QTL Mapping and Candidate Gene Screening for Enhancing Oil Content in Silage Maize
Abstract
1. Introduction
2. Results
2.1. Analysis of Silage Maize Grain Phenotypic Variation
2.2. Sequencing and SNP Identification
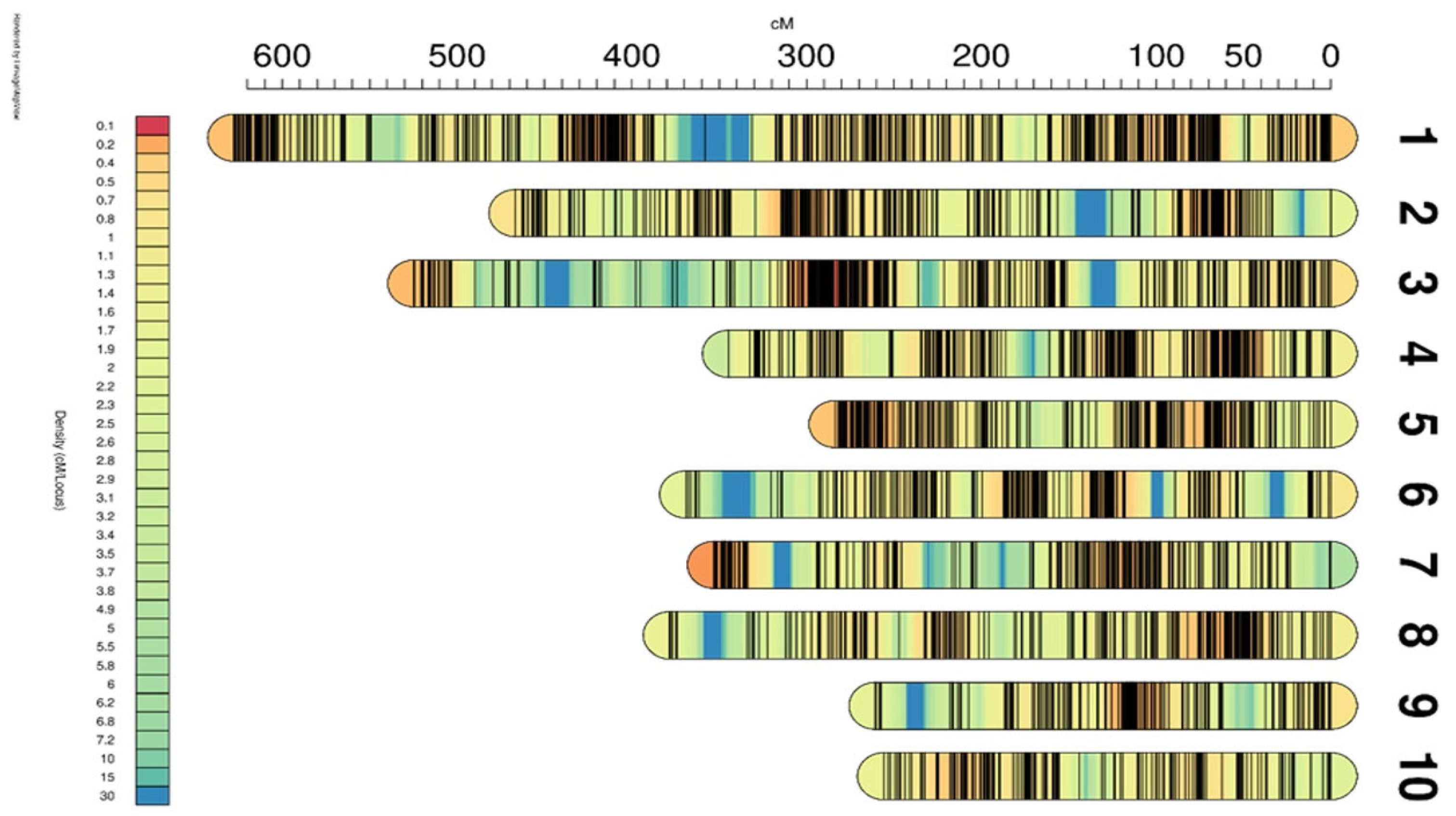
2.3. Construction of Genetic Linkage Map
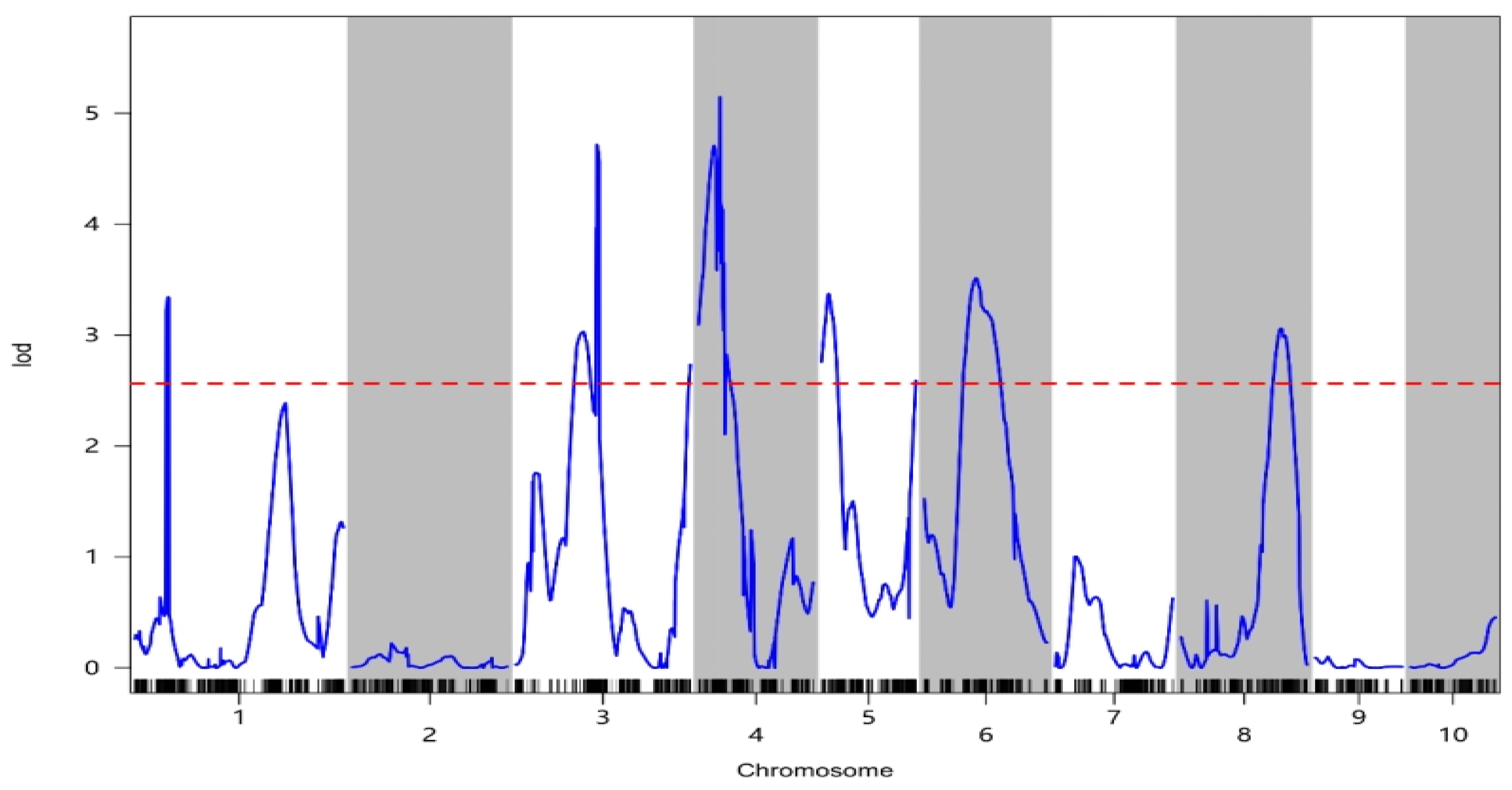
2.4. Mapping of Silage Maize QTLs
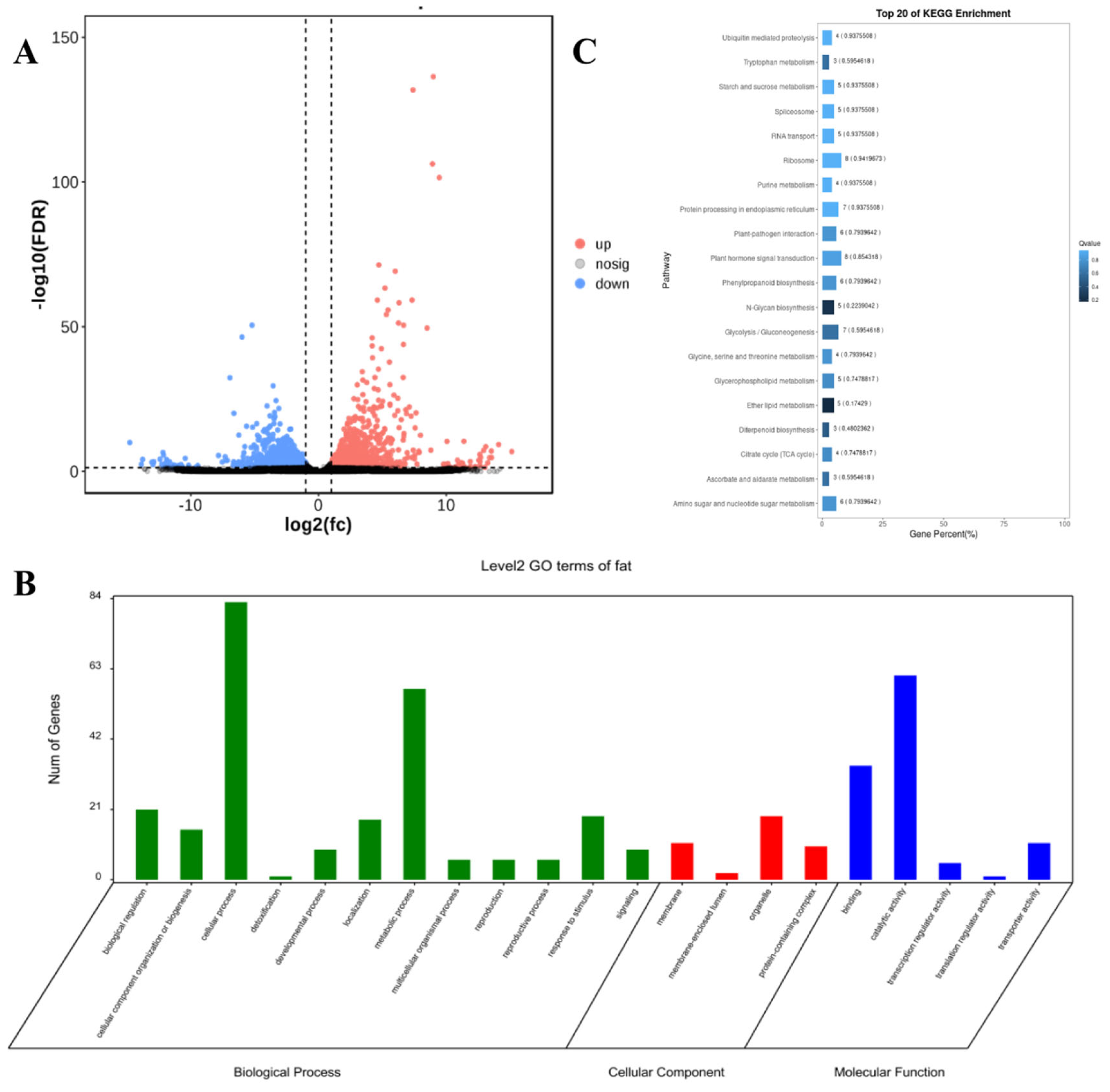
2.5. Prediction of Candidate Genes Within QTL Intervals Associated with Silage Maize Oil Content Traits
2.6. Validation of Oil Content-Associated Differentially Expressed Genes via RT-qPCR
3. Discussion
3.1. Utilizing Genetic Modeling in Silage Maize Breeding
3.2. High-Density Genetic Mapping and QTL Analysis
3.3. Screening of Candidate Genes
4. Materials and Methods
4.1. Plant Material
4.2. Measurement of Oil Content
4.3. Sequencing and Genotyping
4.4. Construction, Evaluation, and QTL Mapping of Genetic Linkage Map
4.5. RNA Sequencing (RNA-Seq) Analysis and RT-qPCR
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| QTL | quantitative trait loci |
| DH | double haploid complex interval mapping (CIM) |
| SNP | single nucleotide polymorphism |
| LOD | logarithm of odds |
| TAG | triacylglycerol |
| PPP | pentose phosphate pathway |
| AIC | Akaike information criterion |
| GBTS | genotyping by target sequencing |
| BWA | Burrows–Wheeler aligner |
| GO | Gene Ontology |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| K-S | Kolmogorov–Smirnov |
| BP | biological process |
| CC | cell component |
| MF | molecular function |
| ABI3 | ABA-INSENSITIVE 3 |
| KASI-β | ketoacyl-[acyl carrier protein] synthase I |
| BRs | brassinosteroids |
| PVE | phenotypic variation explained |
| ABI3 | ABA-INSENSITIVE 3 |
| DAG | diacylglycerol |
| FUS3 | FUSCA3 |
References
- Hristov, A.N.; Harper, M.T.; Roth, G.; Canale, C.; Huhtanen, P.; Richard, T.L.; DiMarco, K. Effects of ensiling time on corn silage neutral detergent fiber degradability and relationship between laboratory fiber analyses and in vivo digestibility—ScienceDirect. J. Dairy Sci. 2020, 103, 2333–2346. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.Z.; Li, S.Y.; Lin, H.; Ma, Y.H.; Pan, L.Y.; Li, D.L.; Sun, D.Q. Genetic Variation and Principal Component Analysis of Silage Maize Quality Traits. Crops 2019, 35, 42–48. [Google Scholar]
- Reith, R.R.; Sieck, R.L.; Grijalva, P.C.; Duffy, E.M.; Swanson, R.M.; Fuller, A.M.; Petersen, J.L. 256 Heat stress and β-adrenergic agonists alter the adipose transcriptome and fatty acid mobilization in ruminant livestock. J. Anim. Sci. 2020, 98, 195–196. [Google Scholar] [CrossRef]
- Li, Z.Q.; Liu, C.L. Nutrition of Silage and Application Research on Ruminant Production. Acta Ecol. Anim. Domastici 2010, 31, 95–98. [Google Scholar]
- Huang, Y.; Wang, H.; Zhu, Y.; Li, S.; Wu, X.; Wu, Y. THP9 enhances seed protein content and nitrogen-use efficiency in maize. Nature 2022, 612, 292–300. [Google Scholar] [CrossRef]
- Yang, X.H.; Guo, Y.Q.; Yan, J.B.; Zhang, J.; Song, T.M.; Li, R.S. Major and minor QTL and epistasis contribute to fatty acid compositions and oil concentration in high-oil maize. Theor. Appl. Genet. 2010, 120, 665–678. [Google Scholar] [CrossRef]
- Li, L.; Li, H.; Li, J.Y.; Xu, S.T.; Yang, X.H.; Li, J.S.; Yan, J.B. A genome-wide survey of maize lipid-related genes: Candidate genes mining, digital gene expression profiling and co-location with QTL for maize kernel oil. Sci. China Life Sci. 2010, 06, 690–700. [Google Scholar] [CrossRef]
- Fang, H.; Fu, X.Y.; Ge, H.Q.; Zhang, A.X.; Wang, B.H. Genetic basis of maize kernel oil-related traits revealed by high-density SNP markers in a recombinant inbred line population. BMC Plant Biol. 2021, 21, 344. [Google Scholar] [CrossRef]
- Yang, Z.; Li, X.; Zhang, Y.N.; Jiang, H.W.; Gao, J.; Kuai, B.K.; Ding, Y.L.; Huang, X.Q. Detection of quantitative trait loci for kernel oil and protein concentration in a B73 and Zheng58 maize cross. Genet. Mol. Res. 2016, 15, 10–4238. [Google Scholar] [CrossRef]
- Guo, Y.; Yang, X.H.; Chander, S.; Yan, J.B.; Zhang, J.; Song, T.M.; Li, J.S. Identification of unconditional and conditional QTL for oil, protein and starch content in maize. Crop J. 2013, 1, 34–42. [Google Scholar] [CrossRef]
- Li, H.; Peng, Z.; Yang, X.; Wang, W.; Fu, J.; Wang, J.; Yan, J. Genome-wide association study dissects the genetic architecture of oil biosynthesis in maize kernels. Nat. Genet. 2013, 45, 43–50. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Zhang, J.; Liu, X.; Wang, W.; Liu, D.; Teng, Z.; Zhang, Z. Fine mapping and RNA-Seq unravels candidate genes for a major QTL controlling multiple fiber quality traits at the T1 region in upland cotton. BMC Genom. 2016, 17, 295. [Google Scholar] [CrossRef] [PubMed]
- Lin, F.; Zhou, L.; He, B.; Zhang, X.; Dai, H.; Qian, Y.; Zhao, H. QTL mapping for maize starch content and candidate gene prediction combined with co-expression network analysis. Theor. Appl. Genet. 2019, 132, 1931–1941. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Hou, L.; Jian, H.; Di, F.; Li, J.; Liu, L. Combined QTL mapping, physiological and transcriptomic analyses to identify candidate genes involved in Brassica napus seed aging. Mol. Genet. Genom. 2018, 293, 1421–1435. [Google Scholar] [CrossRef]
- Zhang, Z.; Dunwell, J.M.; Zhang, Y.M. An integrated omics analysis reveals molecular mechanisms that are associated with differences in seed oil content between Glycine max and Brassica napus. BMC Plant Biol. 2018, 18, 328. [Google Scholar] [CrossRef]
- Buckler, E.S.; Holland, J.B.; Bradbury, P.J.; Acharya, C.B.; Brown, P.J.; Browne, C.; McMullen, M.D. The Genetic Architecture of Maize Flowering Time. Science 2009, 325, 714–718. [Google Scholar] [CrossRef]
- Qin, Z.H.; Yin, Z.X.; Zhong, X.Q. Genetic analysis of agronomic traits of two doubled haploid populations in tobacco. J. Zhejiang Univ. (Agric. Life Sci.) 2004, 30, 477–481. [Google Scholar]
- Zhu, S.; Tang, G.; Yang, Z.; Han, R.; Deng, W.; Shen, X.; Huang, R. Fine mapping of a major QTL, qECQ8, for rice taste quality. BMC Plant Biol. 2024, 24, 1034. [Google Scholar] [CrossRef]
- Lv, G.; Jin, X.; Wang, H.; Wang, Y.; Wu, Q.; Wu, H.; Li, S. Cloning a novel reduced-height (Rht) gene TaOSCA1.4 from a QTL in wheat. Front. Plant Sci. 2024, 15, 1381243. [Google Scholar] [CrossRef]
- Yang, G.H.; Dong, Y.B.; Li, Y.L.; Wang, Q.L.; Zhou, Q. QTL verification of grain protein content and its correlation with oil content by using connected RIL populations of high-oil maize. Genet. Mol. Res. 2014, 13, 881–894. [Google Scholar] [CrossRef]
- Cai, H.; Morishima, H. QTL clusters reflect character associations in wild and cultivated rice. Theor. Appl. Genet. 2002, 104, 1217–1228. [Google Scholar] [CrossRef] [PubMed]
- Zerniak, T.; Saenz, J. Lipid membranes modulate the activity of RNA through sequence-dependent interactions. Proc. Natl. Acad. Sci. USA 2022, 119, e2119235119. [Google Scholar] [CrossRef] [PubMed]
- Merklinger, L.; Bauer, J.; Pedersen, P.A.; Damgaard, R.B.; Morth, J.P. Phospholipids alter activity and stability of mitochondrial membrane-bound ubiquitin ligase MARCH5. Life Sci. Alliance 2022, 5, e202101309. [Google Scholar] [CrossRef] [PubMed]
- Frank, C.; Aebi, M. ALG9 mannosyltransferase is involved in two different steps of lipid-linked oligosaccharide biosynthesis. Glycobiology 2005, 15, 1156–1163. [Google Scholar] [CrossRef]
- Watterson, A.; Tatge, L.; Wajahat, N.; Arneaud, S.L.; Solano, F.R.; Beheshti, S.T.; Douglas, P.M. Intracellular lipid surveillance by small G protein geranylgeranylation. Nature 2022, 605, 736–740. [Google Scholar] [CrossRef]
- Chen, W.L.; Zhang, Q.Q.; Tang, S.H.; Gong, W.; Hong, Y.Y. Glycerol-3-phosphate acyltransferase in lipid metabolism, growth and response to stresses in plants. Plant Physiol. J. 2018, 54, 725–735. [Google Scholar]
- Ma, Y.; Cai, Y.; Ma, X.J.; Cui, G.H.; Tang, J.F.; Zeng, W.; Huang, L.Q. Research progress of P450 in the biosynthesis of bioactive compound of medicinal plants. Acta Pharm. Sin. 2020, 55, 1573–1589. [Google Scholar]
- Mueller, J. Vitamin B6 in fat metabolism. Vitam. Horm. 1964, 22, 787–796. [Google Scholar]
- Gupta, A.; Rai, S.; Bano, A.; Sharma, S.; Kumar, M.; Binsuwaidan, R.; Pathak, N. ACC deaminase produced by PGPR mitigates the adverse effect of osmotic and salinity stresses in Pisum sativum through modulating the antioxidants activities. Plants 2018, 11, 3419. [Google Scholar] [CrossRef]
- Jin, J.; Lu, M.; Li, Q.Y.; Wu, Z.Z.; Zhang, P.; Feng, W.; Jin, Q. Microbial synthesis of sterol-based steroidal compounds: A review. Food Res. Dev. 2018, 39, 205–209. [Google Scholar]
- Chu, S.Y.; Sun, B.H.; Tian, X.L.; Wang, X.X. Research progress on the formation and properties of starch-lipid complexes. Food Res. Dev. 2021, 42, 206–211. [Google Scholar]
- Chen, M.; Liu, R.; Yi, W. The Discovery and Revelation of Tricarboxylie Acid Cycle. Med. Philos. 2012, 33, 71–73. [Google Scholar]
- Li, J.Q.; Shi, X.Z.; Niu, Y.; Ji, B.Z. Acetylcoenzyme A carboxylase: A key metabolic enzyme of fatty acid and progress of its gene clone. Chin. J. Appl. Environ. Biol. 2011, 17, 753–758. [Google Scholar]
- Wang, Y.; Zeng, F.; Zhao, Z.; He, L.; He, X.; Pang, H.; Chang, P. Transmembrane protein 68 functions as an MGAT and DGAT enzyme for triacylglycerol biosynthesis. Int. J. Mol. Sci. 2023, 24, 2012. [Google Scholar] [CrossRef]
- Liu, Z.; Tian, G. Production of acrylic acid and propionic acid by constructing a portion of the 3-hydroxypropionate/4-hydroxybutyrate cycle from Metallosphaera sedula in Escherichia coli. J. Ind. Microbiol. Biotechnol. 2016, 43, 1659–1670. [Google Scholar] [CrossRef]
- Zheng, Y.; Xiang, L.; Kai, W. ABI3 with or without FUS3 highly up-regulates lipid droplet proteins and activates oil accumulation. J. Exp. Bot. 2021, 22, 787–796. [Google Scholar]
- Liu, F.; Schnable, P. Functional specialization of maize mitochondrial aldehyde dehydrogenases. Plant Physiol. 2002, 130, 1657–1674. [Google Scholar] [CrossRef][Green Version]
- Li, M.Y.; Ruth, W.; Wang, X.M. Quantitative profiling of Arabidopsis polar glycerolipids in response to phosphorus starvation. Roles of phospholipases D ζ1 and D ζ2 in phosphatidylcholine hydrolysis and digalactosyldiacylglycerol accumulation in phosphorus-starved plants. Plant Physiol. 2006, 142, 750–761. [Google Scholar] [CrossRef]
- Yang, Z.; Liu, X.; Wang, K.; Li, Z.; Jia, Q.; Zhao, C.; Zhang, M. ABA-INSENSITIVE 3 with or without FUSCA3 highly up-regulates lipid droplet proteins and activates oil accumulation. J. Exp. Bot. 2022, 73, 2077–2092. [Google Scholar] [CrossRef]
- Yang, W.; Wang, G.; Li, J.; Bates, P.D.; Wang, X.; Allen, D.K. Phospholipase Dζ Enhances Diacylglycerol Flux into Triacylglycerol. Plant Physiol. 2017, 174, 110–123. [Google Scholar] [CrossRef]
- Mlíčková, K.; Luo, Y.; d’Andrea, S.; Peč, P.; Chardot, T.; Nicaud, J.M. Acyl-CoA oxidase., a key step for lipid accumulation in the yeast Yarrowia lipolytica. J. Mol. Catal. B Enzym. 2014, 28, 81–85. [Google Scholar] [CrossRef]
- Chen, D.H.; Ronald, P.C. A Rapid DNA Minipreparation Method Suitable for AFLP and Other PCR Applications. Plant Mol. Biol. Report. 1999, 17, 53–57. [Google Scholar] [CrossRef]
- Xu, Y.B.; Yang, Q.; Zheng, H.; Sang, J.; Zhang, Z. Genotyping by target sequencing (GBTS) and its applications. Sci. Agric. Sin. 2020, 53, 2983–3004. [Google Scholar]
- Taylor, J.; Butler, D. R Package ASMap: Efficient Genetic Linkage Map Construction and Diagnosis. J. Stat. Softw. 2017, 79, 1–29. [Google Scholar] [CrossRef]
- Zhang, Y.Y.; Zhao, J.Q.; Jiang, L.J.; Shi, Y.H.; Chen, L.; Guo, S.L.; Song, J.C.; Jameson, P.E. High quality rna isolation from mature seeds of maize based on modified and optimized protocols. J. Maize Sci. 2015, 23, 591–594. [Google Scholar] [CrossRef]



| Traits | Parents | DH Lines | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Maternal Parent | Male Parent | T Value | Median Value | K-S Value | Sig Value | Mean | Standard Deviation | Environmental Variance | Coefficient of Variation | |
| Oil content/% | 9.58 | 3.41 | −34.38 ** | 6.49 | 0.034 | 0.200 | 5.72 | 1.07 | 0.14 | 18.25 |
| Linkage Group | Marker Number | Genetic Distance/cM | Average Map Distance/cM | Maximum Interval/cM |
|---|---|---|---|---|
| LG01 | 1014 | 627.74 | 0.62 | 27.71 |
| LG02 | 609 | 466.70 | 0.77 | 33.42 |
| LG03 | 737 | 524.61 | 0.71 | 42.76 |
| LG04 | 495 | 344.61 | 0.70 | 27.22 |
| LG05 | 573 | 283.57 | 0.49 | 19.55 |
| LG06 | 388 | 369.12 | 0.95 | 41.30 |
| LG07 | 480 | 353.16 | 0.74 | 39.37 |
| LG08 | 474 | 378.41 | 0.80 | 40.02 |
| LG09 | 328 | 260.69 | 0.79 | 39.37 |
| LG10 | 302 | 255.91 | 0.85 | 13.06 |
| ALL | 5400 | 3864.51 | 0.74 | 42.76 |
| Chromosome | QTL | Peak/cM | Physical Interval/Mb | LOD Value | Contribution Rate/% | Additive Effect |
|---|---|---|---|---|---|---|
| 1 | qOIL-1-1 | 100.01 | 240.93~256.57 | 3.34 | 5.06 | −0.25~−0.22 |
| 3 | qOIL-3-1 | 203.68 | 6.53~12.14 | 3.03 | 2.78 | −0.36~−0.32 |
| qOIL-3-2 | 243.70 | 160.12~170.98 | 4.72 | 0.02 | −0.41~−0.40 | |
| qOIL-3-3 | 524.61 | 169.01~178.32 | 2.73 | 1.06 | −0.43~−0.41 | |
| 4 | qOIL-4-1 | 46.48 | 11.40~139.47 | 4.71 | 0.05 | −0.43~−0.33 |
| qOIL-4-2 | 64.25 | 150.66~166.04 | 5.15 | 1.89 | −0.37~−0.34 | |
| qOIL-4-3 | 76.11 | 157.95~177.07 | 3.65 | 0.01 | −0.36~−0.32 | |
| 5 | qOIL-5-1 | 20.78 | 3.82~7.25 | 3.37 | 3.52 | −0.31~−0.28 |
| qOIL-5-2 | 283.58 | 15.21~27.66 | 2.59 | 1.11 | −0.23~−0.22 | |
| 6 | qOIL-6-1 | 155.34 | 88.91~102.89 | 3.51 | 0.47 | −0.40~−0.34 |
| qOIL-6-2 | 186.57 | 109.48~130.94 | 3.22 | 0.48 | −0.36~−0.26 | |
| 8 | qOIL-8-1 | 300.00 | 22.65~116.15 | 3.06 | 0.10 | −0.32~−0.28 |
| qOIL-8-2 | 311.27 | 25.34~141.13 | 3.00 | 0.01 | −0.33~−0.28 |
| Chr. | QTL | Gene ID | Homologous Genes in Arabidopsis | Functional Annotation |
|---|---|---|---|---|
| 3 | qOIL-3-2 | GRMZM2G133398 | AT3G24650 | Regulatory protein viviparous-1 [Zea mays] |
| 3 | qOIL-3-2 | GRMZM2G156861 | AT1G55020 | lipoxygenase [Zea mays] |
| 4 | qOIL-4-2 | GRMZM2G125268 | AT3G48000 | mitochondrial aldehyde dehydrogenase RF2B [Zea mays] |
| 6 | qOIL-6-2 | GRMZM2G002959 | AT3G51840 | acyl-CoA oxidase 4 [Zea mays] |
| 8 | qOIL-8-1 | GRMZM2G343588 | AT3G16785 | phospholipase D16 [Zea mays] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, J.; Wang, Q.; Han, W.; Zhao, Q.; Sun, D.; Shen, Z. QTL Mapping and Candidate Gene Screening for Enhancing Oil Content in Silage Maize. Plants 2025, 14, 1181. https://doi.org/10.3390/plants14081181
Wu J, Wang Q, Han W, Zhao Q, Sun D, Shen Z. QTL Mapping and Candidate Gene Screening for Enhancing Oil Content in Silage Maize. Plants. 2025; 14(8):1181. https://doi.org/10.3390/plants14081181
Chicago/Turabian StyleWu, Jianzhong, Qi Wang, Weibo Han, Qian Zhao, Dequan Sun, and Zhongbao Shen. 2025. "QTL Mapping and Candidate Gene Screening for Enhancing Oil Content in Silage Maize" Plants 14, no. 8: 1181. https://doi.org/10.3390/plants14081181
APA StyleWu, J., Wang, Q., Han, W., Zhao, Q., Sun, D., & Shen, Z. (2025). QTL Mapping and Candidate Gene Screening for Enhancing Oil Content in Silage Maize. Plants, 14(8), 1181. https://doi.org/10.3390/plants14081181







