The Role of MYC2 Transcription Factors in Plant Secondary Metabolism and Stress Response Mechanisms
Abstract
:1. Introduction
2. The Role of MYC2 in the Synthesis of Plant Secondary Metabolites
2.1. MYC2 Transcription Factors in Alkaloid Biosynthesis
2.2. MYC2 Transcription Factors in Terpenoid Biosynthesis
2.3. MYC2 Transcription Factors in Flavonoid Synthesis
2.4. Regulatory Role of MYC2 in VOCs, Essential Oils, and Other Compounds
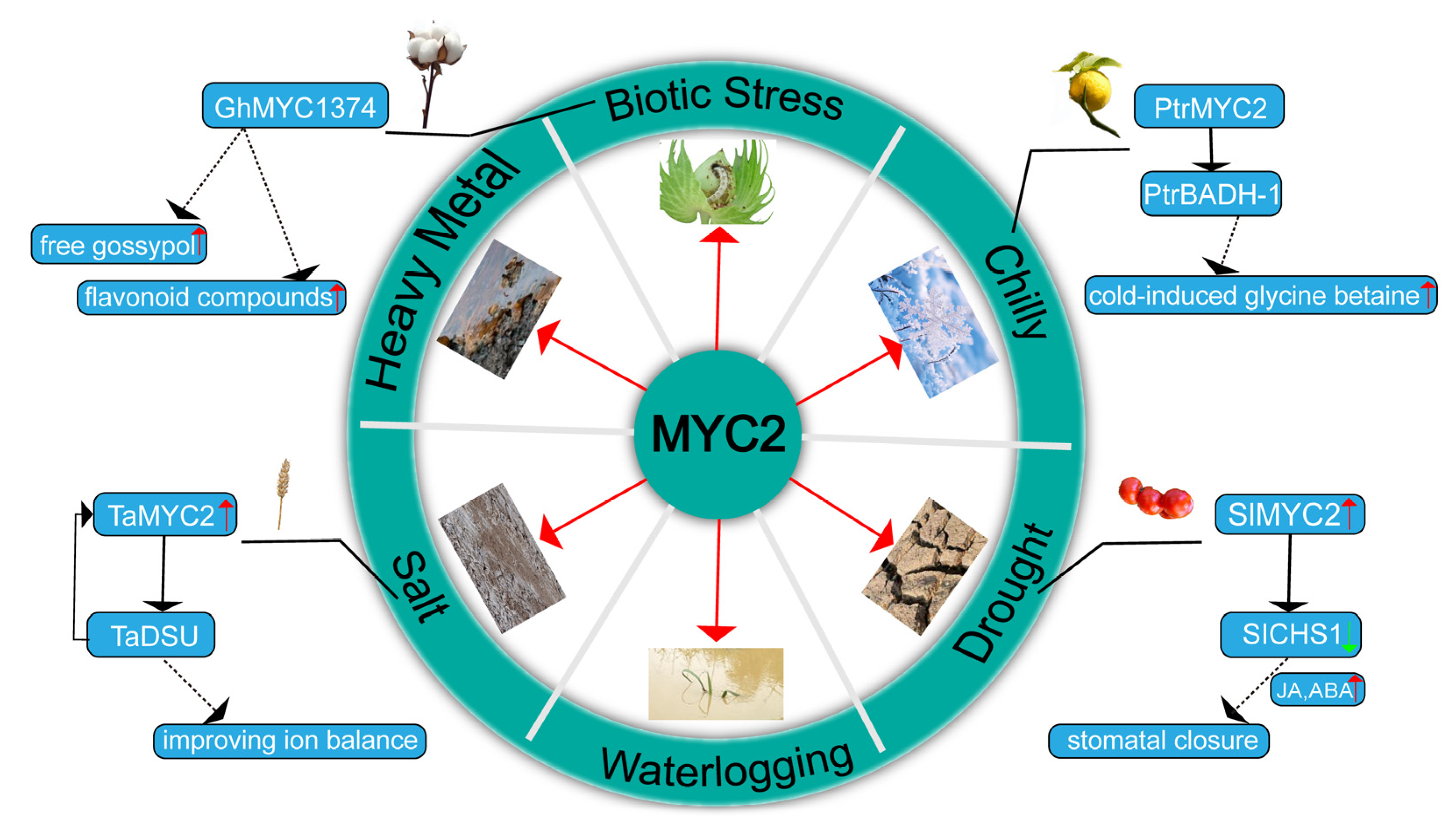
3. Response of MYC2 to Stress
3.1. The Role of MYC2 in Cold Stress
3.2. The Role of MYC2 in Drought Tolerance
3.3. The Role of MYC2 in Water Stress
3.4. Role of MYC2 in Salt Stress
3.5. Role of MYC2 in Heavy Metal Stress
3.6. Role of MYC2 in Biotic Stress
3.7. MYC2 Mediators of Hormonal Interactions and Stress Adaptation in Plants
4. Applications and Prospects of MYC2 Transcription Factors
4.1. MYC2 as a Central Regulatory Hub Gene Across and Within Species
4.2. Balancing Growth and Stress Resistance: The Dilemma of MYC Transcription Factors in Plants
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Peñuelas, M.; Monte, I.; Schweizer, F.; Vallat, A.; Reymond, P.; García-Casado, G. Jasmonate-related MYC transcription factors are functionally conserved in Marchantia polymorpha. Plant Cell 2019, 31, 2491–2509. [Google Scholar] [CrossRef] [PubMed]
- Figueroa, P.; Browse, J. The Arabidopsis JAZ2 promoter contains a G-box and thymidine-rich module that are necessary and sufficient for jasmonate-dependent activation by MYC transcription factors and repression by JAZ proteins. Plant Cell Physiol. 2012, 53, 330–343. [Google Scholar] [CrossRef] [PubMed]
- López-Vidriero, I.; Godoy, M.; Grau, J.; Peñuelas, M.; Solano, R.; Franco-Zorrilla, J.M. DNA features beyond the transcription factor binding site specify target recognition by plant MYC2-related bHLH proteins. Plant Commun. 2021, 2, 100232. [Google Scholar] [CrossRef]
- Lian, T.F.; Xu, Y.P.; Li, L.F.; Su, X.D. Crystal structure of tetrameric Arabidopsis MYC2 reveals the mechanism of enhanced interaction with DNA. Cell Rep. 2017, 19, 1334–1342. [Google Scholar] [CrossRef]
- Lee, Y.S.; Shiu, S.-H.; Grotewold, E. Evolution and diversification of the ACT-like domain associated with plant basic helix–loop–helix transcription factors. Proc. Natl. Acad. Sci. USA 2023, 120, e2219469120. [Google Scholar] [CrossRef]
- Gao, F.; Dubos, C. The Arabidopsis bHLH transcription factor family. Trends Plant Sci. 2024, 29, 668–680. [Google Scholar] [CrossRef] [PubMed]
- Hou, X.; Singh, S.K.; Werkman, J.R.; Liu, Y.; Yuan, Q.; Wu, X.; Patra, B.; Sui, X.; Lyu, R.; Wang, B. Partial desensitization of MYC2 transcription factor alters the interaction with jasmonate signaling components and affects specialized metabolism. Int. J. Biol. Macromol. 2023, 252, 126472. [Google Scholar] [CrossRef]
- Zhai, Q.; Yan, L.; Tan, D.; Chen, R.; Sun, J.; Gao, L.; Dong, M.-Q.; Wang, Y.; Li, C. Phosphorylation-coupled proteolysis of the transcription factor MYC2 is important for jasmonate-signaled plant immunity. PLoS Genet. 2013, 9, e1003422. [Google Scholar] [CrossRef]
- Zhu, J.; Wang, W.-S.; Yan, D.-W.; Hong, L.-W.; Li, T.-T.; Gao, X.; Yang, Y.-H.; Ren, F.; Lu, Y.-T.; Yuan, T.-T. CK2 promotes jasmonic acid signaling response by phosphorylating MYC2 in Arabidopsis. Nucleic Acids Res. 2023, 51, 619–630. [Google Scholar] [CrossRef]
- Zhang, F.; Ke, J.; Zhang, L.; Chen, R.; Sugimoto, K.; Howe, G.A. Structural insights into alternative splicing-mediated desensitization of jasmonate signaling. Proc. Natl. Acad. Sci. USA 2017, 114, 1720–1725. [Google Scholar] [CrossRef]
- Zhai, Q.; Li, C. The plant mediator complex and its role in jasmonate signaling. J. Exp. Bot. 2019, 70, 3415–3424. [Google Scholar] [CrossRef] [PubMed]
- Zhai, Q.; Deng, L.; Li, C. Mediator subunit MED25: At the nexus of jasmonate signaling. Curr. Opin. Plant Biol. 2020, 57, 78–86. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Yao, J.; Ke, J.; Zhang, L.; Lam, V.Q.; Xin, X.F. Structural basis of JAZ repression of MYC transcription factors in jasmonate signaling. Nature 2015, 525, 269–273. [Google Scholar] [CrossRef]
- An, C.; Li, L.; Zhai, Q.; You, Y.; Deng, L.; Wu, F. Mediator subunit MED25 links the jasmonate receptor to transcriptionally active chromatin. Proc. Natl. Acad. Sci. USA 2017, 114, 8930–8939. [Google Scholar] [CrossRef]
- You, Y.; Zhai, Q.; An, C.; Li, C. Leunig_homolog mediates MYC2-dependent transcriptional activation in cooperation with the coactivators HAC1 and MED25. Plant Cell 2019, 31, 2187–2205. [Google Scholar] [CrossRef]
- Chico, J.M.; Lechner, E.; Fernandez-Barbero, G.; Canibano, E.; García-Casado, G.; Franco-Zorrilla, J.M.; Hammann, P.; Zamarreño, A.M.; García-Mina, J.M.; Rubio, V.; et al. CUL3BPM E3 ubiquitin ligases regulate MYC2, MYC3, and MYC4 stability and JA responses. Proc. Natl. Acad. Sci. USA 2020, 117, 6205–6215. [Google Scholar] [CrossRef]
- Song, C.; Cao, Y.; Dai, J.; Li, G.; Manzoor, M.A.; Chen, C.; Deng, H. The multifaceted roles of MYC2 in plants: Toward transcriptional reprogramming and stress tolerance by jasmonate signaling. Front. Plant Sci. 2022, 13, 868874. [Google Scholar] [CrossRef]
- Yang, J.; Duan, G.; Li, C.; Liu, L.; Han, G.; Zhang, Y.; Wang, C. The crosstalks between jasmonic acid and other plant hormone signaling highlight the involvement of jasmonic acid as a core component in plant response to biotic and abiotic stresses. Front. Plant Sci. 2019, 10, 1349. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Huang, Y.; Shen, Z.; Wu, M.; Huang, M.; Hong, S.-B.; Xu, L.; Zang, Y. Advances in functional studies of plant MYC transcription factors. Theor. Appl. Genet. 2024, 137, 195. [Google Scholar] [CrossRef]
- Afrin, S.; Huang, J.J.; Luo, Z.Y. JA-mediated transcriptional regulation of secondary metabolism in medicinal plants. Sci. Bull. 2015, 60, 1062–1072. [Google Scholar] [CrossRef]
- Luo, L.; Wang, Y.; Qiu, L.; Han, X.; Zhu, Y.; Liu, L.; Man, M.; Li, F.; Ren, M.; Xing, Y. MYC2: A master switch for plant physiological processes and specialized metabolite synthesis. Int. J. Mol. Sci. 2023, 24, 3511. [Google Scholar] [CrossRef] [PubMed]
- Shoji, T.; Hashimoto, T. Tobacco MYC2 regulates jasmonate-inducible nicotine biosynthesis genes directly and by way of the NIC2-Locus ERF genes. Plant Cell Physiol. 2011, 52, 1117–1130. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.B.; Bokowiec, M.T.; Rushton, P.J.; Han, S.C.; Timko, M.P. Tobacco transcription factors NtMYC2a and NtMYC2b form nuclear complexes with the NtJAZ1 repressor and regulate multiple jasmonate- inducible steps in nicotine biosynthesis. Mol. Plant 2012, 5, 73–84. [Google Scholar] [CrossRef]
- Sui, X.; He, X.; Song, Z.; Gao, Y.; Zhao, L.; Jiao, F.; Kong, G.; Li, Y.; Han, S.; Wang, B. The gene NtMYC2a acts as a ‘master switch’in the regulation of JA-induced nicotine accumulation in tobacco. Plant Biol. 2021, 23, 317–326. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Z.L.; Yang, W.W.; Yang, A.G.; Deng, L.L.; Geng, R.M.; Xiang, H.Y.; Kong, W.S.; Jiang, C.H.; Li, X.M.; Chen, Z.Q.; et al. CRISPR/Cas9-mediated knockout of NtMYC2a gene involved in resistance to bacterial wilt in tobacco. Gene 2024, 927, 148622. [Google Scholar] [CrossRef]
- Zhang, H.; Hedhili, S.; Montiel, G.; Zhang, Y.; Chatel, G.; Pré, M. The basic helix-loop-helix transcription factor CrMYC2 controls the jasmonate- responsive expression of the ORCA genes that regulate alkaloid biosynthesis in Catharanthus roseus. Plant J. 2011, 67, 61–71. [Google Scholar] [CrossRef]
- Goklany, S.; Rizvi, N.F.; Loring, R.H.; Cram, E.J.; Lee-Parsons, C.W.T. Jasmonate-dependent alkaloid biosynthesis in Catharanthus roseus hairy root cultures is correlated with the relative expression of ORCA and ZCT transcription factors. Biotechnol. Prog. 2013, 29, 1367–1376. [Google Scholar] [CrossRef]
- Sui, X.; Singh, S.K.; Patra, B.; Schluttenhofer, C.; Guo, W.; Pattanaik, S. Cross-family transcription factor interaction between MYC2 and GBFs modulates terpenoid indole alkaloid biosynthesis. J. Exp. Bot. 2018, 69, 4267–4281. [Google Scholar] [CrossRef]
- Patra, B.; Pattanaik, S.; Schluttenhofer, C.; Yuan, L. A network of jasmonate-responsive bHLH factors modulate monoterpenoid indole alkaloid biosynthesis in Catharanthus roseus. New Phytol. 2018, 217, 1566–1581. [Google Scholar] [CrossRef]
- Swinnen, G.; De Meyer, M.; Pollier, J.; Molina-Hidalgo, F.J.; Ceulemans, E.; Venegas-Molina, J.; De Milde, L.; Fernandez-Calvo, P.; Ron, M.; Pauwels, L.; et al. The basic helix-loop-helix transcription factors MYC1 and MYC2 have a dual role in the regulation of constitutive and stress-inducible specialized metabolism in tomato. New Phytol. 2022, 236, 911–928. [Google Scholar] [CrossRef]
- Panda, S.; Jozwiak, A.; Sonawane, P.D.; Szymanski, J.; Kazachkova, Y.; Vainer, A.; Kilambi, H.V.; Almekias-Siegl, E.; Dikaya, V.; Bocobza, S. Steroidal alkaloids defence metabolism and plant growth are modulated by the joint action of gibberellin and jasmonate signalling. New Phytol. 2022, 233, 1220–1237. [Google Scholar] [CrossRef]
- Zhou, Z.; Wu, M.Z.; Sun, B.; Li, J.; Li, J.D.; Liu, Z.T.; Gao, M.; Xue, L.; Xu, S.; Wang, R. Identification of transcription factor genes responsive to MeJA and characterization of a LaMYC2 transcription factor positively regulates lycorine biosynthesis in Lycoris aurea. J. Plant Physiol. 2024, 296, 154218. [Google Scholar] [CrossRef] [PubMed]
- Zeng, T.; Li, J.; Li, J.; Hu, H.; Zhu, L.; Liu, K.; Bai, J.; Jiang, Q.; Wang, C. Pyrethrins in Tanacetum cinerariifolium: Biosynthesis, regulation, and agricultural application. Ornam. Plant Res. 2024, 4, e015. [Google Scholar] [CrossRef]
- Zeng, T.; Li, J.W.; Xu, Z.Z.; Zhou, L.; Li, J.J.; Yu, Q.; Luo, J.; Chan, Z.L.; Jongsma, M.A.; Hu, H.; et al. TcMYC2 regulates pyrethrin biosynthesis in Tanacetum cinerariifolium. Hortic. Res. 2022, 9, uhac178. [Google Scholar] [CrossRef]
- Zeng, T.; Yu, Q.; Shang, J.; Xu, Z.; Zhou, L.; Li, W.; Li, J.; Hu, H.; Zhu, L.; Li, J.; et al. TcbHLH14 a jasmonate associated MYC2-like transcription factor positively regulates pyrethrin biosynthesis in Tanacetum cinerariifolium. Int. J. Mol. Sci. 2023, 24, 7379. [Google Scholar] [CrossRef] [PubMed]
- Lan, Y.A.; Zhang, K.M.; Wang, L.N.; Liang, X.Y.; Liu, H.X.; Zhang, X.Y.; Jiang, N.Q.; Wu, M.; Yan, H.W.; Xiang, Y. The R2R3-MYB transcription factor OfMYB21 positively regulates linalool biosynthesis in Osmanthus fragrans flowers. Int. J. Biol. Macromol. 2023, 249, 126099. [Google Scholar] [CrossRef] [PubMed]
- Aslam, M.Z.; Lin, X.; Li, X.; Yang, N.; Chen, L.Q. Molecular cloning and functional characterization of CpMYC2 and CpBHLH13 transcription factors from wintersweet (Chimonanthus praecox L.). Plants 2020, 9, 785. [Google Scholar] [CrossRef]
- Yang, Z.; Li, Y.; Gao, F.; Jin, W.; Li, S.; Kimani, S.; Yang, S.; Bao, T.; Gao, X.; Wang, L. MYB21 interacts with MYC2 to control the expression of terpene synthase genes in flowers of Freesia hybrida and Arabidopsis thaliana. J. Exp. Bot. 2020, 71, 4140–4158. [Google Scholar] [CrossRef]
- Yang, Z.Z.; Jin, W.; Luo, Q.; Li, X.L.; Wei, Y.M.; Lin, Y.L. FhMYB108 regulates the expression of linalool synthase gene in Freesia hybrida and Arabidopsis. Biol. 2024, 13, 556. [Google Scholar] [CrossRef]
- Li, J.; Luo, Y.; Li, M.; Li, J.; Zeng, T.; Luo, J.; Chang, X.; Wang, M.; Jongsma, M.A.; Hu, H.; et al. Nocturnal burst emissions of germacrene D from the open disk florets of pyrethrum flowers induce moths to oviposit on a nonhost and improve pollination success. New Phytol. 2024, 244, nph.20081. [Google Scholar] [CrossRef]
- Li, J.J.; Hu, H.; Mao, J.; Yu, L.; Stoopen, G.; Wang, M.Q.; Mumm, R.; de Ruijter, N.C.A.; Dicke, M.; Jongsma, M.A.; et al. Defense of pyrethrum flowers: Repelling herbivores and recruiting carnivores by producing aphid alarm pheromone. New Phytol. 2019, 223, 1607–1620. [Google Scholar] [CrossRef] [PubMed]
- Shen, S.; Yin, M.; Zhou, Y.; Huan, C.; Zheng, X.; Chen, K. The role of CitMYC3 in regulation of valencene synthesis-related CsTPS1 and CitAP2.10 in Newhall sweet orange. Sci. Hortic. 2024, 334, 113338. [Google Scholar] [CrossRef]
- Hong, G.J.; Xue, X.Y.; Mao, Y.B.; Wang, L.J.; Chen, X.Y. Arabidopsis MYC2 interacts with DELLA proteins in regulating sesquiterpene synthase gene expression. Plant Cell 2012, 24, 2635–2648. [Google Scholar] [CrossRef]
- Olsson, M.E.; Olofsson, L.M.; Lindahl, A.L.; Lundgren, A.; Brodelius, M.; Brodelius, P.E. Localization of enzymes of artemisinin biosynthesis to the apical cells of glandular secretory trichomes of Artemisia annua L. Phytochemistry 2009, 70, 1123–1128. [Google Scholar] [CrossRef] [PubMed]
- Shen, Q.; Lu, X.; Yan, T.X.; Fu, X.Q.; Lv, Z.Y.; Zhang, F.Y.; Pan, Q.F.; Wang, G.F.; Sun, X.F.; Tang, K.X. The jasmonate-responsive AaMYC2 transcription factor positively regulates artemisinin biosynthesis in Artemisia annua. New Phytol. 2016, 210, 1269–1281. [Google Scholar] [CrossRef] [PubMed]
- Ji, Y.P.; Xiao, J.W.; Shen, Y.L.; Ma, D.M.; Li, Z.Q.; Pu, G.B.; Li, X.; Huang, L.L.; Liu, B.Y.; Ye, H.C.; et al. Cloning and characterization of AabHLH1, a bHLH transcription factor that positively regulates artemisinin biosynthesis in Artemisia annua. Plant Cell Physiol. 2014, 55, 1592–1604. [Google Scholar] [CrossRef]
- Yuan, M.Y.; Sheng, Y.G.; Bao, J.J.; Wu, W.K.; Nie, G.B.; Wang, L.J.; Cao, J.F. AaMYC3 bridges the regulation of glandular trichome density and artemisinin biosynthesis in Artemisia annua. Plant Biotechnol. J. 2024, 23, 315–332. [Google Scholar] [CrossRef]
- Shen, Q.; Huang, H.; Xie, L.; Hao, X.; Kayani, S.I.; Liu, H.; Qin, W.; Chen, T.; Pan, Q.; Liu, P. Basic helix-loop-helix transcription factors AabHLH2 and AabHLH3 function antagonistically with AaMYC2 and are negative regulators in artemisinin biosynthesis. Front. Plant Sci. 2022, 13, 885622. [Google Scholar] [CrossRef]
- Xu, Y.H.; Liao, Y.C.; Lv, F.F.; Zhang, Z.; Sun, P.W.; Gao, Z.H.; Hu, K.P.; Sui, C.; Jin, Y.; Wei, J.H. Transcription factor AsMYC2 controls the jasmonate-responsive expression of ASS1 regulating sesquiterpene biosynthesis in Aquilaria sinensis (Lour.) Gilg. Plant Cell Physiol. 2017, 58, 1924–1933. [Google Scholar] [CrossRef]
- Han, X.; Xing, Y.; Zhu, Y.; Luo, L.; Liu, L.; Zhai, Y.; Wang, W.; Shao, R.; Ren, M.; Li, F. GhMYC2 activates cytochrome P450 gene CYP71BE79 to regulate gossypol biosynthesis in cotton. Planta 2022, 256, 63. [Google Scholar] [CrossRef]
- Dong, Y.; Zhang, W.; Li, J.; Wang, D.; Bai, H.; Li, H.; Shi, L. The transcription factor LaMYC4 from lavender regulates volatile terpenoid biosynthesis. BMC Plant Biol. 2022, 22, 289. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.Y.; Wu, S.Y.; Lan, K.; Wang, Q.; Ye, T.Y.; Jin, H.N.; Hu, T.Y.; Xie, T.; Wei, Q.H.; Yin, X.P. An investigation of the JAZ family and the CwMYC2-like protein to reveal their regulatory roles in the MeJA-induced biosynthesis of β-elemene in Curcuma wenyujin. Int. J. Mol. Sci. 2023, 24, 15004. [Google Scholar] [CrossRef]
- Wang, L.L.; Xu, G.J.; Li, L.H.; Ruan, M.Y.; Bennion, A.; Wang, G.L.; Li, R.; Qu, S.H. The OsBDR1-MPK3 module negatively regulates blast resistance by suppressing the jasmonate signaling and terpenoid biosynthesis pathway. Proc. Natl. Acad. Sci. USA 2023, 120, e2211102120. [Google Scholar] [CrossRef] [PubMed]
- Yin, J.Y.; Lai, M.; Yu, X.Y.; Su, D.D.; Xiong, X.Y.; Li, Y.L. Comprehensive strategies for paclitaxel production: Insights from plant cell culture, endophytic microorganisms, and synthetic biology. Horticulture Research 2025, 12, uhae346. [Google Scholar] [CrossRef]
- Cui, Y.; Mao, R.; Chen, J.; Guo, Z. Regulation mechanism of MYC family transcription factors in jasmonic acid signalling pathway on taxol biosynthesis. Int. J. Mol. Sci. 2019, 20, 1843. [Google Scholar] [CrossRef]
- Zhang, M.; Jin, X.F.; Chen, Y.; Wei, M.; Liao, W.F.; Zhao, S.Y.; Fu, C.H.; Yu, L.J. TcMYC2a, a basic helix loop helix transcription factor, transduces ja-signals and regulates taxol biosynthesis in Taxus chinensis. Front. Plant Sci. 2018, 9, 863. [Google Scholar] [CrossRef]
- Zhou, Y.; Sun, W.; Chen, J.; Tan, H.; Xiao, Y.; Li, Q. SmMYC2a and SmMYC2b played similar but irreplaceable roles in regulating the biosynthesis of tanshinones and phenolic acids in Salvia miltiorrhiza. Sci. Rep. 2016, 6, 22852. [Google Scholar] [CrossRef] [PubMed]
- Du, T.; Niu, J.; Su, J.; Li, S.; Guo, X.; Li, L.; Cao, X.; Kang, J. SmbHLH37 functions antagonistically with SmMYC2 in regulating jasmonate-mediated biosynthesis of phenolic acids in Salvia miltiorrhiza. Front. Plant Sci. 2018, 9, 1720. [Google Scholar] [CrossRef]
- Cao, R.; Lv, B.; Shao, S.; Zhao, Y.; Yang, M.; Zuo, A.; Wei, J.; Dong, J.; Ma, P. The SmMYC2-SmMYB36 complex is involved in methyl jasmonate-mediated tanshinones biosynthesis in Salvia miltiorrhiza. Plant J. 2024, 119, 746–761. [Google Scholar] [CrossRef]
- Liu, S.; Wang, Y.; Shi, M.; Maoz, I.; Gao, X.; Sun, M.; Yuan, T.; Li, K.; Zhou, W.; Guo, X. SmbHLH60 and SmMYC2 antagonistically regulate phenolic acids and anthocyanins biosynthesis in Salvia miltiorrhiza. J. Adv. Res. 2022, 42, 205–219. [Google Scholar] [CrossRef]
- Alfieri, M.; Vaccaro, M.C.; Cappetta, E.; Ambrosone, A.; De Tommasi, N.; Leone, A. Coactivation of MEP-biosynthetic genes and accumulation of abietane diterpenes in Salvia sclarea by heterologous expression of WRKY and MYC2 transcription factors. Sci. Rep. 2018, 8, 11009. [Google Scholar] [CrossRef]
- Shi, M.; Wang, Y.; Wang, X.; Deng, C.; Cao, W.; Hua, Q.; Kai, G. Simultaneous promotion of tanshinone and phenolic acid biosynthesis in Salvia miltiorrhiza hairy roots by overexpressing Arabidopsis MYC2. Ind. Crops Prod. 2020, 155, 112826. [Google Scholar] [CrossRef]
- Huo, Y.; Zhang, J.; Zhang, B.; Chen, L.; Zhang, X.; Zhu, C. MYC2 transcription factors TwMYC2a and TwMYC2b negatively regulate triptolide biosynthesis in Tripterygium wilfordii hairy roots. Plants 2021, 10, 679. [Google Scholar] [CrossRef] [PubMed]
- Du, J.F.; Zhao, Z.; Xu, W.B.; Wang, Q.L.; Li, P.; Lu, X. Comprehensive analysis of JAZ family members in Ginkgo biloba reveals the regulatory role of the GbCOI1/GbJAZs/GbMYC2 module in ginkgolide biosynthesis. Tree Physiol. 2024, 44, tpad121. [Google Scholar] [CrossRef] [PubMed]
- Zheng, J.; Liao, Y.; Ye, J.; Xu, F.; Zhang, W.; Zhou, X.; Wang, L.; He, X.; Cao, Z.; Yi, Y.; et al. The transcription factor MYC2 positively regulates terpene trilactone biosynthesis through activating GbGGPPS expression in Ginkgo biloba. Hortic. Res. 2024, 11, uhae228. [Google Scholar] [CrossRef]
- Yue, P.; Jiang, Z.; Sun, Q.; Wei, R.; Yin, Y.; Xie, Z.; Larkin, R.M.; Ye, J.; Chai, L.; Deng, X. Jasmonate activates a CsMPK6-CsMYC2 module that regulates the expression of β-citraurin biosynthetic genes and fruit coloration in orange (Citrus sinensis). Plant Cell 2023, 35, 1167–1185. [Google Scholar] [CrossRef]
- Gao, W.; Meng, Q.; Wang, X.; Chen, F.; Zhou, Y.; He, M. Overexpression of CiMYC2 transcription factor from Chrysanthemum indicum var. aromaticum resulted in modified trichome formation and terpenoid biosynthesis in transgenic tobacco. J. Plant Growth Regul. 2023, 42, 4161–4175. [Google Scholar] [CrossRef]
- Guo, D.; Li, H.L.; Wang, Y.; Zhu, J.H.; Peng, S.Q. A myelocytomatosis transcription factor from Hevea brasiliensis positively regulates the expression of the small rubber particle protein gene. Ind. Crops Prod. 2019, 133, 90–97. [Google Scholar] [CrossRef]
- Wu, Y.L.; Dong, G.Q.; Luo, F.Q.; Xie, H.; Li, X.D.; Yan, J. TkJAZs-TkMYC2-TkSRPP/REF regulates the biosynthesis of natural rubber in Taraxacum kok-saghyz. Plants 2024, 13, 2034. [Google Scholar] [CrossRef]
- Huang, D.; Li, J.M.; Chen, J.H.; Yao, S.C.; Li, L.B.; Huang, R.S.; Tan, Y.; Ming, R.H.; Huang, Y. Genome-wide identification and characterization of the JAZ gene family in Gynostemma pentaphyllum reveals the COI1/JAZ/MYC2 complex potential involved in the regulation of the MeJA-induced gypenoside biosynthesis. Plant Physiol. Biochem. 2024, 214, 108952. [Google Scholar] [CrossRef]
- Liu, T.; Liao, J.; Shi, M.; Li, L.; Liu, Q.; Cui, X.; Ning, W.; Kai, G. A jasmonate-responsive bHLH transcription factor TaMYC2 positively regulates triterpenes biosynthesis in Taraxacum antungense Kitag. Plant Sci. 2023, 326, 111506. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Hu, S.; Li, T.; Liu, C. Regulation of MYC2 in Bupleurum chinense under simulated drought stress. Acta Bot. Boreali-Occident. Sin. 2022, 41, 1747–1754. [Google Scholar] [CrossRef]
- Wang, D.D.; Li, P.; Chen, Q.Y.; Chen, X.Y.; Yan, Z.W.; Wang, M.Y.; Mao, Y.B. Differential contributions of MYCs to insect defense reveal flavonoids alleviating growth inhibition caused by wounding in Arabidopsis. Front. Plant Sci. 2021, 12, 700555. [Google Scholar] [CrossRef]
- An, J.P.; Li, H.H.; Song, L.Q.; Su, L.; Liu, X.; You, C.X.; Wang, X.F.; Hao, Y.J. The molecular cloning and functional characterization of MdMYC2, a bHLH transcription factor in apple. Plant Physiol. Biochem. 2016, 108, 24–31. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Xu, Y.Z.; Lu, Y.Q. A regulatory mechanism on pathways: Modulating roles of MYC2 and BBX21 in the flavonoid network. Plants 2024, 13, 1156. [Google Scholar] [CrossRef] [PubMed]
- Ogawa, S.; Miyamoto, K.; Nemoto, K.; Sawasaki, T.; Yamane, H.; Nojiri, H.; Okada, K. OsMYC2, an essential factor for JA-inductive sakuranetin production in rice, interacts with MYC2-like proteins that enhance its transactivation ability. Sci. Rep. 2017, 7, 40175. [Google Scholar] [CrossRef]
- Hu, Y.F.; Zhao, H.Y.; Xue, L.Y.; Nie, N.; Zhang, H.; Zhao, N.; He, S.Z.; Liu, Q.C.; Gao, S.P.; Zhai, H. IbMYC2 contributes to salt and drought stress tolerance via modulating anthocyanin accumulation and ROS-scavenging system in sweet potato. Int. J. Mol. Sci. 2024, 25, 2096. [Google Scholar] [CrossRef]
- Zhang, C.; Dai, Z.W.; Ferrier, T.; Orduna, L.; Santiago, A.; Peris, A.; Wong, D.C.J.; Kappel, C.; Savoi, S.; Loyola, R.; et al. MYB24 orchestrates terpene and flavonol metabolism as light responses to anthocyanin depletion in variegated grape berries. Plant Cell 2023, 35, 4238–4265. [Google Scholar] [CrossRef]
- Wang, X.; Wang, J.P.; Cui, H.T.; Yang, W.L.; Yu, B.; Zhang, C.; Wen, J.Q.; Kang, J.M.; Wang, Z.; Yang, Q.C. The UDP-glycosyltransferase MtUGT84A1 regulates anthocyanin accumulation and plant growth via JA signaling in Medicago truncatula. Environ. Exp. Bot. 2022, 201, 104972. [Google Scholar] [CrossRef]
- Fu, J.Y.; Liu, L.J.; Liu, Q.; Shen, Q.Q.; Wang, C.; Yang, P.P.; Zhu, C.Y.; Wang, Q. ZmMYC2 exhibits diverse functions and enhances JA signaling in transgenic Arabidopsis. Plant Cell Rep. 2020, 39, 273–288. [Google Scholar] [CrossRef]
- Zhu, J.Y.; Yan, X.M.; Liu, S.R.; Xia, X.B.; An, Y.L.; Xu, Q.S.; Zhao, S.Q.; Liu, L.; Guo, R.; Zhang, Z.L.; et al. Alternative splicing of CsJAZ1 negatively regulates flavan-3-ol biosynthesis in tea plants. Plant J. 2022, 110, 243–261. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Jiao, Y.; Zhao, Y.; Gao, M.; Wu, L.; Wang, S.; Yang, J.; Wang, J.; Chen, Y.; Wang, Y. Phytohormone and transcriptome of pericarp reveals jasmonate and LcMYC2 are involved in neral and geranial biosynthesis in Litsea cubeba. Ind. Crops Prod. 2022, 177, 114423. [Google Scholar] [CrossRef]
- Hao, X.; Wang, S.; Fu, Y.; Liu, Y.; Shen, H.; Jiang, L.; McLamore, E.S.; Shen, Y. The WRKY46-MYC2 module plays a critical role in E-2-hexenal-induced anti-herbivore responses by promoting flavonoid accumulation. Plant Commun. 2024, 5, 100734. [Google Scholar] [CrossRef]
- Teng, Z.; Zheng, W.; Yu, Y.; Hong, S.-B.; Zhu, Z.; Zang, Y. Effects of BrMYC2/3/4 on plant development, glucosinolate metabolism, and Sclerotinia sclerotiorum resistance in transgenic Arabidopsis thaliana. Front. Plant Sci. 2021, 12, 707054. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.J.; Feng, Y.F.; Li, S.S.; Li, D.M.; Yu, J.; Zhao, Z.Y. Jasmonate-induced MdMYC2 improves fruit aroma during storage of Ruixue’ apple based on transcriptomic, metabolic and functional analyses. Lwt-Food Sci. Technol. 2023, 185, 115168. [Google Scholar] [CrossRef]
- Wang, Y.; Xu, H.; Liu, W.; Wang, N.; Qu, C.; Jiang, S. Methyl jasmonate enhances apple’ cold tolerance through the JAZ-MYC2 pathway. Plant Cell Tissue Organ Cult. 2019, 136, 75–84. [Google Scholar] [CrossRef]
- Yu, X.; Li, W.; Li, Z.; Ruan, M. Cold resistance function analysis of cassava MeMYC2.2. Biotechnol. Bull. 2023, 39, 224–231. [Google Scholar]
- Chen, M.S.; Guo, H.M.; Chen, S.Q.; Li, T.T.; Li, M.Q.; Rashid, A.; Xu, C.J.; Wang, K. Methyl jasmonate promotes phospholipid remodeling and jasmonic acid signaling to alleviate chilling injury in peach fruit. J. Agric. Food Chem. 2019, 67, 9958–9966. [Google Scholar] [CrossRef]
- Wang, R.; Yu, M.; Xia, J.; Xing, J.; Fan, X.; Xu, Q.; Cang, J.; Zhang, D. Overexpression of TaMYC2 confers freeze tolerance by ICE-CBF-COR module in Arabidopsis thaliana. Front. Plant Sci. 2022, 13, 1042889. [Google Scholar] [CrossRef]
- Ming, R.; Zhang, Y.; Wang, Y.; Khan, M.; Dahro, B.; Liu, J.H. The JA-responsive MYC2-BADH-like transcriptional regulatory module in Poncirus trifoliata contributes to cold tolerance by modulation of glycine betaine biosynthesis. New Phytol. 2021, 229, 2730–2750. [Google Scholar] [CrossRef]
- Zhao, Y.; Yang, Y.; Jiang, J.; Zhang, X.; Ma, Z.; Meng, L.; Cui, G.; Yin, X. The caucasian clover gene TaMYC2 responds to abiotic stress and improves tolerance by increasing the activity of antioxidant enzymes. Genes 2022, 13, 329. [Google Scholar] [CrossRef]
- Ding, F.; Wang, C.; Xu, N.; Wang, M.; Zhang, S. Jasmonic acid-regulated putrescine biosynthesis attenuates cold-induced oxidative stress in tomato plants. Sci. Hortic. 2021, 288, 110373. [Google Scholar] [CrossRef]
- Li, Z.; Min, D.; Fu, X.; Zhao, X.; Wang, J.; Zhang, X.; Li, F.; Li, X. The roles of SlMYC2 in regulating ascorbate-glutathione cycle mediated by methyl jasmonate in postharvest tomato fruits under cold stress. Sci. Hortic. 2021, 288, 110406. [Google Scholar] [CrossRef]
- Ding, F.; Wang, C.; Zhang, S.X.; Wang, M.L. A jasmonate-responsive glutathione S-transferase gene SlGSTU24 mitigates cold-induced oxidative stress in tomato plants. Sci. Hortic. 2022, 303, 111231. [Google Scholar] [CrossRef]
- Fan, X.L.; Lin, H.R.; Ding, F.; Wang, M.L. Jasmonates promote β-amylase-mediated starch degradation to confer cold tolerance in tomato plants. Plants 2024, 13, 1055. [Google Scholar] [CrossRef] [PubMed]
- Ding, F.; Wang, X.Z.; Li, Z.Y.; Wang, M.L. Jasmonate positively regulates cold tolerance by promoting ABA biosynthesis in tomato. Plants 2023, 12, 60. [Google Scholar] [CrossRef]
- Ding, F.; Wang, C.; Xu, N.; Wang, M.L. The ethylene response factor SlERF.B8 triggers jasmonate biosynthesis to promote cold tolerance in tomato. Environ. Exp. Bot. 2022, 203, 105073. [Google Scholar] [CrossRef]
- Min, D.D.; Zhou, J.X.; Li, J.Z.; Ai, W.; Li, Z.L.; Zhang, X.H.; Fu, X.D.; Zhao, X.M.; Li, F.J.; Li, X.A.; et al. SlMYC2 targeted regulation of polyamines biosynthesis contributes to methyl jasmonate-induced chilling tolerance in tomato fruit. Postharvest Biol. Technol. 2021, 174, 111443. [Google Scholar] [CrossRef]
- Ding, F.; Ren, L.M.; Xie, F.; Wang, M.L.; Zhang, S.X. Jasmonate and melatonin act synergistically to potentiate cold tolerance in tomato plants. Front. Plant Sci. 2022, 12, 763284. [Google Scholar] [CrossRef]
- Li, Y.; Yang, X.; Li, X. Role of jasmonate signaling pathway in resistance to dehydration stress in Arabidopsis. Acta Physiol. Plant. 2019, 41, 100. [Google Scholar] [CrossRef]
- Zhao, W.; Huang, H.; Wang, J.; Wang, X.; Xu, B.; Yao, X.; Sun, L.; Yang, R.; Wang, J.; Sun, A. Jasmonic acid enhances osmotic stress responses by MYC2-mediated inhibition of protein phosphatase 2C1 and response regulators 26 transcription factor in tomato. Plant J. 2023, 113, 546–561. [Google Scholar] [CrossRef] [PubMed]
- Xu, B.-Q.; Wang, J.-J.; Peng, Y.; Huang, H.; Sun, L.-L.; Yang, R.; Suo, L.-N.; Wang, S.-H.; Zhao, W.-C. SlMYC2 mediates stomatal movement in response to drought stress by repressing SlCHS1 expression. Front. Plant Sci. 2022, 13, 952758. [Google Scholar] [CrossRef]
- Wang, W.; Shi, X.; Chen, D.; Wang, F.; Zhang, H. The Brassica napus MYC2 regulates drought tolerance by monitoring stomatal closure. Eur. J. Hortic. Sci. 2020, 85, 226–231. [Google Scholar]
- Wang, G.L.; Long, Y.F.; Jin, X.Y.; Yang, Z.; Dai, L.Y.; Yang, Y.H.; Lu, G.H.; Sun, B. SbMYC2 mediates jasmonic acid signaling to improve drought tolerance via directly activating SbGR1 in sorghum. Theor. Appl. Genet. 2024, 137, 72. [Google Scholar] [CrossRef] [PubMed]
- Shamloo-Dashtpagerdi, R.; Lindlöf, A.; Nouripour-Sisakht, J. Unraveling the regulatory role of MYC2 on ASMT gene expression in wheat: Implications for melatonin biosynthesis and drought tolerance. Physiol. Plant. 2023, 175, e14015. [Google Scholar] [CrossRef] [PubMed]
- Shamloo-Dashtpagerdi, R.; Shahriari, A.G.; Tahmasebi, A.; Vetukuri, R.R. Potential role of the regulatory miR1119-MYC2 module in wheat (Triticum aestivum L.) drought tolerance. Front. Plant Sci. 2023, 14, 1161245. [Google Scholar] [CrossRef]
- Xia, Y.F.; Jiang, S.X.; Wu, W.Q.; Du, K.; Kang, X.Y. MYC2 regulates stomatal density and water use efficiency via targeting EPF2/EPFL4/EPFL9 in poplar. New Phytol. 2024, 241, 2506–2522. [Google Scholar] [CrossRef] [PubMed]
- Aliakbari, M.; Tahmasebi, S.; Sisakht, J.N. Jasmonic acid improves barley photosynthetic efficiency through a possible regulatory module, MYC2-RcaA, under combined drought and salinity stress. Photosynth. Res. 2024, 159, 69–78. [Google Scholar] [CrossRef]
- Van Moerkercke, A.; Duncan, O.; Zander, M.; Šimura, J.; Broda, M.; Vanden Bossche, R.; Lewsey, M.G.; Lama, S.; Singh, K.B.; Ljung, K.; et al. A MYC2/MYC3/MYC4-dependent transcription factor network regulates water spray-responsive gene expression and jasmonate levels. Proc. Natl. Acad. Sci. USA 2019, 116, 23345–23356. [Google Scholar] [CrossRef]
- Zhu, J.; Chen, H.; Liu, L.; Xia, X.; Yan, X.; Mi, X.; Liu, S.; Wei, C. JA-mediated MYC2/LOX/AOS feedback loop regulates osmotic stress response in tea plant. Hortic. Plant J. 2024, 10, 931–946. [Google Scholar] [CrossRef]
- Andrade, A.; Escalante, M.; Ramirez, F.; Vigliocco, A.; Alemano, S. Phytohormones and related genes function as physiological and molecular switches regulating water stress response in the sunflower. Physiol. Mol. Biol. Plants 2024, 30, 1277–1295. [Google Scholar] [CrossRef]
- Xie, L.J.; Wang, J.H.; Liu, H.S.; Yuan, L.B.; Tan, Y.F.; Tan, W.J.; Zhou, Y.; Chen, Q.F.; Qi, H.; Li, J.F.; et al. MYB30 integrates light signals with antioxidant biosynthesis to regulate plant responses during postsubmergence recovery. New Phytol. 2023, 237, 2238–2254. [Google Scholar] [CrossRef] [PubMed]
- Song, R.-F.; Li, T.-T.; Liu, W.-C. Jasmonic acid impairs Arabidopsis seedling salt stress tolerance through MYC2-mediated repression of CAT2 expression. Front. Plant Sci. 2021, 12, 730228. [Google Scholar] [CrossRef] [PubMed]
- Verma, D.; Jalmi, S.K.; Bhagat, P.K.; Verma, N.; Sinha, A.K. A bHLH transcription factor, MYC2, imparts salt intolerance by regulating proline biosynthesis in Arabidopsis. FEBS J. 2020, 287, 2560–2576. [Google Scholar] [CrossRef]
- Wang, X.Y.; Zhu, N.N.; Yang, J.S.; Zhou, D.; Yuan, S.T.; Pan, X.J.; Jiang, C.X.; Wu, Z.G. CwJAZ4/9 negatively regulates jasmonate-mediated biosynthesis of terpenoids through interacting with CwMYC2 and confers salt tolerance in Curcuma wenyujin. Plant Cell Environ. 2024, 47, 3090–3110. [Google Scholar] [CrossRef]
- Deng, H.; Li, Q.; Cao, R.; Ren, Y.; Wang, G.; Guo, H.; Bu, S.; Liu, J.; Ma, P. Overexpression of SmMYC2 enhances salt resistance in Arabidopsis thaliana and Salvia miltiorrhiza hairy roots. J. Plant Physiol. 2023, 280, 153862. [Google Scholar] [CrossRef]
- Xiao, G.L.; Jiang, Z.N.; Xing, T.; Chen, Y.; Zhang, H.J.; Qian, J.J.; Wang, X.T.; Wang, Y.X.; Xia, G.M.; Wang, M.C. Small ubiquitin-like modifier protease gene TaDSU enhances salt tolerance of wheat. New Phytol. 2024, 245, 2540–2552. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Huang, D.M.; Lin, X.F. Interlinked regulator loops of ABA and JA respond to salt and drought stress in Caragana korshinskii. Environ. Exp. Bot. 2024, 225, 105829. [Google Scholar] [CrossRef]
- Liu, H.; Cui, P.; Zhang, B.; Zhu, J.; Liu, C.; Li, Q. Binding of the transcription factor MYC2-like to the ABRE of the OsCYP2 promoter enhances salt tolerance in Oryza sativa. PLoS ONE 2022, 17, 0276075. [Google Scholar] [CrossRef]
- Cao, L.; Liu, L.; Zhang, C.; Ren, W.; Zheng, J.; Tao, C.; Zhu, W.; Xiang, M.; Wang, L.; Liu, Y.; et al. The MYC2 and MYB43 transcription factors cooperate to repress HMA2 and HMA4 expression, altering cadmium tolerance in Arabidopsis thaliana. J. Hazard. Mater. 2024, 479, 135703. [Google Scholar] [CrossRef]
- Du, X.; Fang, L.; Li, J.; Chen, T.; Cheng, Z.; Zhu, B.; Gu, L.; Wang, H. The TabHLH094–TaMYC8 complex mediates the cadmium response in wheat. Mol. Breed. 2023, 43, 57. [Google Scholar] [CrossRef] [PubMed]
- Hua, B.; Chang, J.; Wu, M.; Xu, Z.; Zhang, F.; Yang, M.; Xu, H.; Wang, L.J.; Chen, X.Y.; Wu, S. Mediation of JA signalling in glandular trichomes by the woolly/SlMYC1 regulatory module improves pest resistance in tomato. Plant Biotechnol. J. 2021, 19, 375–393. [Google Scholar] [CrossRef]
- Guan, Y.Q.; Jiang, L.; Wang, Y.; Liu, G.H.; Wu, J.Y.; Luo, H.; Chen, S.M.; Chen, F.D.; Niinemets, U.; Chen, F.; et al. CmMYC2-CmMYBML1 module orchestrates the resistance to herbivory by synchronously regulating the trichome development and constitutive terpene biosynthesis in Chrysanthemum. New Phytol. 2024, 244, 914–933. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.K.; Nie, G.B.; Lin, J.L.; Huang, J.F.; Guo, X.X.; Chen, M.; Fang, X.; Mao, Y.B.; Li, Y.; Wang, L.J.; et al. Regulation of glandular size and phytoalexin biosynthesis by a negative feedback loop in cotton. Adv. Sci. 2024, 11, 2403059. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Wang, Y.X.; Liu, T.; Luo, X.C.; Wang, Y.; Chu, L.Y.; Li, J.P.; An, H.L.; Wan, P.; Xu, D.; et al. GhMYC1374 regulates the cotton defense response to cotton aphids by mediating the production of flavonoids and free gossypol. Plant Physiol. Biochem. 2023, 205, 108162. [Google Scholar] [CrossRef]
- Sun, A.; Yu, B.; Zhang, Q.; Peng, Y.; Yang, J.; Sun, Y.; Qin, P.; Jia, T.; Smeekens, S.; Teng, S. MYC2-activated TRICHOME BIREFRINGENCE-LIKE37 acetylates cell walls and enhances herbivore resistance. Plant Physiol. 2020, 184, 1083–1096. [Google Scholar] [CrossRef]
- Jiang, X.; Yang, Y.; Guo, S.-x.; Wu, Y.-c.; Li, Z.-a. Study of BnMYC2 cloning and resistance to insect stress. Acta Bot. Boreali-Occident. Sin. 2022, 42, 201–209. [Google Scholar] [CrossRef]
- Cao, Y.Y.; Liu, L.; Ma, K.S.; Wang, W.J.; Lv, H.M.; Gao, M.; Wang, X.M.; Zhang, X.C.; Ren, S.X.; Zhang, N.; et al. The jasmonate-induced bHLH gene SlJIG functions in terpene biosynthesis and resistance to insects and fungus. J. Integr. Plant Biol. 2022, 64, 1102–1115. [Google Scholar] [CrossRef]
- Chen, Y.M.; Jin, G.C.; Liu, M.Y.; Wang, L.L.; Lou, Y.G.; Baldwin, I.; Li, R. Multiomic analyses reveal key sectors of jasmonate-mediated defense responses in rice. Plant Cell 2024, 36, 3362–3377. [Google Scholar] [CrossRef]
- Wu, S.F.; Hu, C.Y.; Zhu, C.A.; Fan, Y.F.; Zhou, J.; Xia, X.J.; Shi, K.; Zhou, Y.H.; Foyer, C.H.; Yu, J.Q. The MYC2-PUB22-JAZ4 module plays a crucial role in jasmonate signaling in tomato. Mol. Plant 2024, 17, 598–613. [Google Scholar] [CrossRef]
- Lv, W.Y.; Jiang, H.; Cao, Q.H.; Ren, H.Z.; Wang, X.C.; Wang, Y.C. A tau class glutathione S-transferase in tea plant, CsGSTU45, facilitates tea plant susceptibility to Colletotrichum camelliae infection mediated by the jasmonate signaling pathway. Plant J. 2024, 117, 1356–1376. [Google Scholar] [CrossRef]
- Cao, H.Z.; Zhang, K.; Li, W.; Pang, X.; Liu, P.F.; Si, H.L.; Zang, J.P.; Xing, J.H.; Dong, J.G. ZmMYC7 directly regulates ZmERF147 to increase maize resistance to Fusarium graminearum. Crop J. 2023, 11, 79–88. [Google Scholar] [CrossRef]
- Ma, C.R.; Li, R.Y.; Sun, Y.; Zhang, M.; Li, S.; Xu, Y.X.; Song, J.; Li, J.; Qi, J.F.; Wang, L.; et al. ZmMYC2s play important roles in maize responses to simulated herbivory and jasmonate. J. Integr. Plant Biol. 2023, 65, 1041–1058. [Google Scholar] [CrossRef] [PubMed]
- Chen, R.; Jiang, H.; Li, L.; Zhai, Q.; Qi, L.; Zhou, W.; Liu, X.; Li, H.; Zheng, W.; Sun, J.; et al. The Arabidopsis mediator subunit MED25 differentially regulates jasmonate and abscisic acid signaling through interacting with the MYC2 and ABI5 transcription factors. Plant Cell 2012, 24, 2898–2916. [Google Scholar] [CrossRef] [PubMed]
- Verma, D.; Bhagat, P.K.; Sinha, A.K. MKK3-MPK6-MYC2 module positively regulates ABA biosynthesis and signalling in Arabidopsis. J. Plant Biochem. Biotechnol. 2020, 29, 785–795. [Google Scholar] [CrossRef]
- Nomoto, M.; Skelly, M.J.; Itaya, T.; Mori, T.; Suzuki, T.; Matsushita, T.; Tokizawa, M.; Kuwata, K.; Mori, H.; Yamamoto, Y.Y. Suppression of MYC transcription activators by the immune cofactor NPR1 fine-tunes plant immune responses. Cell Rep. 2021, 37, 110125. [Google Scholar] [CrossRef]
- Gautam, J.K.; Giri, M.K.; Singh, D.; Chattopadhyay, S.; Nandi, A.K. MYC2 influences salicylic acid biosynthesis and defense against bacterial pathogens in Arabidopsis thaliana. Physiol. Plant. 2021, 173, 2248–2261. [Google Scholar] [CrossRef]
- Liu, Y.X.; Han, W.H.; Wang, J.X.; Zhang, F.B.; Ji, S.X.; Zhong, Y.W.; Liu, S.S.; Wang, X.W. Differential induction of JA/SA determines plant defense against successive leaf-chewing and phloem-feeding insects. J. Pest Sci. 2024, 1–16. [Google Scholar] [CrossRef]
- Jin, G.; Qi, J.; Zu, H.; Liu, S.; Gershenzon, J.; Lou, Y.; Baldwin, I.T.; Li, R. Jasmonate-mediated gibberellin catabolism constrains growth during herbivore attack in rice. Plant Cell 2023, 35, 3828–3844. [Google Scholar] [CrossRef]
- Pérez-Llorca, M.; Pollmann, S.; Müller, M. Ethylene and jasmonates signaling network mediating secondary metabolites under abiotic stress. Int. J. Mol. Sci. 2023, 24, 5990. [Google Scholar] [CrossRef]
- Aerts, N.; Pereira Mendes, M.; Van Wees, S.C. Multiple levels of crosstalk in hormone networks regulating plant defense. Plant J. 2021, 105, 489–504. [Google Scholar] [CrossRef] [PubMed]
- Shi, R.X.; Yu, J.L.; Chang, X.R.; Qiao, L.P.; Liu, X.; Lu, L.F. Recent advances in research into jasmonate biosynthesis and signaling pathways in agricultural crops and products. Processes 2023, 11, 736. [Google Scholar] [CrossRef]
- Liu, Y.; Du, M.; Deng, L.; Shen, J.; Fang, M.; Chen, Q.; Lu, Y.; Wang, Q.; Li, C.; Zhai, Q. MYC2 Regulates the Termination of Jasmonate Signaling via an Autoregulatory Negative Feedback Loop. Plant Cell 2019, 31, 106–127. [Google Scholar] [CrossRef] [PubMed]
- Fukazawa, J.; Mori, K.; Ando, H.; Mori, R.; Kanno, Y.; Seo, M.; Takahashi, Y. Jasmonate inhibits plant growth and reduces gibberellin levels via microRNA5998 and transcription factor MYC2. Plant Physiol. 2023, 193, 2197–2214. [Google Scholar] [CrossRef] [PubMed]
- Guo, Q.; Major, I.T.; Kapali, G.; Howe, G.A. MYC transcription factors coordinate tryptophan-dependent defence responses and compromise seed yield in Arabidopsis. New Phytol. 2022, 236, 132–145. [Google Scholar] [CrossRef]
- Gao, C.; Qi, S.; Liu, K.; Li, D.; Jin, C.; Li, Z.; Huang, G.; Hai, J.; Zhang, M.; Chen, M. MYC2, MYC3, and MYC4 function redundantly in seed storage protein accumulation in Arabidopsis. Plant Physiol. Biochem. 2016, 108, 63–70. [Google Scholar] [CrossRef]
- Hu, S.; Yang, H.; Gao, H.; Yan, J.; Xie, D. Control of seed size by jasmonate. Sci. China Life Sci. 2021, 64, 1215–1226. [Google Scholar] [CrossRef]
- Liu, Z.; Li, N.; Zhang, Y.; Li, Y. Transcriptional repression of GIF1 by the KIX-PPD-MYC repressor complex controls seed size in Arabidopsis. Nat. Commun. 2020, 11, 1846. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zeng, T.; Su, H.; Wang, M.; He, J.; Gu, L.; Wang, H.; Du, X.; Wang, C.; Zhu, B. The Role of MYC2 Transcription Factors in Plant Secondary Metabolism and Stress Response Mechanisms. Plants 2025, 14, 1255. https://doi.org/10.3390/plants14081255
Zeng T, Su H, Wang M, He J, Gu L, Wang H, Du X, Wang C, Zhu B. The Role of MYC2 Transcription Factors in Plant Secondary Metabolism and Stress Response Mechanisms. Plants. 2025; 14(8):1255. https://doi.org/10.3390/plants14081255
Chicago/Turabian StyleZeng, Tuo, Han Su, Meiyang Wang, Jiefang He, Lei Gu, Hongcheng Wang, Xuye Du, Caiyun Wang, and Bin Zhu. 2025. "The Role of MYC2 Transcription Factors in Plant Secondary Metabolism and Stress Response Mechanisms" Plants 14, no. 8: 1255. https://doi.org/10.3390/plants14081255
APA StyleZeng, T., Su, H., Wang, M., He, J., Gu, L., Wang, H., Du, X., Wang, C., & Zhu, B. (2025). The Role of MYC2 Transcription Factors in Plant Secondary Metabolism and Stress Response Mechanisms. Plants, 14(8), 1255. https://doi.org/10.3390/plants14081255






