16, S170–S180. [Google Scholar] [CrossRef] [PubMed]
- Seymour, G.; Poole, M.; Manning, K.; King, G.J. Genetics and epigenetics of fruit development and ripening. Curr. Opin. Plant Biol. 2008, 11, 58–63. [Google Scholar] [CrossRef] [PubMed]
- Pnueli, L.; Hareven, D.; Rounsley, S.D.; Yanofsky, M.F.; Lifschitz, E. Isolation of the tomato AGAMOUS gene TAG1 and analysis of its homeotic role in transgenic plants. Plant Cell 1994, 6, 163–173. [Google Scholar] [PubMed]
- Mahmood, U.; Fan, Y.; Wei, S.; Niu, Y.; Li, Y.; Huang, H.; Chen, Y.; Tang, Z.; Liu, L.; Qu, C. Comprehensive analysis of polygalacturonase genes offers new insights into their origin and functional evolution in land plants. Genomics 2021, 113, 1096–1108. [Google Scholar] [CrossRef]
- Xiang, Y.; Zhang, T.; Zhao, Y.; Dong, H.; Chen, H.; Hu, Y.; Huang, C.H.; Xiang, J.; Ma, H. Angiosperm-wide analysis of fruit and ovary evolution aided by a new nuclear phylogeny supports association of the same ovary type with both dry and fleshy fruits. J. Integr. Plant Biol. 2024, 66, 228–251. [Google Scholar] [CrossRef]
- Gong, L.; Paris, H.S.; Nee, M.H.; Stift, G.; Pachner, M.; Vollmann, J.; Lelley, T. Genetic relationships and evolution in Cucurbita pepo (pumpkin, squash, gourd) as revealed by simple sequence repeat polymorphisms. Theor. Appl. Genet. 2012, 124, 875–891. [Google Scholar] [CrossRef]
- Liu, X.; Pan, Y.; Liu, C.; Ding, Y.; Wang, X.; Cheng, Z.; Meng, H. Cucumber fruit size and shape variations explored from the aspects of morphology, histology, and endogenous hormones. Plants 2020, 9, 772. [Google Scholar] [CrossRef]
- Jiang, S.; An, H.; Luo, J.; Wang, X.; Shi, C.; Xu, F. Comparative analysis of transcriptomes to identify genes associated with fruit size in the early stage of fruit development in Pyrus pyrifolia. Int. J. Mol. Sci. 2018, 19, 2342. [Google Scholar] [CrossRef]
- Corpas, F.J.; Freschi, L.; Palma, J.M. ROS metabolism and ripening of fleshy fruits. In Advances in Botanical Research; Elsevier: Amsterdam, The Netherlands, 2023; Volume 105, pp. 205–238. [Google Scholar]
- Fuentes, L.; Figueroa, C.R.; Valdenegro, M. Recent advances in hormonal regulation and cross-talk during non-climacteric fruit development and ripening. Horticulturae 2019, 5, 45. [Google Scholar] [CrossRef]
- Batista-Silva, W.; Nascimento, V.L.; Medeiros, D.B.; Nunes-Nesi, A.; Ribeiro, D.M.; Zsögön, A.; Araújo, W.L. Modifications in organic acid profiles during fruit development and ripening: Correlation or causation? Front. Plant Sci. 2018, 9, 1689. [Google Scholar] [CrossRef]
- Janick, J.; Paull, R.E. The Encyclopedia of Fruit & Nuts; CABI: Wallingford, UK, 2008. [Google Scholar]
- Morton, J.F. Fruits of Warm Climates; Florida Flair Books: Boynton Beach, FL, USA, 1987. [Google Scholar]
- Whitaker, T.W.; Davis, G.N. Cucurbits; Leonard Hill Ltd.: London, UK, 1962. [Google Scholar]
- Hancock, J. Strawberries; CABI: Wallingford, UK, 1999. [Google Scholar]
- Paull, R.; Duarte, O. Annonas: Soursop and Rollinia. In Tropical Fruits; CABI: Wallingford, UK, 2012; Volume 2, pp. 1–24. [Google Scholar]
- Liu, Z.; Ma, H.; Jung, S.; Main, D.; Guo, L. Developmental mechanisms of fleshy fruit diversity in Rosaceae. Annu. Rev. Plant Biol. 2020, 71, 547–573. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Xu, Y.; Fang, P.; Guo, Q.; Huang, W.; Hou, J.; Wan, H.; Zhang, S. Genetic Regulation of Fruit Shape in Horticultural Crops: A Review. Horticulturae 2024, 10, 1151. [Google Scholar] [CrossRef]
- Brewer, M.T.; Lang, L.; Fujimura, K.; Dujmovic, N.; Gray, S.; van der Knaap, E. Development of a controlled vocabulary and software application to analyze fruit shape variation in tomato and other plant species. Plant Physiol. 2006, 141, 15–25. [Google Scholar] [PubMed]
- Rodríguez, G.R.; Muños, S.; Anderson, C.; Sim, S.-C.; Michel, A.; Causse, M.; Gardener, B.B.M.; Francis, D.; van Der Knaap, E. Distribution of SUN, OVATE, LC, and FAS in the tomato germplasm and the relationship to fruit shape diversity. Plant Physiol. 2011, 156, 275–285. [Google Scholar]
- Pan, Y.; Wang, Y.; McGregor, C.; Liu, S.; Luan, F.; Gao, M.; Weng, Y. Genetic architecture of fruit size and shape variation in cucurbits: A comparative perspective. Theor. Appl. Genet. 2020, 133, 1–21. [Google Scholar]
- Kitagawa, M.; Jackson, D. Control of meristem size. Annu. Rev. Plant Biol. 2019, 70, 269–291. [Google Scholar] [CrossRef]
- Wang, C.; Cao, J.; Hao, N.; Wu, T. Genetic and molecular regulation mechanisms in the formation and development of vegetable fruit shape. Appl. Sci. 2022, 12, 1514. [Google Scholar] [CrossRef]
- Galli, M.; Gallavotti, A. Expanding the regulatory network for meristem size in plants. Trends Genet. 2016, 32, 372–383. [Google Scholar]
- Che, G.; Gu, R.; Zhao, J.; Liu, X.; Song, X.; Zi, H.; Cheng, Z.; Shen, J.; Wang, Z.; Liu, R. Gene regulatory network controlling carpel number variation in cucumber. Development 2020, 147, dev184788. [Google Scholar]
- Yuste-Lisbona, F.J.; Fernández-Lozano, A.; Pineda, B.; Bretones, S.; Ortíz-Atienza, A.; García-Sogo, B.; Müller, N.A.; Angosto, T.; Capel, J.; Moreno, V. ENO regulates tomato fruit size through the floral meristem development network. Proc. Natl. Acad. Sci. USA 2020, 117, 8187–8195. [Google Scholar]
- Xu, P.; Cao, S.; Hu, K.; Wang, X.; Huang, W.; Wang, G.; Lv, Z.; Liu, Z.; Wen, J.; Yi, B. Trilocular phenotype in Brassica juncea L. resulted from interruption of CLAVATA1 gene homologue (BjMc1) transcription. Sci. Rep. 2017, 7, 3498. [Google Scholar] [CrossRef]
- Lippman, Z.; Tanksley, S.D. Dissecting the genetic pathway to extreme fruit size in tomato using a cross between the small-fruited wild species Lycopersicon pimpinellifolium and L. esculentum var. Giant Heirloom. Genetics 2001, 158, 413–422. [Google Scholar] [CrossRef] [PubMed]
- Niu, H.; Liu, X.; Tong, C.; Wang, H.; Li, S.; Lu, L.; Pan, Y.; Zhang, X.; Weng, Y.; Li, Z. The WUSCHEL-related homeobox1 gene of cucumber regulates reproductive organ development. J. Exp. Bot. 2018, 69, 5373–5387. [Google Scholar] [CrossRef] [PubMed]
- Chu, Y.H.; Jang, J.C.; Huang, Z.; van der Knaap, E. Tomato locule number and fruit size controlled by natural alleles of lc and fas. Plant Direct 2019, 3, e00142. [Google Scholar] [CrossRef] [PubMed]
- Correa, J.P.d.O.; Silva, E.M.; Nogueira, F.T. Molecular control by non-coding RNAs during fruit development: From gynoecium patterning to fruit ripening. Front. Plant Sci. 2018, 9, 1760. [Google Scholar] [CrossRef]
- Grandillo, S.; Termolino, P.; van der Knaap, E. Molecular mapping of complex traits in tomato. In Genetics, Genomics, and Breeding of Tomato; CRC Press: Boca Raton, FL, USA, 2013; pp. 150–227. [Google Scholar]
- Wu, S.; Clevenger, J.P.; Sun, L.; Visa, S.; Kamiya, Y.; Jikumaru, Y.; Blakeslee, J.; van der Knaap, E. The control of tomato fruit elongation orchestrated by sun, ovate and fs8. 1 in a wild relative of tomato. Plant Sci. 2015, 238, 95–104. [Google Scholar] [CrossRef]
- Wu, S.; Zhang, B.; Keyhaninejad, N.; Rodríguez, G.R.; Kim, H.J.; Chakrabarti, M.; Illa-Berenguer, E.; Taitano, N.K.; Gonzalo, M.J.; Díaz, A. A common genetic mechanism underlies morphological diversity in fruits and other plant organs. Nat. Commun. 2018, 9, 4734. [Google Scholar] [CrossRef]
- Yu, T.; Ai, G.; Xie, Q.; Wang, W.; Song, J.; Wang, J.; Tao, J.; Zhang, X.; Hong, Z.; Lu, Y. Regulation of tomato fruit elongation by transcription factor BZR1. 7 through promotion of SUN gene expression. Hortic. Res. 2022, 9, uhac121. [Google Scholar] [CrossRef]
- Barry, C.S.; McQuinn, R.P.; Chung, M.-Y.; Besuden, A.; Giovannoni, J.J. Amino acid substitutions in homologs of the STAY-GREEN protein are responsible for the green-flesh and chlorophyll retainer mutations of tomato and pepper. Plant Physiol. 2008, 147, 179–187. [Google Scholar] [CrossRef]
- Guo, J.-E. Histone deacetylase gene SlHDT1 regulates tomato fruit ripening by affecting carotenoid accumulation and ethylene biosynthesis. Plant Sci. 2022, 318, 111235. [Google Scholar] [CrossRef]
- Weng, Y.; Colle, M.; Wang, Y.; Yang, L.; Rubinstein, M.; Sherman, A.; Ophir, R.; Grumet, R. QTL mapping in multiple populations and development stages reveals dynamic quantitative trait loci for fruit size in cucumbers of different market classes. Theor. Appl. Genet. 2015, 128, 1747–1763. [Google Scholar] [CrossRef] [PubMed]
- Wei, Q.-z.; Fu, W.-y.; Wang, Y.-z.; Qin, X.-d.; Wang, J.; Li, J.; Lou, Q.-f.; Chen, J.-f. Rapid identification of fruit length loci in cucumber (Cucumis sativus L.) using next-generation sequencing (NGS)-based QTL analysis. Sci. Rep. 2016, 6, 27496. [Google Scholar] [CrossRef] [PubMed]
- Borovsky, Y.; Paran, I. Characterization of fs10. 1, a major QTL controlling fruit elongation in Capsicum. Theor. Appl. Genet. 2011, 123, 657–665. [Google Scholar] [CrossRef] [PubMed]
- Xin, T.; Zhang, Z.; Li, S.; Zhang, S.; Li, Q.; Zhang, Z.-H.; Huang, S.; Yang, X. Genetic regulation of ethylene dosage for cucumber fruit elongation. Plant Cell 2019, 31, 1063–1076. [Google Scholar] [CrossRef]
- Zhao, J.; Jiang, L.; Che, G.; Pan, Y.; Li, Y.; Hou, Y.; Zhao, W.; Zhong, Y.; Ding, L.; Yan, S. A functional allele of CsFUL1 regulates fruit length through repressing CsSUP and inhibiting auxin transport in cucumber. Plant Cell 2019, 31, 1289–1307, Erratum in Plant Cell 2020, 32, 2048–2055. [Google Scholar] [CrossRef]
- Wang, H.; Sun, J.; Yang, F.; Weng, Y.; Chen, P.; Du, S.; Wei, A.; Li, Y. CsKTN1 for a katanin p60 subunit is associated with the regulation of fruit elongation in cucumber (Cucumis sativus L.). Theor. Appl. Genet. 2021, 134, 2429–2441. [Google Scholar] [CrossRef]
- Liu, J.; Van Eck, J.; Cong, B.; Tanksley, S.D. A new class of regulatory genes underlying the cause of pear-shaped tomato fruit. Proc. Natl. Acad. Sci. USA 2002, 99, 13302–13306. [Google Scholar] [CrossRef]
- Zhou, S.; Hu, Z.; Li, F.; Tian, S.; Zhu, Z.; Li, A.; Chen, G. Overexpression of SlOFP20 affects floral organ and pollen development. Hortic. Res. 2019, 6, 125. [Google Scholar]
- Rodríguez, G.R.; Kim, H.J.; Van Der Knaap, E. Mapping of two suppressors of OVATE (sov) loci in tomato. Heredity 2013, 111, 256–264. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, Q.; Hao, W.; Sun, H.; Zhang, L. Characterization of the OFP gene family and its putative involvement of tuberous root shape in radish. Int. J. Mol. Sci. 2020, 21, 1293. [Google Scholar] [CrossRef]
- Li, B.-J.; Grierson, D.; Shi, Y.; Chen, K.-S. Roles of abscisic acid in regulating ripening and quality of strawberry, a model non-climacteric fruit. Hortic. Res. 2022, 9, uhac089. [Google Scholar] [PubMed]
- Li, Q.; Luo, S.; Zhang, L.; Feng, Q.; Song, L.; Sapkota, M.; Xuan, S.; Wang, Y.; Zhao, J.; Van Der Knaap, E. Molecular and genetic regulations of fleshy fruit shape and lessons from Arabidopsis and rice. Hortic. Res. 2023, 10, uhad108. [Google Scholar]
- Snouffer, A.; Kraus, C.; van der Knaap, E. The shape of things to come: Ovate family proteins regulate plant organ shape. Curr. Opin. Plant Biol. 2020, 53, 98–105. [Google Scholar] [PubMed]
- Hamant, O.; Heisler, M.G.; Jonsson, H.; Krupinski, P.; Uyttewaal, M.; Bokov, P.; Corson, F.; Sahlin, P.; Boudaoud, A.; Meyerowitz, E.M. Developmental patterning by mechanical signals in Arabidopsis. Science 2008, 322, 1650–1655. [Google Scholar] [CrossRef] [PubMed]
- Lazzaro, M.D.; Wu, S.; Snouffer, A.; Wang, Y.; Van der Knaap, E. Plant organ shapes are regulated by protein interactions and associations with microtubules. Front. Plant Sci. 2018, 9, 1766. [Google Scholar]
- Wu, S.; Xiao, H.; Cabrera, A.; Meulia, T.; van der Knaap, E. SUN regulates vegetative and reproductive organ shape by changing cell division patterns. Plant Physiol. 2011, 157, 1175–1186. [Google Scholar]
- Pang, Y.; Zang, X.; Pang, F.; Zhou, T.; Tian, F. Changes of CTK and few nitrogen index during development of flower and fruit in Zhanhua jujube. J. N. China Agric. 2017, 5, 101–104. [Google Scholar]
- Perpiñá, G.; Esteras, C.; Gibon, Y.; Monforte, A.J.; Picó, B. A new genomic library of melon introgression lines in a cantaloupe genetic background for dissecting desirable agronomical traits. BMC Plant Biol. 2016, 16, 154. [Google Scholar]
- Abel, S.; Bürstenbinder, K.; Müller, J. The emerging function of IQD proteins as scaffolds in cellular signaling and trafficking. Plant Signal. Behav. 2013, 8, e24369. [Google Scholar]
- Abel, S.; Savchenko, T.; Levy, M. Genome-wide comparative analysis of the IQD gene families in Arabidopsis thaliana and Oryza sativa. BMC Evol. Biol. 2005, 5, 72. [Google Scholar]
- Clevenger, J.P.; Van Houten, J.; Blackwood, M.; Rodríguez, G.R.; Jikumaru, Y.; Kamiya, Y.; Kusano, M.; Saito, K.; Visa, S.; Van Der Knaap, E. Network analyses reveal shifts in transcript profiles and metabolites that accompany the expression of SUN and an elongated tomato fruit. Plant Physiol. 2015, 168, 1164–1178. [Google Scholar] [PubMed]
- Jin, B.; Kim, J.; Jung, J.; Kim, D.; Park, Y. Characterization of IQ domain gene homologs as common candidate genes for elongated fruit shape in cucurbits. Hortic. Sci. Technol. 2018, 36, 85–97. [Google Scholar]
- Yalovsky, S. Protein lipid modifications and the regulation of ROP GTPase function. J. Exp. Bot. 2015, 66, 1617–1624. [Google Scholar] [PubMed]
- Dou, J.; Zhao, S.; Lu, X.; He, N.; Zhang, L.; Ali, A.; Kuang, H.; Liu, W. Genetic mapping reveals a candidate gene (ClFS1) for fruit shape in watermelon (Citrullus lanatus L.). Theor. Appl. Genet. 2018, 131, 947–958. [Google Scholar]
- Legendre, R.; Kuzy, J.; McGregor, C. Markers for selection of three alleles of ClSUN25-26-27a (Cla011257) associated with fruit shape in watermelon. Mol. Breed. 2020, 40, 19. [Google Scholar]
- Wang, X.; Li, Y.; Zhang, H.; Sun, G.; Zhang, W.; Qiu, L. Evolution and association analysis of GmCYP78A10 gene with seed size/weight and pod number in soybean. Mol. Biol. Rep. 2015, 42, 489–496. [Google Scholar]
- Łangowski, Ł.; Stacey, N.; Østergaard, L. Diversification of fruit shape in the Brassicaceae family. Plant Reprod. 2016, 29, 149–163. [Google Scholar]
- Yang, J.; Liu, Y.; Liang, B.; Yang, Q.; Li, X.; Chen, J.; Li, H.; Lyu, Y.; Lin, T. Genomic basis of selective breeding from the closest wild relative of large-fruited tomato. Hortic. Res. 2023, 10, uhad142. [Google Scholar] [CrossRef]
- Ruggieri, V.; Francese, G.; Sacco, A.; D’Alessandro, A.; Rigano, M.M.; Parisi, M.; Milone, M.; Cardi, T.; Mennella, G.; Barone, A. An association mapping approach to identify favourable alleles for tomato fruit quality breeding. BMC Plant Biol. 2014, 14, 337. [Google Scholar]
- Obel, H.O.; Cheng, C.; Tian, Z.; Li, J.; Lou, Q.; Yu, X.; Wang, Y.; Ogweno, J.O.; Chen, J. Molecular Research Progress on Xishuangbanna Cucumber (Cucumis sativus L. var. Xishuangbannesis Qi et Yuan): Current Status and Future Prospects. Agronomy 2022, 12, 300. [Google Scholar] [CrossRef]
- Fernandez-Silva, I.; Moreno, E.; Essafi, A.; Fergany, M.; Garcia-Mas, J.; Martín-Hernandez, A.M.; Álvarez, J.M.; Monforte, A.J. Shaping melons: Agronomic and genetic characterization of QTLs that modify melon fruit morphology. Theor. Appl. Genet. 2010, 121, 931–940. [Google Scholar] [CrossRef]
- Zhao, X.; Muhammad, N.; Zhao, Z.; Yin, K.; Liu, Z.; Wang, L.; Luo, Z.; Wang, L.; Liu, M. Molecular regulation of fruit size in horticultural plants: A review. Sci. Hortic. 2021, 288, 110353. [Google Scholar] [CrossRef]
- Shi, L.; Song, J.; Guo, C.; Wang, B.; Guan, Z.; Yang, P.; Chen, X.; Zhang, Q.; King, G.J.; Wang, J. A CACTA-like transposable element in the upstream region of BnaA9. CYP 78A9 acts as an enhancer to increase silique length and seed weight in rapeseed. Plant J. 2019, 98, 524–539. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Chakrabarti, M.; Taitano, N.K.; Okazaki, Y.; Saito, K.; Al-Abdallat, A.M.; van der Knaap, E. Differential expression of SlKLUH controlling fruit and seed weight is associated with changes in lipid metabolism and photosynthesis-related genes. J. Exp. Bot. 2021, 72, 1225–1244. [Google Scholar] [CrossRef] [PubMed]
- Ali, S.; Baloch, A.M. Overview of sustainable plant growth and differentiation and the role of hormones in controlling growth and development of plants under various stresses. Recent Pat. Food Nutr. Agric. 2020, 11, 105–114. [Google Scholar] [CrossRef]
- Li, S.; Chen, K.; Grierson, D. Molecular and hormonal mechanisms regulating fleshy fruit ripening. Cells 2021, 10, 1136. [Google Scholar] [CrossRef]
- Wang, Y.; Clevenger, J.P.; Illa-Berenguer, E.; Meulia, T.; van der Knaap, E.; Sun, L. A comparison of sun, ovate, fs8. 1 and auxin application on tomato fruit shape and gene expression. Plant Cell Physiol. 2019, 60, 1067–1081. [Google Scholar] [CrossRef]
- Khan, M.N.; Nabi, G. Role of Auxin in vegetative growth, flowering, yield and fruit quality of Horticultural crops—A review. Pure Appl. Biol. (PAB) 2023, 12, 1234–1241. [Google Scholar]
- Lin, Y.-C. Genetic Characterization of Resistance to Phytophthora capsici and Morphological Diversity in Cucumber; Michigan State University: East Lansing, MI, USA, 2023. [Google Scholar]
- Hu, J.; Israeli, A.; Ori, N.; Sun, T.-P. The interaction between DELLA and ARF/IAA mediates crosstalk between gibberellin and auxin signaling to control fruit initiation in tomato. Plant Cell 2018, 30, 1710–1728. [Google Scholar] [CrossRef]
- Pattison, R.J.; Catalá, C. Evaluating auxin distribution in tomato (Solanum lycopersicum) through an analysis of the PIN and AUX/LAX gene families. Plant J. 2012, 70, 585–598. [Google Scholar] [CrossRef]
- Hwang, I.; Sheen, J.; Müller, B. Cytokinin signaling networks. Annu. Rev. Plant Biol. 2012, 63, 353–380. [Google Scholar] [CrossRef] [PubMed]
- Bohner, J.; Bangerth, F. Effects of fruit set sequence and defoliation on cell number, cell size and hormone levels of tomato fruits (Lycopersicon esculentum Mill.) within a truss. J. Plant Growth Regul. 1988, 7, 141–155. [Google Scholar] [CrossRef]
- Nardozza, S.; Cooney, J.; Boldingh, H.L.; Hewitt, K.G.; Trower, T.; Jones, D.; Thrimawithana, A.H.; Allan, A.C.; Richardson, A.C. Phytohormone and transcriptomic analysis reveals endogenous cytokinins affect kiwifruit growth under restricted carbon supply. Metabolites 2020, 10, 23. [Google Scholar] [CrossRef] [PubMed]
- Fos, M.; Nuez, F.; Garcıa-Martınez, J.L. The gene pat-2, which induces natural parthenocarpy, alters the gibberellin content in unpollinated tomato ovaries. Plant Physiol. 2000, 122, 471–480. [Google Scholar] [CrossRef]
- Serrani, J.C.; Fos, M.; Atarés, A.; García-Martínez, J.L. Effect of gibberellin and auxin on parthenocarpic fruit growth induction in the cv Micro-Tom of tomato. J. Plant Growth Regul. 2007, 26, 211–221. [Google Scholar] [CrossRef]
- Srivastava, A.; Handa, A.K. Hormonal regulation of tomato fruit development: A molecular perspective. J. Plant Growth Regul. 2005, 24, 67–82. [Google Scholar] [CrossRef]
- Vandenbussche, F.; Vaseva, I.; Vissenberg, K.; Van Der Straeten, D. Ethylene in vegetative development: A tale with a riddle. New Phytol. 2012, 194, 895–909. [Google Scholar] [CrossRef]
- Boualem, A.; Troadec, C.; Camps, C.; Lemhemdi, A.; Morin, H.; Sari, M.-A.; Fraenkel-Zagouri, R.; Kovalski, I.; Dogimont, C.; Perl-Treves, R. A cucurbit androecy gene reveals how unisexual flowers develop and dioecy emerges. Science 2015, 350, 688–691. [Google Scholar] [CrossRef]
- Nitsch, L.; Kohlen, W.; Oplaat, C.; Charnikhova, T.; Cristescu, S.; Michieli, P.; Wolters-Arts, M.; Bouwmeester, H.; Mariani, C.; Vriezen, W. ABA-deficiency results in reduced plant and fruit size in tomato. J. Plant Physiol. 2012, 169, 878–883. [Google Scholar] [CrossRef]
- Cutler, S.R.; Rodriguez, P.L.; Finkelstein, R.R.; Abrams, S.R. Abscisic acid: Emergence of a core signaling network. Annu. Rev. Plant Biol. 2010, 61, 651–679. [Google Scholar] [CrossRef]
- Liao, X.; Li, M.; Liu, B.; Yan, M.; Yu, X.; Zi, H.; Liu, R.; Yamamuro, C. Interlinked regulatory loops of ABA catabolism and biosynthesis coordinate fruit growth and ripening in woodland strawberry. Proc. Natl. Acad. Sci. USA 2018, 115, E11542–E11550. [Google Scholar] [PubMed]
- Davies, P.J. Plant Hormones: Biosynthesis, Signal Transduction, Action; Springer Science & Business Media: Berlin/Heidelberg, Germany, 2004. [Google Scholar]
- Hwang, I.; Sheen, J. Two-component circuitry in Arabidopsis cytokinin signal transduction. Nature 2001, 413, 383–389. [Google Scholar]
- Hedden, P.; Thomas, S.G. Gibberellin biosynthesis and its regulation. Biochem. J. 2012, 444, 11–25. [Google Scholar] [PubMed]
- Gupta, K.; Wani, S.H.; Razzaq, A.; Skalicky, M.; Samantara, K.; Gupta, S.; Pandita, D.; Goel, S.; Grewal, S.; Hejnak, V. Abscisic acid: Role in fruit development and ripening. Front. Plant Sci. 2022, 13, 817500. [Google Scholar]
- Lelièvre, J.M.; Latchè, A.; Jones, B.; Bouzayen, M.; Pech, J.C. Ethylene and fruit ripening. Physiol. Plant. 1997, 101, 727–739. [Google Scholar]
- Heuvelink, E.; Okello, R.C.; Peet, M.; Giovannoni, J.J.; Dorais, M. Tomato. In The Physiology of Vegetable Crops; CABI: Wallingford, UK, 2020; pp. 138–178. [Google Scholar]
- Chatterjee, A.; Dhal, S.; Pal, H. Insight into the regulatory network of miRNA to unravel the ripening physiology of climacteric and non-climacteric fruits. Plant Gene 2021, 28, 100329. [Google Scholar]
- Böttcher, C.; Keyzers, R.A.; Boss, P.K.; Davies, C. Sequestration of auxin by the indole-3-acetic acid-amido synthetase GH3-1 in grape berry (Vitis vinifera L.) and the proposed role of auxin conjugation during ripening. J. Exp. Bot. 2010, 61, 3615–3625. [Google Scholar]
- van der Knaap, E.; Chakrabarti, M.; Chu, Y.H.; Clevenger, J.P.; Illa-Berenguer, E.; Huang, Z.; Keyhaninejad, N.; Mu, Q.; Sun, L.; Wang, Y. What lies beyond the eye: The molecular mechanisms regulating tomato fruit weight and shape. Front. Plant Sci. 2014, 5, 227. [Google Scholar]
- Kim, S.; Park, M.; Yeom, S.-I.; Kim, Y.-M.; Lee, J.M.; Lee, H.-A.; Seo, E.; Choi, J.; Cheong, K.; Kim, K.-T. Genome sequence of the hot pepper provides insights into the evolution of pungency in Capsicum species. Nat. Genet. 2014, 46, 270–278. [Google Scholar]
- Castro-Camba, R.; Sánchez, C.; Vidal, N.; Vielba, J.M. Plant development and crop yield: The role of gibberellins. Plants 2022, 11, 2650. [Google Scholar] [CrossRef]
- Su, L.; Rahat, S.; Ren, N.; Kojima, M.; Takebayashi, Y.; Sakakibara, H.; Wang, M.; Chen, X.; Qi, X. Cytokinin and auxin modulate cucumber parthenocarpy fruit development. Sci. Hortic. 2021, 282, 110026. [Google Scholar]
- Shahab, M.; Roberto, S.R.; Ahmed, S.; Colombo, R.C.; Silvestre, J.P.; Koyama, R.; de Souza, R.T. Relationship between anthocyanins and skin color of table grapes treated with abscisic acid at different stages of berry ripening. Sci. Hortic. 2020, 259, 108859. [Google Scholar]
- Gao, J.; Zhang, Y.; Li, Z.; Liu, M. Role of ethylene response factors (ERFs) in fruit ripening. Food Qual. Saf. 2020, 4, 15–20. [Google Scholar]
- Kondo, S. Roles of jasmonates in fruit ripening and environmental stress. In Proceedings of the XI International Symposium on Plant Bioregulators in Fruit Production 884, Bologna, Italy, 20–24 September 2009; pp. 711–716. [Google Scholar]
- Garrido-Auñón, F.; Puente-Moreno, J.; García-Pastor, M.E.; Serrano, M.; Valero, D. Brassinosteroids: An Innovative Compound Family That Could Affect the Growth, Ripening, Quality, and Postharvest Storage of Fleshy Fruits. Plants 2024, 13, 3082. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Allan, A.C.; Wang, W.-q.; Yin, X.-r. The effects of salicylic acid on quality control of horticultural commodities. N. Z. J. Crop Hortic. Sci. 2022, 50, 99–117. [Google Scholar]
- Manzoor, M.A.; Xu, Y.; Xu, J.; Wang, Y.; Sun, W.; Liu, X.; Wang, L.; Wang, J.; Liu, R.; Whiting, M.D. Fruit crop abiotic stress management: A comprehensive review of plant hormones mediated responses. Fruit Res. 2023, 3, 30. [Google Scholar]
- Pilati, S.; Bagagli, G.; Sonego, P.; Moretto, M.; Brazzale, D.; Castorina, G.; Simoni, L.; Tonelli, C.; Guella, G.; Engelen, K. Abscisic acid is a major regulator of grape berry ripening onset: New insights into ABA signaling network. Front. Plant Sci. 2017, 8, 1093. [Google Scholar]
- Lin, Y.; Wang, C.; Cao, S.; Sun, Z.; Zhang, Y.; Li, M.; He, W.; Wang, Y.; Chen, Q.; Zhang, Y. Proanthocyanidins delay fruit coloring and softening by repressing related gene expression during strawberry (Fragaria x ananassa Duch.) Ripening. Int. J. Mol. Sci. 2023, 24, 3139. [Google Scholar]
- Luo, Y.; Ge, C.; Ling, Y.; Mo, F.; Yang, M.; Jiang, L.; Chen, Q.; Lin, Y.; Sun, B.; Zhang, Y. ABA and sucrose co-regulate strawberry fruit ripening and show inhibition of glycolysis. Mol. Genet. Genom. 2020, 295, 421–438. [Google Scholar]
- Salama, A.-M.; Abdelsalam, M.A.; Rehan, M.; Elansary, M.; El-Shereif, A. Anthocyanin accumulation and its corresponding gene expression, total phenol, antioxidant capacity, and fruit quality of ‘Crimson Seedless’ Grapevine (Vitis vinifera L.) in response to grafting and pre-harvest applications. Horticulturae 2023, 9, 1001. [Google Scholar] [CrossRef]
- Bai, Q.; Huang, Y.; Shen, Y. The physiological and molecular mechanism of abscisic acid in regulation of fleshy fruit ripening. Front. Plant Sci. 2021, 11, 619953. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Mou, W.; Xia, R.; Li, L.; Zawora, C.; Ying, T.; Mao, L.; Liu, Z.; Luo, Z. Integrated analysis of high-throughput sequencing data shows abscisic acid-responsive genes and miRNAs in strawberry receptacle fruit ripening. Hortic. Res. 2019, 6, 26. [Google Scholar] [CrossRef] [PubMed]
- Jia, H.; Li, C.; Chai, Y.; Xing, Y.; Shen, Y. Sucrose promotes strawberry fruit ripening by stimulation of abscisic acid biosynthesis. Pak. J. Bot. 2013, 45, 169–175. [Google Scholar]
- Fujii, H.; Chinnusamy, V.; Rodrigues, A.; Rubio, S.; Antoni, R.; Park, S.-Y.; Cutler, S.R.; Sheen, J.; Rodriguez, P.L.; Zhu, J.-K. In vitro reconstitution of an abscisic acid signalling pathway. Nature 2009, 462, 660–664. [Google Scholar]
- Santiago, J.; Rodrigues, A.; Saez, A.; Rubio, S.; Antoni, R.; Dupeux, F.; Park, S.Y.; Márquez, J.A.; Cutler, S.R.; Rodriguez, P.L. Modulation of drought resistance by the abscisic acid receptor PYL5 through inhibition of clade A PP2Cs. Plant J. 2009, 60, 575–588. [Google Scholar]
- Umezawa, T.; Nakashima, K.; Miyakawa, T.; Kuromori, T.; Tanokura, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Molecular basis of the core regulatory network in ABA responses: Sensing, signaling and transport. Plant Cell Physiol. 2010, 51, 1821–1839. [Google Scholar]
- Liu, Z.-Q.; Yan, L.; Wu, Z.; Mei, C.; Lu, K.; Yu, Y.-T.; Liang, S.; Zhang, X.-F.; Wang, X.-F.; Zhang, D.-P. Cooperation of three WRKY-domain transcription factors WRKY18, WRKY40, and WRKY60 in repressing two ABA-responsive genes ABI4 and ABI5 in Arabidopsis. J. Exp. Bot. 2012, 63, 6371–6392. [Google Scholar]
- Shang, Y.; Yan, L.; Liu, Z.-Q.; Cao, Z.; Mei, C.; Xin, Q.; Wu, F.-Q.; Wang, X.-F.; Du, S.-Y.; Jiang, T. The Mg-chelatase H subunit of Arabidopsis antagonizes a group of WRKY transcription repressors to relieve ABA-responsive genes of inhibition. Plant Cell 2010, 22, 1909–1935. [Google Scholar]
- Yan, L.; Liu, Z.-Q.; Xu, Y.-H.; Lu, K.; Wang, X.-F.; Zhang, D.-P. Auto-and cross-repression of three Arabidopsis WRKY transcription factors WRKY18, WRKY40, and WRKY60 negatively involved in ABA signaling. J. Plant Growth Regul. 2013, 32, 399–416. [Google Scholar] [CrossRef]
- Gao, Z.; Li, Q.; Li, J.; Chen, Y.; Luo, M.; Li, H.; Wang, J.; Wu, Y.; Duan, S.; Wang, L. Characterization of the ABA receptor VlPYL1 that regulates anthocyanin accumulation in grape berry skin. Front. Plant Sci. 2018, 9, 592. [Google Scholar]
- Zhang, S.; Hou, B.; Chai, L.; Yang, A.; Yu, X.; Shen, Y. Sigma factor FaSigE positively regulates strawberry fruit ripening by ABA. Plant Growth Regul. 2017, 83, 417–427. [Google Scholar]
- Pathak, G. Coordinated Regulation: Interplay between Ethylene and ABA in Fruit Ripening and Development. Adv. Res. 2024, 25, 513–527. [Google Scholar] [CrossRef]
- He, H.; Yamamuro, C. Interplays between auxin and GA signaling coordinate early fruit development. Hortic. Res. 2022, 9, uhab078. [Google Scholar] [CrossRef] [PubMed]
- Gomes, G.; Scortecci, K. Auxin and its role in plant development: Structure, signalling, regulation and response mechanisms. Plant Biol. 2021, 23, 894–904. [Google Scholar]
- Zhang, Y.; Yu, J.; Xu, X.; Wang, R.; Liu, Y.; Huang, S.; Wei, H.; Wei, Z. Molecular mechanisms of diverse auxin responses during plant growth and development. Int. J. Mol. Sci. 2022, 23, 12495. [Google Scholar] [CrossRef]
- Estrada-Johnson, E.; Csukasi, F.; Pizarro, C.M.; Vallarino, J.G.; Kiryakova, Y.; Vioque, A.; Brumos, J.; Medina-Escobar, N.; Botella, M.A.; Alonso, J.M. Transcriptomic analysis in strawberry fruits reveals active auxin biosynthesis and signaling in the ripe receptacle. Front. Plant Sci. 2017, 8, 889. [Google Scholar]
- Medina-Puche, L.; Martínez-Rivas, F.J.; Molina-Hidalgo, F.J.; Mercado, J.A.; Moyano, E.; Rodríguez-Franco, A.; Caballero, J.L.; Muñoz-Blanco, J.; Blanco-Portales, R. An atypical HLH transcriptional regulator plays a novel and important role in strawberry ripened receptacle. BMC Plant Biol. 2019, 19, 586. [Google Scholar] [CrossRef]
- Ziliotto, F.; Corso, M.; Rizzini, F.M.; Rasori, A.; Botton, A.; Bonghi, C. Grape berry ripening delay induced by a pre-véraison NAA treatment is paralleled by a shift in the expression pattern of auxin-and ethylene-related genes. BMC Plant Biol. 2012, 12, 185. [Google Scholar] [CrossRef]
- Kühn, N.; Serrano, A.; Abello, C.; Arce, A.; Espinoza, C.; Gouthu, S.; Deluc, L.; Arce-Johnson, P. Regulation of polar auxin transport in grapevine fruitlets (Vitis vinifera L.) and the proposed role of auxin homeostasis during fruit abscission. BMC Plant Biol. 2016, 16, 234. [Google Scholar] [CrossRef]
- Tennessen, J.A.; Govindarajulu, R.; Ashman, T.-L.; Liston, A. Evolutionary origins and dynamics of octoploid strawberry subgenomes revealed by dense targeted capture linkage maps. Genome Biol. Evol. 2014, 6, 3295–3313. [Google Scholar] [CrossRef]
- Tadiello, A.; Ziosi, V.; Negri, A.S.; Noferini, M.; Fiori, G.; Busatto, N.; Espen, L.; Costa, G.; Trainotti, L. On the role of ethylene, auxin and a GOLVEN-like peptide hormone in the regulation of peach ripening. BMC Plant Biol. 2016, 16, 44. [Google Scholar]
- Kou, X.; Feng, Y.; Yuan, S.; Zhao, X.; Wu, C.; Wang, C.; Xue, Z. Different regulatory mechanisms of plant hormones in the ripening of climacteric and non-climacteric fruits: A review. Plant Mol. Biol. 2021, 107, 477–497. [Google Scholar] [PubMed]
- Giovannoni, J. Ripening activator turned repressor. Nat. Plants 2017, 3, 920–921. [Google Scholar] [PubMed]
- Pimentel, P.; Salvatierra, A.; Moya-León, M.A.; Herrera, R. Isolation of genes differentially expressed during development and ripening of Fragaria chiloensis fruit by suppression subtractive hybridization. J. Plant Physiol. 2010, 167, 1179–1187. [Google Scholar] [CrossRef]
- Csukasi, F.; Osorio, S.; Gutierrez, J.R.; Kitamura, J.; Giavalisco, P.; Nakajima, M.; Fernie, A.R.; Rathjen, J.P.; Botella, M.A.; Valpuesta, V. Gibberellin biosynthesis and signalling during development of the strawberry receptacle. New Phytol. 2011, 191, 376–390. [Google Scholar]
- Olszewski, N.; Sun, T.-p.; Gubler, F. Gibberellin signaling: Biosynthesis, catabolism, and response pathways. Plant Cell 2002, 14, S61–S80. [Google Scholar]
- Yamaguchi, S. Gibberellin metabolism and its regulation. Annu. Rev. Plant Biol. 2008, 59, 225–251. [Google Scholar]
- Bustamante, C.A.; Civello, P.M.; Martínez, G.A. Cloning of the promoter region of β-xylosidase (FaXyl1) gene and effect of plant growth regulators on the expression of FaXyl1 in strawberry fruit. Plant Sci. 2009, 177, 49–56. [Google Scholar]
- Moyano-Canete, E.; Bellido, M.L.; García-Caparrós, N.; Medina-Puche, L.; Amil-Ruiz, F.; González-Reyes, J.A.; Caballero, J.L.; Munoz-Blanco, J.; Blanco-Portales, R. FaGAST2, a strawberry ripening-related gene, acts together with FaGAST1 to determine cell size of the fruit receptacle. Plant Cell Physiol. 2013, 54, 218–236. [Google Scholar]
- Yang, D.; Li, Z.; Li, J.; Chen, J.; Wang, J.; Jing, X.; Guan, X. Effect of pre-flowering gibberellic acid applications on tartaric acid content in grape berries. Sci. Hortic. 2024, 325, 112659. [Google Scholar]
- Han, J.; Fang, J.; Wang, C.; Yin, Y.; Sun, X.; Leng, X.; Song, C. Grapevine microRNAs responsive to exogenous gibberellin. BMC Genom. 2014, 15, 111. [Google Scholar] [CrossRef] [PubMed]
- Cheng, C.; Xu, X.; Singer, S.D.; Li, J.; Zhang, H.; Gao, M.; Wang, L.; Song, J.; Wang, X. Effect of GA3 treatment on seed development and seed-related gene expression in grape. PLoS ONE 2013, 8, e80044. [Google Scholar] [CrossRef] [PubMed]
- Nilsen, G.; Martinussen, I.; Erntsen, A.; Bhuvaneswari, T.; Junttila, O. Involvement of gibberellins during fruit development in cloudberry (Rubus chamaemorus). In Proceedings of the VIII International Rubus and Ribes Symposium 585, Dundee, UK, 4–11 July 2001; pp. 517–519. [Google Scholar]
- Palonen, P.; Mouhu, K. Vegetative Growth and Flowering of Primocane Raspberry ‘Ariadne’as Affected by Prohexadione–calcium Treatments. HortScience 2009, 44, 529–531. [Google Scholar] [CrossRef]
- Li, D.; Li, L.; Luo, Z.; Mou, W.; Mao, L.; Ying, T. Comparative transcriptome analysis reveals the influence of abscisic acid on the metabolism of pigments, ascorbic acid and folic acid during strawberry fruit ripening. PLoS ONE 2015, 10, e0130037. [Google Scholar] [CrossRef]
- Monsalve, L.; Bernales, M.; Ayala-Raso, A.; Álvarez, F.; Valdenegro, M.; Alvaro, J.-E.; Figueroa, C.R.; Defilippi, B.G.; Fuentes, L. Relationship between endogenous ethylene production and firmness during the ripening and cold storage of raspberry (Rubus idaeus ‘Heritage’) Fruit. Horticulturae 2022, 8, 262. [Google Scholar] [CrossRef]
- Chervin, C.; Tira-umphon, A.; Terrier, N.; Zouine, M.; Severac, D.; Roustan, J.P. Stimulation of the grape berry expansion by ethylene and effects on related gene transcripts, over the ripening phase. Physiol. Plant. 2008, 134, 534–546. [Google Scholar] [CrossRef]
- Navarro-Calderon, A.; Falagan, N.; Terry, L.A.; Alamar, M.C. Biomarkers of postharvest resilience: Unveiling the role of abscisic acid in table grapes during cold storage. Front. Plant Sci. 2023, 14, 1266807. [Google Scholar] [CrossRef]
- Li, L.; Lichter, A.; Chalupowicz, D.; Gamrasni, D.; Goldberg, T.; Nerya, O.; Ben-Arie, R.; Porat, R. Effects of the ethylene-action inhibitor 1-methylcyclopropene on postharvest quality of non-climacteric fruit crops. Postharvest Biol. Technol. 2016, 111, 322–329. [Google Scholar] [CrossRef]
- Zenoni, S.; Savoi, S.; Busatto, N.; Tornielli, G.B.; Costa, F. Molecular regulation of apple and grape ripening: Exploring common and distinct transcriptional aspects of representative climacteric and non-climacteric fruits. J. Exp. Bot. 2023, 74, 6207–6223. [Google Scholar] [CrossRef]
- Ayub, R.A.; Pessenti, I.L.; Pereira, A.B. Ethylene responses in non-climacteric horticultural crops. CABI Rev. 2020. [Google Scholar] [CrossRef]
- Perkins-Veazie, P.; Clark, J.; Huber, D.; Baldwin, E. Ripening physiology in’Navaho’thornless blackberries: Color, respiration, ethylene production, softening, and compositional changes. J. Am. Soc. Hortic. Sci. 2000, 125, 357–363. [Google Scholar]
- Merchante, C.; Vallarino, J.G.; Osorio, S.; Aragüez, I.; Villarreal, N.; Ariza, M.T.; Martínez, G.A.; Medina-Escobar, N.; Civello, M.P.; Fernie, A.R. Ethylene is involved in strawberry fruit ripening in an organ-specific manner. J. Exp. Bot. 2013, 64, 4421–4439. [Google Scholar] [PubMed]
- Chen, Y.; Grimplet, J.; David, K.; Castellarin, S.D.; Terol, J.; Wong, D.C.; Luo, Z.; Schaffer, R.; Celton, J.-M.; Talon, M. Ethylene receptors and related proteins in climacteric and non-climacteric fruits. Plant Sci. 2018, 276, 63–72. [Google Scholar] [PubMed]
- Ju, C.; Chang, C. Mechanistic insights in ethylene perception and signal transduction. Plant Physiol. 2015, 169, 85–95. [Google Scholar] [PubMed]
- Sun, J.-H.; Luo, J.-J.; Tian, L.; Li, C.-L.; Xing, Y.; Shen, Y.-Y. New evidence for the role of ethylene in strawberry fruit ripening. J. Plant Growth Regul. 2013, 32, 461–470. [Google Scholar]
- Bhatla, S.C.; Lal, M.A. Fruit Development and Ripening. In Plant Physiology, Development and Metabolism; Springer: Berlin/Heidelberg, Germany, 2023; pp. 607–624. [Google Scholar]
- Zhang, Y.; Deng, M.; Gu, X.; Guo, C.; Chen, Y.; Lin, Y.; Chen, Q.; Wang, Y.; Zhang, Y.; Luo, Y. Ethylene Signaling Pathway Genes in Strawberry and Their Expression Patterns during Fruit Ripening. Agronomy 2023, 13, 1930. [Google Scholar] [CrossRef]
- Wasternack, C.; Strnad, M. Jasmonates: News on occurrence, biosynthesis, metabolism and action of an ancient group of signaling compounds. Int. J. Mol. Sci. 2018, 19, 2539. [Google Scholar] [CrossRef]
- Garrido-Bigotes, A.; Figueroa, N.E.; Figueroa, P.M.; Figueroa, C.R. Jasmonate signalling pathway in strawberry: Genome-wide identification, molecular characterization and expression of JAZ s and MYC s during fruit development and ripening. PLoS ONE 2018, 13, e0197118. [Google Scholar]
- Böttcher, C.; Burbidge, C.A.; di Rienzo, V.; Boss, P.K.; Davies, C. Jasmonic acid-isoleucine formation in grapevine (Vitis vinifera L.) by two enzymes with distinct transcription profiles. J. Integr. Plant Biol. 2015, 57, 618–627. [Google Scholar]
- Günther, C.S.; Plunkett, B.J.; Cooney, J.M.; Jensen, D.J.; Trower, T.M.; Elborough, C.; Nguyen, H.M.; Deng, C.H.; Lafferty, D.J.; Albert, N.W. Biotic stress-induced and ripening-related anthocyanin biosynthesis are regulated by alternate phytohormone signals in blueberries. Environ. Exp. Bot. 2022, 203, 105065. [Google Scholar]
- Delgado, L.D.; Zúñiga, P.E.; Figueroa, N.E.; Pastene, E.; Escobar-Sepúlveda, H.F.; Figueroa, P.M.; Garrido-Bigotes, A.; Figueroa, C.R. Application of a JA-Ile biosynthesis inhibitor to methyl jasmonate-treated strawberry fruit induces upregulation of specific MBW complex-related genes and accumulation of proanthocyanidins. Molecules 2018, 23, 1433. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.-Y.; Liu, X.-F.; Fu, B.-l.; Zhang, Q.-Y.; Tong, Y.; Wang, J.; Wang, W.-Q.; Grierson, D.; Yin, X.-R. Methyl jasmonate enhances ethylene synthesis in kiwifruit by inducing NAC genes that activate ACS1. J. Agric. Food Chem. 2020, 68, 3267–3276. [Google Scholar] [PubMed]
- Zuñiga, P.E.; Castañeda, Y.; Arrey-Salas, O.; Fuentes, L.; Aburto, F.; Figueroa, C.R. Methyl jasmonate applications from flowering to ripe fruit stages of strawberry (Fragaria × ananassa ‘Camarosa’) reinforce the fruit antioxidant response at post-harvest. Front. Plant Sci. 2020, 11, 538. [Google Scholar]
- Tao, X.; Wu, Q.; Li, J.; Huang, S.; Cai, L.; Mao, L.; Luo, Z.; Li, L.; Ying, T. Exogenous methyl jasmonate regulates phenolic compounds biosynthesis during postharvest tomato ripening. Postharvest Biol. Technol. 2022, 184, 111760. [Google Scholar]
- Ferrandino, A.; Pagliarani, C. Secondary metabolites in grapevine stress response-women in plant science series. Front. Plant Sci. 2023, 14, 1272668. [Google Scholar]
- Clouse, S.D. Brassinosteroid signal transduction: From receptor kinase activation to transcriptional networks regulating plant development. Plant Cell 2011, 23, 1219–1230. [Google Scholar]
- Chai, Y.-m.; Zhang, Q.; Tian, L.; Li, C.-L.; Xing, Y.; Qin, L.; Shen, Y.-Y. Brassinosteroid is involved in strawberry fruit ripening. J. Plant Growth Regul. 2013, 69, 63–69. [Google Scholar]
- Yang, N.; Zhou, Y.; Wang, Z.; Zhang, Z.; Xi, Z.; Wang, X. Emerging roles of brassinosteroids and light in anthocyanin biosynthesis and ripeness of climacteric and non-climacteric fruits. Crit. Rev. Food Sci. Nutr. 2023, 63, 4541–4553. [Google Scholar]
- Roh, J.; Moon, J.; Youn, J.-H.; Seo, C.; Park, Y.J.; Kim, S.-K. Establishment of biosynthetic pathways to generate castasterone as the biologically active brassinosteroid in Brachypodium distachyon. J. Agric. Food Chem. 2020, 68, 3912–3923. [Google Scholar]
- Ayub, R.; Reis, L.; Bosetto, L.; Lopes, P.; Galvão, C.; Etto, R. Brassinosteroid plays a role on pink stage for receptor and transcription factors involved in strawberry fruit ripening. Plant Growth Regul. 2018, 84, 159–167. [Google Scholar]
- Espinoza, A.; Contreras, R.; Zúñiga, G.E.; Herrera, R.; Moya-León, M.A.; Norambuena, L.; Handford, M. FcLDP1, a gene encoding a late embryogenesis abundant (LEA) domain protein, responds to brassinosteroids and abscisic acid during the development of fruits in Fragaria chiloensis. Front. Plant Sci. 2016, 7, 788. [Google Scholar] [CrossRef] [PubMed]
- Bombarely, A.; Moser, M.; Amrad, A.; Bao, M.; Bapaume, L.; Barry, C.S.; Bliek, M.; Boersma, M.R.; Borghi, L.; Bruggmann, R. Insight into the evolution of the Solanaceae from the parental genomes of Petunia hybrida. Nat. Plants 2016, 2, 16074. [Google Scholar] [CrossRef] [PubMed]
- Kang, C.; Darwish, O.; Geretz, A.; Shahan, R.; Alkharouf, N.; Liu, Z. Genome-scale transcriptomic insights into early-stage fruit development in woodland strawberry Fragaria vesca. Plant Cell 2013, 25, 1960–1978. [Google Scholar] [CrossRef] [PubMed]
- Gan, L.; Song, M.; Wang, X.; Yang, N.; Li, H.; Liu, X.; Li, Y. Cytokinins are involved in regulation of tomato pericarp thickness and fruit size. Hortic. Res. 2022, 9, uhab041. [Google Scholar] [CrossRef]
- Du, C.-L.; Cai, C.-L.; Lu, Y.; Li, Y.-M.; Xie, Z.-S. Identification and expression analysis of invertase family genes during grape (Vitis vinifera L.) berry development under CPPU and GA treatment. Mol. Genet. Genom. 2023, 298, 777–789. [Google Scholar] [CrossRef]
- Pérez-Rojas, M.; Díaz-Ramírez, D.; Ortíz-Ramírez, C.I.; Galaz-Ávalos, R.M.; Loyola-Vargas, V.M.; Ferrándiz, C.; Abraham-Juárez, M.d.R.; Marsch-Martínez, N. The role of cytokinins during the development of strawberry flowers and receptacles. Plants 2023, 12, 3672. [Google Scholar] [CrossRef]
- Zahid, G.; Iftikhar, S.; Shimira, F.; Ahmad, H.M.; Kaçar, Y.A. An overview and recent progress of plant growth regulators (PGRs) in the mitigation of abiotic stresses in fruits: A review. Sci. Hortic. 2023, 309, 111621. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).