Analysis of the Genes from Gibberellin, Jasmonate, and Auxin Signaling Under Drought Stress: A Genome-Wide Approach in Castor Bean (Ricinus communis L.)
Abstract
1. Introduction
2. Results
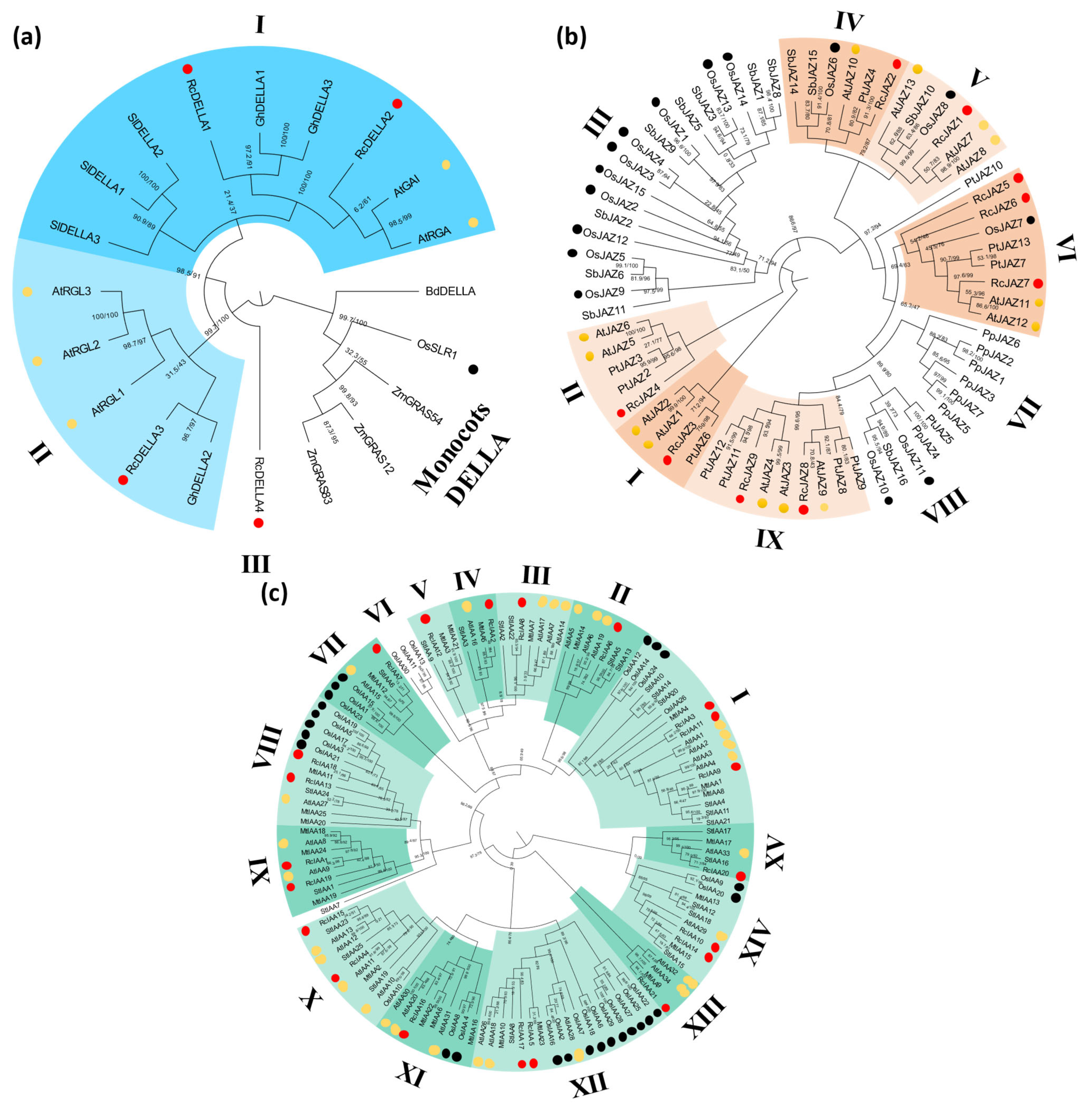
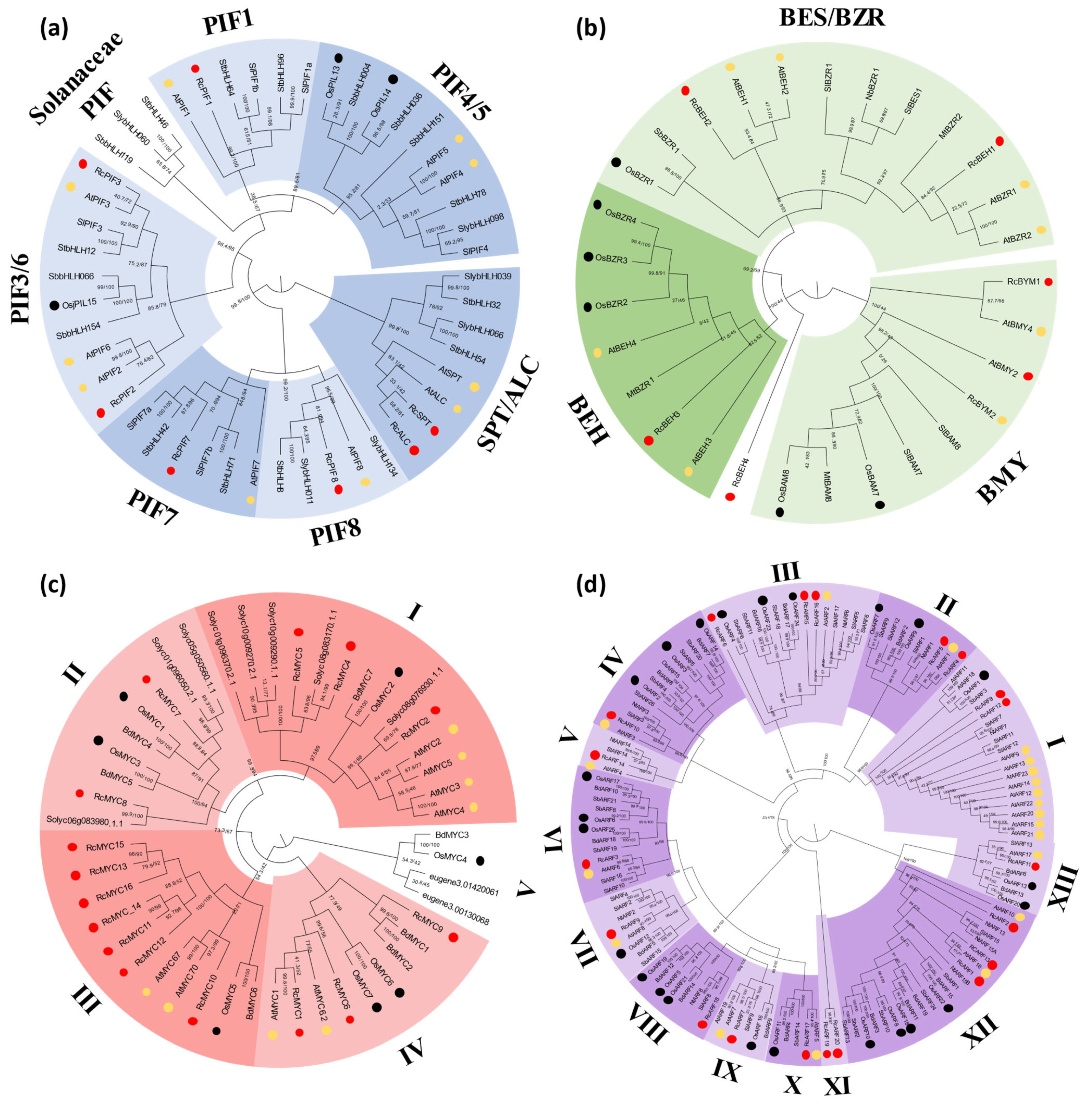
2.1. Genome-Wide Identification and Phylogenetic Analysis of GAs, JA, and Aux Signaling Genes
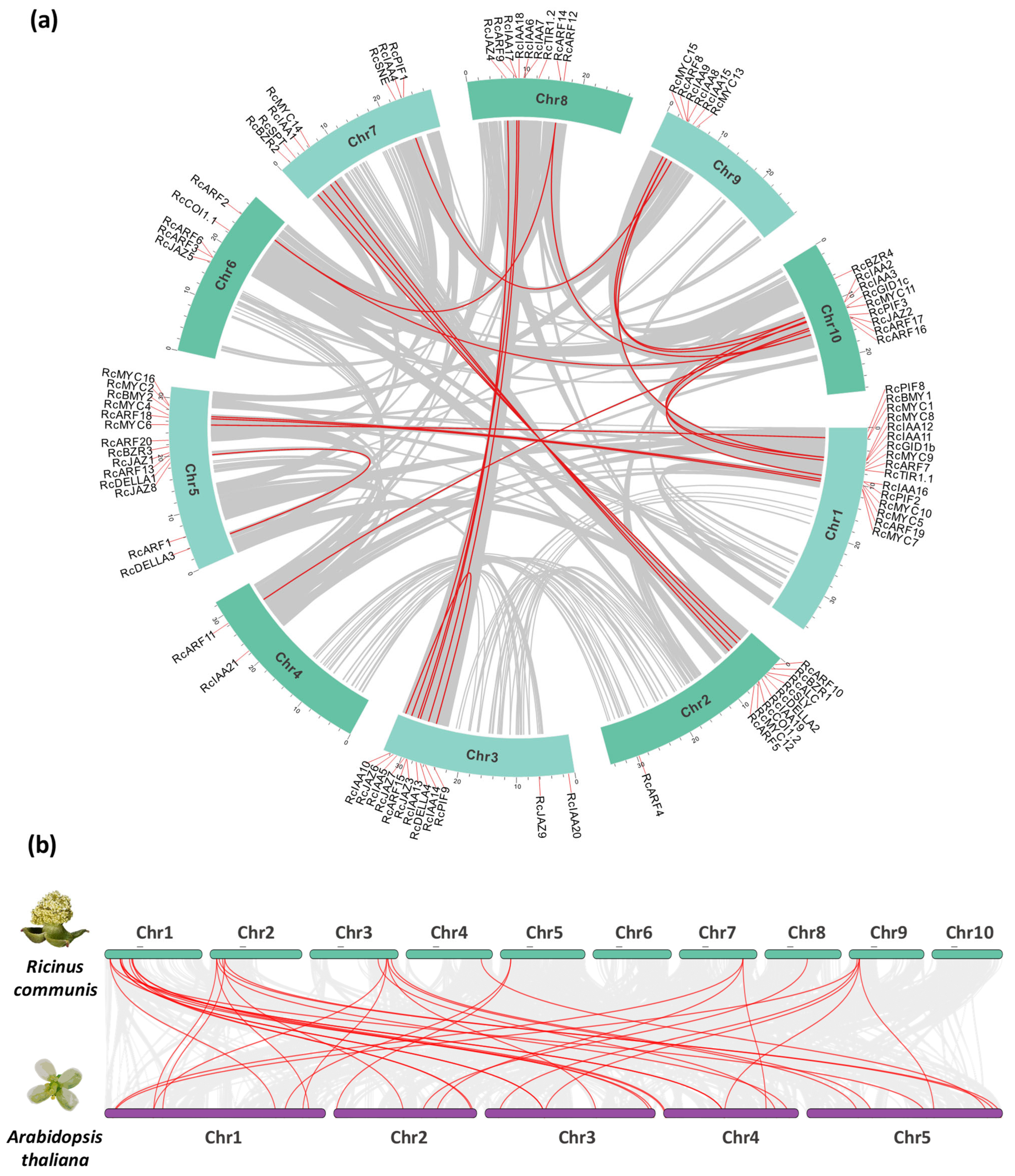
2.2. Chromosomal Positions, Synteny, Collinearity, and Duplication Analysis
2.3. Gene Structure and Distribution of Conserved Motifs
2.4. Cis-Regulatory Elements and microRNAs Predicted to Target the Identified Genes
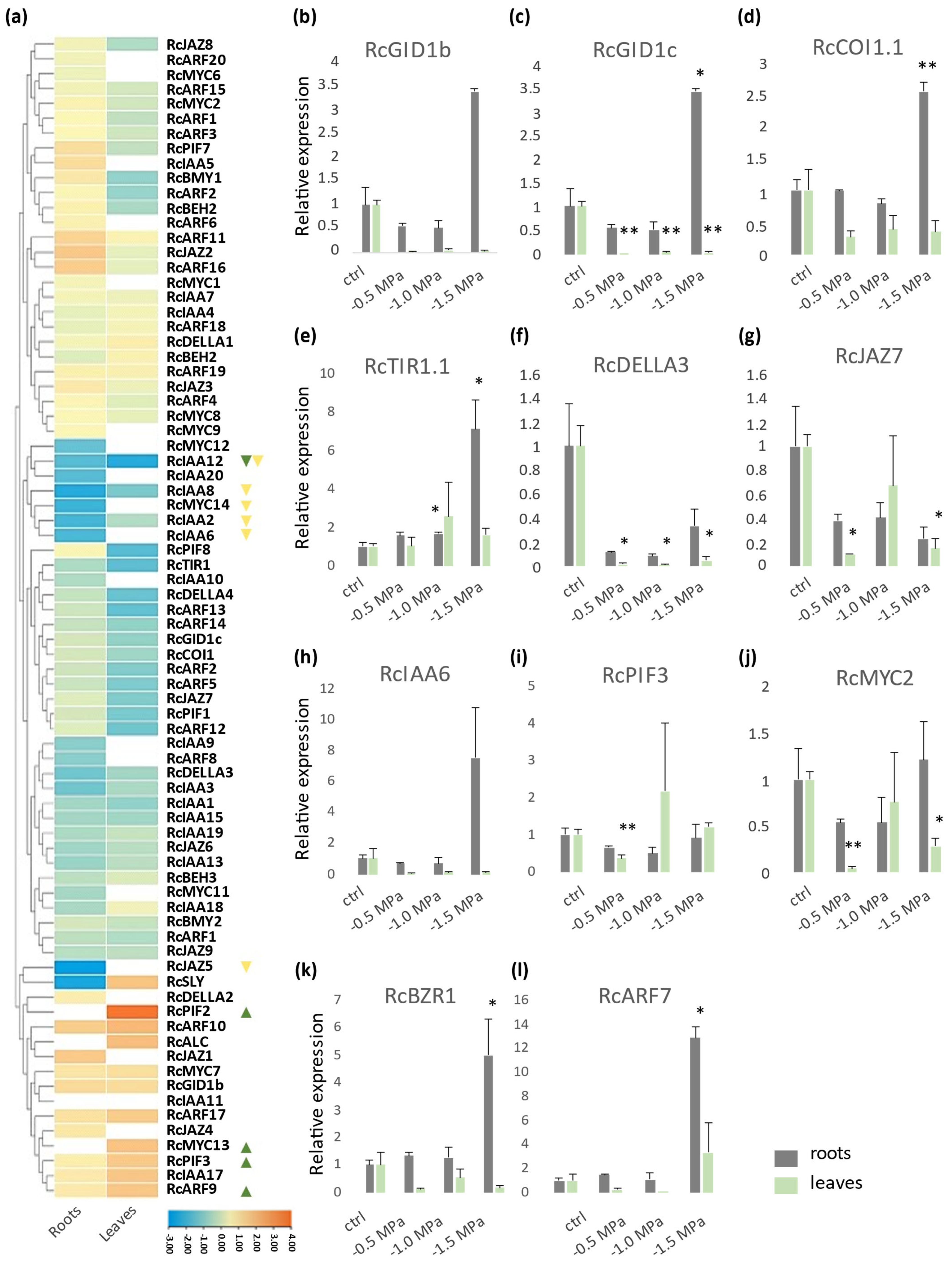
2.5. Expression Profiles of Castor Bean GA, JA, and Aux Signaling Genes Under Drought Stress
3. Discussion
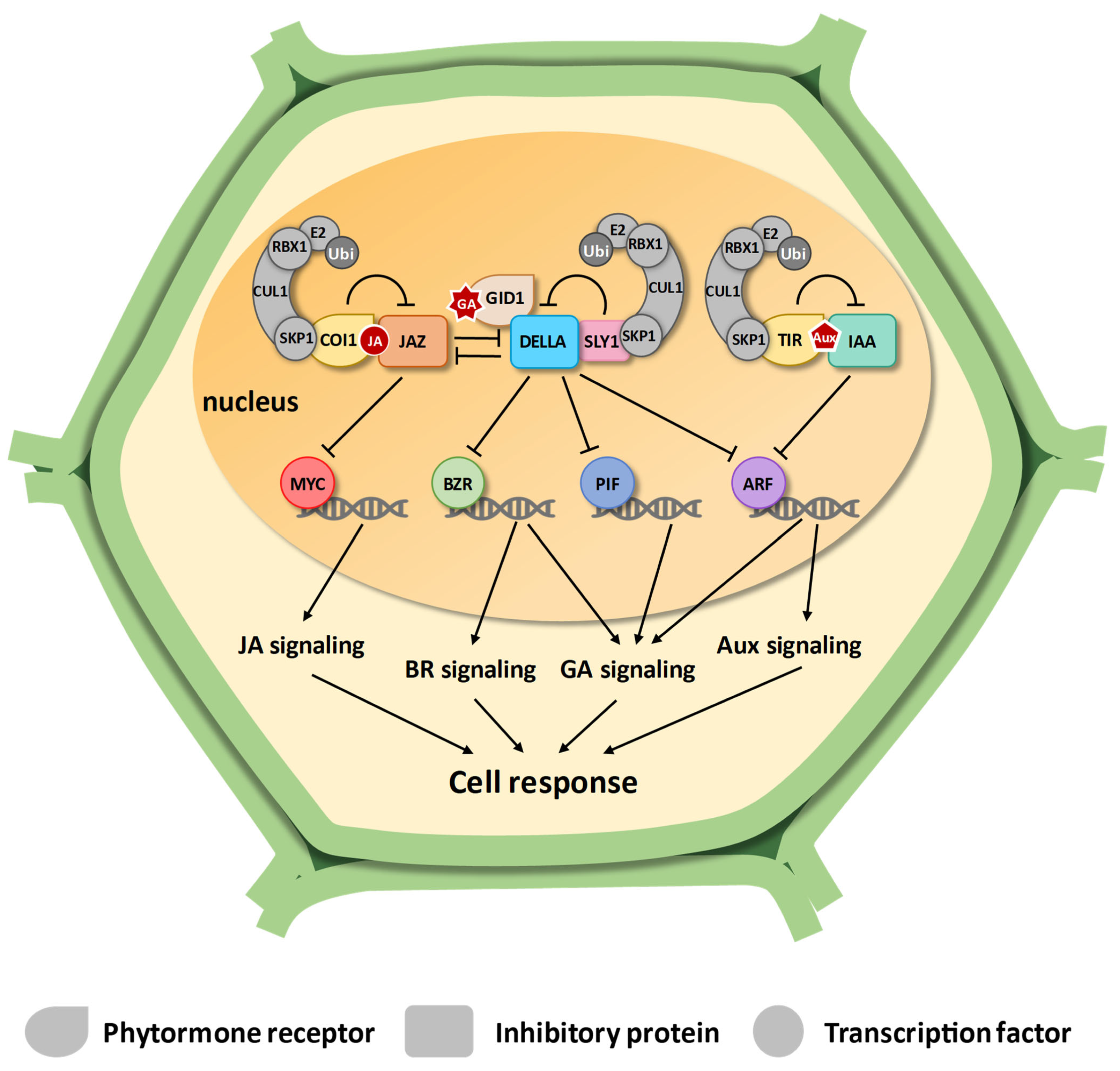
3.1. Gibberellin Signaling
3.2. Jasmonate Signaling
3.3. Auxin Signaling
3.4. Potential Regulation of Drought-Responsive Genes by miRNAs
4. Conclusions
5. Materials and Methods
5.1. Gene Identification
5.2. Phylogenetic Analysis
5.3. Gene Structure, Chromosomal Positions, and Gene Duplications Analysis
5.4. Prediction of Cis-Regulatory Elements
5.5. Identification of miRNA Targets
5.6. Protein Analysis In Silico
5.7. Plant Material and RNA-Seq Analysis
5.8. Reverse Transcriptase and Quantitative PCR (RT-qPCR) Analysis
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- FAO. The State of the World’s Land and Water Resources for Food and Agriculture—Systems at Breaking Point (SOLAW 2021); FAO: Rome, Italy, 2021; ISBN 978-92-5-135327-1. [Google Scholar]
- Gupta, A.; Rico-Medina, A.; Caño-Delgado, A.I. The Physiology of Plant Responses to Drought. Science 2020, 368, 266–269. [Google Scholar] [CrossRef] [PubMed]
- Calvin, K.; Dasgupta, D.; Krinner, G.; Mukherji, A.; Thorne, P.W.; Trisos, C.; Romero, J.; Aldunce, P.; Barrett, K.; Blanco, G.; et al. IPCC, 2023: Climate Change 2023: Synthesis Report. Contribution of Working Groups I, II and III to the Sixth Assessment Report of the Intergovernmental Panel on Climate Change; IPCC: Geneva, Switzerland, 2023. [Google Scholar] [CrossRef]
- Salvi, P.; Manna, M.; Kaur, H.; Thakur, T.; Gandass, N.; Bhatt, D.; Muthamilarasan, M. Phytohormone Signaling and Crosstalk in Regulating Drought Stress Response in Plants. Plant Cell Rep. 2021, 40, 1305–1329. [Google Scholar] [CrossRef]
- Browse, J.; Wallis, J.G. Arabidopsis Flowers Unlocked the Mechanism of Jasmonate Signaling. Plants 2019, 8, 285. [Google Scholar] [CrossRef] [PubMed]
- Nelson, S.K.; Steber, C.M. Transcriptional Mechanisms Associated with Seed Dormancy and Dormancy Loss in the Gibberellin-Insensitive Sly1-2 Mutant of Arabidopsis thaliana. PLoS ONE 2017, 12, e0179143. [Google Scholar] [CrossRef]
- Yang, D.L.; Yao, J.; Mei, C.S.; Tong, X.H.; Zeng, L.J.; Li, Q.; Xiao, L.T.; Sun, T.P.; Li, J.; Deng, X.W.; et al. Plant Hormone Jasmonate Prioritizes Defense over Growth by Interfering with Gibberellin Signaling Cascade. Proc. Natl. Acad. Sci. USA 2012, 109, E1192–E1200. [Google Scholar] [CrossRef] [PubMed]
- Jardim-Messeder, D.; Cassol, D.; Souza-Vieira, Y.; Ehlers Loureiro, M.; Girke, T.; Boroni, M.; Lopes Corrêa, R.; Coelho, A.; Sachetto-Martins, G. Genome-Wide Identification of Core Components of ABA Signaling and Transcriptome Analysis Reveals Gene Circuits Involved in Castor Bean (Ricinus communis L.) Response to Drought. Gene 2023, 883, 147668. [Google Scholar] [CrossRef]
- Jardim-Messeder, D.; de Souza-Vieira, Y.; Sachetto-Martins, G. Dressed Up to the Nines: The Interplay of Phytohormones Signaling and Redox Metabolism During Plant Response to Drought. Plants 2025, 14, 208. [Google Scholar] [CrossRef]
- Olszewski, N.; Sun, T.P.; Gubler, F. Gibberellin Signaling: Biosynthesis, Catabolism, and Response Pathways. Plant Cell 2002, 14, S61–S80. [Google Scholar] [CrossRef]
- Gazara, R.K.; Moharana, K.C.; Bellieny-Rabelo, D.; Venancio, T.M. Expansion and Diversification of the Gibberellin Receptor GIBBERELLIN INSENSITIVE DWARF1 (GID1) Family in Land Plants. Plant Mol. Biol. 2018, 97, 435–449. [Google Scholar] [CrossRef]
- Murase, K.; Hirano, Y.; Sun, T.P.; Hakoshima, T. Gibberellin-Induced DELLA Recognition by the Gibberellin Receptor GID1. Nature 2008, 456, 459–463. [Google Scholar] [CrossRef]
- Ariizumi, T.; Lawrence, P.K.; Steber, C.M. The Role of Two F-Box Proteins, SLEEPY1 and SNEEZY, in Arabidopsis Gibberellin Signaling. Plant Physiol. 2011, 155, 765–775. [Google Scholar] [CrossRef] [PubMed]
- Hartweck, L.M. Gibberellin Signaling. Planta 2008, 229, 1–13. [Google Scholar] [CrossRef]
- Liu, K.; Li, Y.; Chen, X.; Li, L.; Liu, K.; Zhao, H.; Wang, Y.; Han, S. ERF72 Interacts with ARF6 and BZR1 to Regulate Hypocotyl Elongation in Arabidopsis. J. Exp. Bot. 2018, 69, 3933–3947. [Google Scholar] [CrossRef]
- Colebrook, E.H.; Thomas, S.G.; Phillips, A.L.; Hedden, P. The Role of Gibberellin Signalling in Plant Responses to Abiotic Stress. J. Exp. Biol. 2014, 217, 67–75. [Google Scholar] [CrossRef]
- Jogawat, A. Crosstalk Among Phytohormone Signaling Pathways During Abiotic Stress. In Molecular Plant Abiotic Stress: Biology and Biotechnology; Wiley: Hoboken, NJ, USA, 2019; pp. 209–220. [Google Scholar]
- Lee, H.Y.; Seo, J.S.; Cho, J.H.; Jung, H.; Kim, J.K.; Lee, J.S.; Rhee, S.; Do Choi, Y. Oryza Sativa COI Homologues Restore Jasmonate Signal Transduction in Arabidopsis Coi1-1 Mutants. PLoS ONE 2013, 8, e0052802. [Google Scholar] [CrossRef]
- Sheard, L.B.; Tan, X.; Mao, H.; Withers, J.; Ben-Nissan, G.; Hinds, T.R.; Kobayashi, Y.; Hsu, F.F.; Sharon, M.; Browse, J.; et al. Jasmonate Perception by Inositol-Phosphate-Potentiated COI1-JAZ Co-Receptor. Nature 2010, 468, 400–407. [Google Scholar] [CrossRef] [PubMed]
- Yan, J.; Yao, R.; Chen, L.; Li, S.; Gu, M.; Nan, F.; Xie, D. Dynamic Perception of Jasmonates by the F-Box Protein COI1. Mol. Plant 2018, 11, 1237–1247. [Google Scholar] [CrossRef] [PubMed]
- Major, I.T.; Yoshida, Y.; Campos, M.L.; Kapali, G.; Xin, X.F.; Sugimoto, K.; de Oliveira Ferreira, D.; He, S.Y.; Howe, G.A. Regulation of Growth–Defense Balance by the JASMONATE ZIM-DOMAIN (JAZ)-MYC Transcriptional Module. New Phytol. 2017, 215, 1533–1547. [Google Scholar] [CrossRef]
- Ishiguro, S.; Kawai-Oda, A.; Ueda, J.; Nishida, I.; Okada, K. The Defective in Anther Dehiscence1 gene Encodes a Novel Phospholipase A1 Catalyzing the Initial Step of Jasmonic Acid Biosynthesis, Which Synchronizes Pollen Maturation, Anther Dehiscence, and Flower Opening in Arabidopsis. Plant Cell 2001, 13, 2191–2209. [Google Scholar] [CrossRef]
- Howe, G.A.; Major, I.T.; Koo, A.J. Modularity in Jasmonate Signaling for Multistress Resilience. Annu. Rev. Plant Biol. 2018, 8, 25–45. [Google Scholar] [CrossRef]
- Kazan, K.; Manners, J.M. The Interplay between Light and Jasmonate Signalling during Defence and Development. J. Exp. Bot. 2011, 62, 4087–4100. [Google Scholar] [CrossRef]
- Hou, X.; Lee, L.Y.C.; Xia, K.; Yan, Y.; Yu, H. DELLAs Modulate Jasmonate Signaling via Competitive Binding to JAZs. Dev. Cell 2010, 19, 884–894. [Google Scholar] [CrossRef] [PubMed]
- Lavy, M.; Estelle, M. Mechanisms of Auxin Signaling. Dev. Camb. 2016, 143, 3226–3229. [Google Scholar] [CrossRef]
- Takatsuka, H.; Umeda, M. Hormonal Control of Cell Division and Elongation along Differentiation Trajectories in Roots. J. Exp. Bot. 2014, 65, 2633–2643. [Google Scholar] [CrossRef] [PubMed]
- Sharma, E.; Sharma, R.; Borah, P.; Jain, M.; Khurana, J.P. Emerging Roles of Auxin in Abiotic Stress Responses. In Elucidation of Abiotic Stress Signaling in Plants: Functional Genomics Perspectives; Springer: New York, NY, USA, 2015; Volume 1, pp. 299–328. [Google Scholar]
- Wolf, J. Oilseed Crops, 2nd ed.; Weiss, E.A., Ed.; Blackwell Science: Oxford, UK, 2000. [Google Scholar]
- Audran-Delalande, C.; Bassa, C.; Mila, I.; Regad, F.; Zouine, M.; Bouzayen, M. Genome-Wide Identification, Functional Analysis and Expression Profiling of the Aux/IAA Gene Family in Tomato. Plant Cell Physiol. 2012, 53, 659–672. [Google Scholar] [CrossRef] [PubMed]
- Dreher, K.A.; Brown, J.; Saw, R.E.; Callis, J. The Arabidopsis Aux/IAA Protein Family Has Diversified in Degradation and Auxin Responsiveness. Plant Cell 2006, 18, 699–714. [Google Scholar] [CrossRef]
- Guo, Y.; Wu, H.; Li, X.; Li, Q.; Zhao, X.; Duan, X.; An, Y.; Lv, W.; An, H. Identification and Expression of GRAS Family Genes in Maize (Zea mays L.). PLoS ONE 2017, 12, e0185418. [Google Scholar] [CrossRef]
- Jain, M.; Kaur, N.; Garg, R.; Thakur, J.K.; Tyagi, A.K.; Khurana, J.P. Structure and Expression Analysis of Early Auxin-Responsive Aux/IAA Gene Family in Rice (Oryza sativa). Funct. Integr. Genom. 2006, 6, 47–59. [Google Scholar] [CrossRef]
- Niu, Y.; Figueroa, P.; Browse, J. Characterization of JAZ-Interacting bHLH Transcription Factors That Regulate Jasmonate Responses in Arabidopsis. J. Exp. Bot. 2011, 62, 2143–2154. [Google Scholar] [CrossRef]
- Tian, C.; Wan, P.; Sun, S.; Li, J.; Chen, M. Genome-Wide Analysis of the GRAS Gene Family in Rice and Arabidopsis. Plant Mol. Biol. 2004, 54, 519–532. [Google Scholar] [CrossRef]
- Toledo-Ortiz, G.; Huq, E.; Quail, P.H. The Arabidopsis Basic/Helix-Loop-Helix Transcription Factor Family. Plant Cell 2003, 15, 1749–1770. [Google Scholar] [CrossRef] [PubMed]
- Um, T.Y.; Lee, H.Y.; Lee, S.; Chang, S.H.; Chung, P.J.; Oh, K.B.; Kim, J.K.; Jang, G.; Choi, Y.D. Jasmonate Zim-Domain Protein 9 Interacts with Slender Rice 1 to Mediate the Antagonistic Interaction between Jasmonic and Gibberellic Acid Signals in Rice. Front. Plant Sci. 2018, 871, 1866. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Qiao, L.; Bai, J.; Wang, P.; Duan, W.; Yuan, S.; Yuan, G.; Zhang, F.; Zhang, L.; Zhao, C. Genome-Wide Characterization of JASMONATE-ZIM DOMAIN Transcription Repressors in Wheat (Triticum aestivum L.). BMC Genom. 2017, 18, 152. [Google Scholar] [CrossRef]
- Ye, M.; Luo, S.M.; Xie, J.F.; Li, Y.F.; Xu, T.; Liu, Y.; Song, Y.Y.; Zhu-Salzman, K.; Zeng, R.S. Silencing COI1 in Rice Increases Susceptibility to Chewing Insects and Impairs Inducible Defense. PLoS ONE 2012, 7, e0036214. [Google Scholar] [CrossRef]
- Zhang, Y.; Mayba, O.; Pfeiffer, A.; Shi, H.; Tepperman, J.M.; Speed, T.P.; Quail, P.H. A Quartet of PIF bHLH Factors Provides a Transcriptionally Centered Signaling Hub That Regulates Seedling Morphogenesis through Differential Expression-Patterning of Shared Target Genes in Arabidopsis. PLoS Genet. 2013, 9, e1003244. [Google Scholar] [CrossRef]
- Shrirame, H.Y.; Panwar, N.L.; Bamniya, B.R. Bio Diesel from Castor Oil—A Green Energy Option. Low Carbon Econ. 2011, 2, 1–6. [Google Scholar] [CrossRef]
- Baldwin, B.S.; Cossar, R.D. Castor Yield in Response to Planting Date at Four Locations in the South-Central United States. Ind. Crops Prod. 2009, 29, 316–319. [Google Scholar] [CrossRef]
- Feng, L.; Li, G.; He, Z.; Han, W.; Sun, J.; Huang, F.; Di, J.; Chen, Y. The ARF, GH3, and Aux/IAA Gene Families in Castor Bean (Ricinus communis L.): Genome-Wide Identification and Expression Profiles in High-Stalk and Dwarf Strains. Ind. Crops Prod. 2019, 141, 111804. [Google Scholar] [CrossRef]
- Fernández-Calvo, P.; Chini, A.; Fernández-Barbero, G.; Chico, J.M.; Gimenez-Ibanez, S.; Geerinck, J.; Eeckhout, D.; Schweizer, F.; Godoy, M.; Franco-Zorrilla, J.M.; et al. The Arabidopsis bHLH Transcription Factors MYC3 and MYC4 Are Targets of JAZ Repressors and Act Additively with MYC2 in the Activation of Jasmonate Responses. Plant Cell 2011, 23, 701–715. [Google Scholar] [CrossRef]
- Gagne, J.M.; Downes, B.P.; Shiu, S.-H.; Durski, A.M.; Vierstra, R.D. The F-Box Subunit of the SCF E3 Complex Is Encoded by a Diverse Superfamily of Genes in Arabidopsis. Proc. Natl. Acad. Sci. USA 2002, 99, 11519–11524. [Google Scholar] [CrossRef]
- Garrido-Bigotes, A.; Valenzuela-Riffo, F.; Figueroa, C.R. Evolutionary Analysis of JAZ Proteins in Plants: An Approach in Search of the Ancestral Sequence. Int. J. Mol. Sci. 2019, 20, 5060. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.H.; Kim, B.; Song, S.K.; Heo, J.O.; Yu, N.I.; Lee, S.A.; Kim, M.; Kim, D.G.; Sohn, S.O.; Lim, C.E.; et al. Large-Scale Analysis of the GRAS Gene Family in Arabidopsis thaliana. Plant Mol. Biol. 2008, 67, 659–670. [Google Scholar] [CrossRef]
- Ji, Z.; Belfield, E.J.; Zhang, S.; Bouvier, J.; Li, S.; Schnell, J.; Fu, X.; Harberd, N.P. Evolution of a Plant Growth-Regulatory Protein Interaction Specificity. Nat. Plants 2023, 9, 2059–2070. [Google Scholar] [CrossRef]
- Reinhold, H.; Soyk, S.; Šimková, K.; Hostettler, C.; Marafino, J.; Mainiero, S.; Vaughan, C.K.; Monroe, J.D.; Zeeman, S.C. β-Amylase–Like Proteins Function as Transcription Factors in Arabidopsis, Controlling Shoot Growth and Development[C][W][OA]. Plant Cell 2011, 23, 1391–1403. [Google Scholar] [CrossRef]
- Mutte, S.K.; Kato, H.; Rothfels, C.; Melkonian, M.; Wong, G.K.-S.; Weijers, D. Origin and Evolution of the Nuclear Auxin Response System. Available online: https://elifesciences.org/articles/33399/figures (accessed on 15 January 2025).
- Yoshida, H.; Tanimoto, E.; Hirai, T.; Miyanoiri, Y.; Mitani, R.; Kawamura, M.; Takeda, M.; Takehara, S.; Hirano, K.; Kainosho, M.; et al. Evolution and Diversification of the Plant Gibberellin Receptor GID1. Proc. Natl. Acad. Sci. USA 2018, 115, E7844–E7853. [Google Scholar] [CrossRef] [PubMed]
- Phokas, A.; Coates, J.C. Evolution of DELLA Function and Signaling in Land Plants. In Evolution and Development; Blackwell Publishing Inc.: Oxford, UK, 2021; Volume 23, pp. 137–154. [Google Scholar]
- Sun, T. The Molecular Mechanism and Evolution of the GA–GID1–DELLA Signaling Module in Plants. Curr. Biol. 2011, 21, R338–R345. [Google Scholar] [CrossRef] [PubMed]
- Qi, L.; Kwiatkowski, M.; Chen, H.; Hoermayer, L.; Sinclair, S.; Zou, M.; del Genio, C.I.; Kubeš, M.F.; Napier, R.; Jaworski, K.; et al. Adenylate Cyclase Activity of TIR1/AFB Auxin Receptors in Plants. Nature 2022, 611, 133–138. [Google Scholar] [CrossRef]
- Santner, A.; Estelle, M. The Ubiquitin-Proteasome System Regulates Plant Hormone Signaling. Plant J. 2010, 61, 1029–1040. [Google Scholar] [CrossRef]
- Pauwels, L.; Goossens, A. The JAZ Proteins: A Crucial Interface in the Jasmonate Signaling Cascade. Plant Cell 2011, 23, 3089–3100. [Google Scholar] [CrossRef]
- Calderón Villalobos, L.I.A.; Lee, S.; De Oliveira, C.; Ivetac, A.; Brandt, W.; Armitage, L.; Sheard, L.B.; Tan, X.; Parry, G.; Mao, H.; et al. A Combinatorial TIR1/AFB–Aux/IAA Co-Receptor System for Differential Sensing of Auxin. Nat. Chem. Biol. 2012, 8, 477–485. [Google Scholar] [CrossRef]
- Nelson, S.K.; Steber, C.M. Gibberellin Hormone Signal Perception: Down-Regulating DELLA Repressors of Plant Growth and Development. In Annual Plant Reviews: The Gibberellins; John Wiley and Sons, Ltd.: Hoboken, NJ, USA, 2016; Volume 49, pp. 153–188. [Google Scholar]
- Yang, Y.; Guang, Y.; Wang, F.; Chen, Y.; Yang, W.; Xiao, X.; Luo, S.; Zhou, Y. Characterization of Phytochrome-Interacting Factor Genes in Pepper and Functional Analysis of CaPIF8 in Cold and Salt Stress. Front. Plant Sci. 2021, 12, 746517. [Google Scholar] [CrossRef] [PubMed]
- Yin, Y.; Vafeados, D.; Tao, Y.; Yoshida, S.; Asami, T.; Chory, J. A New Class of Transcription Factors Mediates Brassinosteroid-Regulated Gene Expression in Arabidopsis. Cell 2005, 120, 249–259. [Google Scholar] [CrossRef] [PubMed]
- Song, C.; Cao, Y.; Dai, J.; Li, G.; Manzoor, M.A.; Chen, C.; Deng, H. The Multifaceted Roles of MYC2 in Plants: Toward Transcriptional Reprogramming and Stress Tolerance by Jasmonate Signaling. Front. Plant Sci. 2022, 13, 868874. [Google Scholar] [CrossRef]
- Wang, R.; Estelle, M. Diversity and Specificity: Auxin Perception and Signaling through the TIR1/AFB Pathway. Curr. Opin. Plant Biol. 2014, 21, 51–58. [Google Scholar] [CrossRef]
- Song, S.; Qi, T.; Huang, H.; Xie, D. Regulation of Stamen Development by Coordinated Actions of Jasmonate, Auxin, and Gibberellin in Arabidopsis. Mol. Plant 2013, 6, 1065–1073. [Google Scholar] [CrossRef] [PubMed]
- Bai, M.Y.; Shang, J.X.; Oh, E.; Fan, M.; Bai, Y.; Zentella, R.; Sun, T.P.; Wang, Z.Y. Brassinosteroid, Gibberellin and Phytochrome Impinge on a Common Transcription Module in Arabidopsis. Nat. Cell Biol. 2012, 14, 810–817. [Google Scholar] [CrossRef]
- Ahmad, H.M.; Wang, X.; Ijaz, M.; Mahmood-Ur-Rahman; Oranab, S.; Ali, M.A.; Fiaz, S. Molecular Aspects of MicroRNAs and Phytohormonal Signaling in Response to Drought Stress: A Review. Curr. Issues Mol. Biol. 2022, 44, 3695–3710. [Google Scholar] [CrossRef]
- Jardim-Messeder, D.; de Souza-Vieira, Y.; Lavaquial, L.C.; Cassol, D.; Galhego, V.; Bastos, G.A.; Felix-Cordeiro, T.; Corrêa, R.L.; Zámocký, M.; Margis-Pinheiro, M.; et al. Ascorbate-Glutathione Cycle Genes Families in Euphorbiaceae: Characterization and Evolutionary Analysis. Biology 2022, 12, 19. [Google Scholar] [CrossRef]
- de Souza-Vieira, Y.; Felix-Mendes, E.; Galhego, V.; Bastos, G.A.; Felix-Cordeiro, T.; Ding, X.; Zhang, Y.; Corrêa, R.L.; Wang, X.; Sachetto-Martins, G.; et al. Euphorbiaceae Superoxide Dismutase, Catalase, and Glutathione Peroxidase as Clues to Better Comprehend High Drought Tolerance in Castor Bean. Ind. Crops Prod. 2024, 222, 119510. [Google Scholar] [CrossRef]
- Illouz-Eliaz, N.; Nissan, I.; Nir, I.; Ramon, U.; Shohat, H.; Weiss, D. Mutations in the Tomato Gibberellin Receptors Suppress Xylem Proliferation and Reduce Water Loss under Water-Deficit Conditions. J. Exp. Bot. 2020, 71, 3603–3612. [Google Scholar] [CrossRef]
- Xu, W.; Chen, Z.; Ahmed, N.; Han, B.; Cui, Q.; Liu, A. Genome-Wide Identification, Evolutionary Analysis, and Stress Responses of the GRAS Gene Family in Castor Beans. Int. J. Mol. Sci. 2016, 17, 1004. [Google Scholar] [CrossRef] [PubMed]
- Yasumura, Y.; Crumpton-Taylor, M.; Fuentes, S.; Harberd, N.P. Step-by-Step Acquisition of the Gibberellin-DELLA Growth-Regulatory Mechanism during Land-Plant Evolution. Curr. Biol. 2007, 17, 1225–1230. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Tian, H.; Park, J.; Oh, D.H.; Hu, J.; Zentella, R.; Qiao, H.; Dassanayake, M.; Sun, T.P. The Master Growth Regulator DELLA Binding to Histone H2A Is Essential for DELLA-Mediated Global Transcription Regulation. Nat. Plants 2023, 9, 1291–1305. [Google Scholar] [CrossRef] [PubMed]
- Saud, S.; Shi, Z.; Xiong, L.; Danish, S.; Datta, R.; Ahmad, I.; Fahad, S.; Banout, J. Recognizing the Basics of Phytochrome-Interacting Factors in Plants for Abiotic Stress Tolerance. Plant Stress 2022, 3, 100050. [Google Scholar] [CrossRef]
- Cordeiro, A.M.; Andrade, L.; Monteiro, C.C.; Leitão, G.; Wigge, P.A.; Saibo, N.J.M. Phytochrome-Interacting Factors: A Promising Tool to Improve Crop Productivity. J. Exp. Bot. 2022, 73, 3881–3897. [Google Scholar] [CrossRef]
- Wang, X.R.; Wang, Y.H.; Jia, M.; Zhang, R.R.; Liu, H.; Xu, Z.S.; Xiong, A.S. The Phytochrome-Interacting Factor DcPIF3 of Carrot Plays a Positive Role in Drought Stress by Increasing Endogenous ABA Level in Arabidopsis. Plant Sci. 2022, 322, 111367. [Google Scholar] [CrossRef]
- Kudo, M.; Kidokoro, S.; Yoshida, T.; Mizoi, J.; Todaka, D.; Fernie, A.R.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Double Overexpression of DREB and PIF Transcription Factors Improves Drought Stress Tolerance and Cell Elongation in Transgenic Plants. Plant Biotechnol. J. 2017, 15, 458–471. [Google Scholar] [CrossRef]
- Liu, S.; Zhang, Y.; Pan, X.; Li, B.; Yang, Q.; Yang, C.; Zhang, J.; Wu, F.; Yang, A.; Li, Y. PIF1, a Phytochrome-Interacting Factor Negatively Regulates Drought Tolerance and Carotenoids Biosynthesis in Tobacco. Int. J. Biol. Macromol. 2023, 247, 125693. [Google Scholar] [CrossRef]
- Cui, X.Y.; Gao, Y.; Guo, J.; Yu, T.F.; Zheng, W.J.; Liu, Y.W.; Chen, J.; Xu, Z.S.; Ma, Y.Z. BES/BZR Transcription Factor TaBZR2 Positively Regulates Drought Responses by Activation of TaGST1. Plant Physiol. 2019, 180, 605–620. [Google Scholar] [CrossRef]
- Ye, H.; Liu, S.; Tang, B.; Chen, J.; Xie, Z.; Nolan, T.M.; Jiang, H.; Guo, H.; Lin, H.Y.; Li, L.; et al. RD26 Mediates Crosstalk between Drought and Brassinosteroid Signalling Pathways. Nat. Commun. 2017, 8, 14573. [Google Scholar] [CrossRef]
- Wang, L.; Lin, M.; Zou, L.; Zhang, S.; Lan, Y.; Yan, H.; Xiang, Y. Comprehensive Investigation of BZR Gene Family in Four Dicots and the Function of PtBZR9 and PtBZR12 under Drought Stress. Plant Physiol. Biochem. 2024, 207, 108360. [Google Scholar] [CrossRef] [PubMed]
- Thines, B.; Katsir, L.; Melotto, M.; Niu, Y.; Mandaokar, A.; Liu, G.; Nomura, K.; He, S.Y.; Howe, G.A.; Browse, J. JAZ Repressor Proteins Are Targets of the SCFCOI1 Complex during Jasmonate Signalling. Nature 2007, 448, 661–665. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.M.; To, T.K.; Matsui, A.; Tanoi, K.; Kobayashi, N.I.; Matsuda, F.; Habu, Y.; Ogawa, D.; Sakamoto, T.; Matsunaga, S.; et al. Acetate-Mediated Novel Survival Strategy against Drought in Plants. Nat. Plants 2017, 3, 17097. [Google Scholar] [CrossRef]
- Waadt, R.; Seller, C.A.; Hsu, P.K.; Takahashi, Y.; Munemasa, S.; Schroeder, J.I. Plant Hormone Regulation of Abiotic Stress Responses. Nat. Rev. Mol. Cell Biol. 2022, 23, 680–694. [Google Scholar] [CrossRef] [PubMed]
- Fu, J.; Wu, H.; Ma, S.; Xiang, D.; Liu, R.; Xiong, L. OSJAZ1 Attenuates Drought Resistance by Regulating JA and ABA Signaling in Rice. Front. Plant Sci. 2017, 8, 02108. [Google Scholar] [CrossRef]
- Liu, B.; Seong, K.; Pang, S.; Song, J.; Gao, H.; Wang, C.; Zhai, J.; Zhang, Y.; Gao, S.; Li, X.; et al. Functional Specificity, Diversity, and Redundancy of Arabidopsis JAZ Family Repressors in Jasmonate and COI1-Regulated Growth, Development, and Defense. New Phytol. 2021, 231, 1525–1545. [Google Scholar] [CrossRef]
- Aleman, F.; Yazaki, J.; Lee, M.; Takahashi, Y.; Kim, A.Y.; Li, Z.; Kinoshita, T.; Ecker, J.R.; Schroeder, J.I. An ABA-Increased Interaction of the PYL6 ABA Receptor with MYC2 Transcription Factor: A Putative Link of ABA and JA Signaling. Sci. Rep. 2016, 6, 28941. [Google Scholar] [CrossRef]
- Kazan, K.; Manners, J.M. MYC2: The Master in Action. Mol. Plant 2013, 6, 686–703. [Google Scholar] [CrossRef]
- Jing, H.; Wilkinson, E.G.; Sageman-Furnas, K.; Strader, L.C. Auxin and Abiotic Stress Responses. J. Exp. Bot. 2023, 74, 7000–7014. [Google Scholar] [CrossRef]
- Du, W.; Lu, Y.; Li, Q.; Luo, S.; Shen, S.; Li, N.; Chen, X. TIR1/AFB Proteins: Active Players in Abiotic and Biotic Stress Signaling. Front. Plant Sci. 2022, 13, 1083409. [Google Scholar] [CrossRef]
- Salehin, M.; Li, B.; Tang, M.; Katz, E.; Song, L.; Ecker, J.R.; Kliebenstein, D.J.; Estelle, M. Auxin-Sensitive Aux/IAA Proteins Mediate Drought Tolerance in Arabidopsis by Regulating Glucosinolate Levels. Nat. Commun. 2019, 10, 4021. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.-P.; Montgomery, T.A.; Fahlgren, N.; Kasschau, K.D.; Nonogaki, H.; Carrington, J.C. Repression of AUXIN RESPONSE FACTOR10 by microRNA160 Is Critical for Seed Germination and Post-Germination Stages. Plant J. 2007, 52, 133–146. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.-H.; Tian, X.; Li, Y.-J.; Wu, C.-A.; Zheng, C.-C. Microarray-Based Analysis of Stress-Regulated microRNAs in Arabidopsis Thaliana. RNA 2008, 14, 836–843. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Yang, J.; Zhang, N.; Wu, J.; Si, H. Roles of microRNAs in Abiotic Stress Response and Characteristics Regulation of Plant. Front. Plant Sci. 2022, 13, 919243. [Google Scholar] [CrossRef]
- Arshad, M.; Gruber, M.Y.; Hannoufa, A. Transcriptome Analysis of microRNA156 Overexpression Alfalfa Roots under Drought Stress. Sci. Rep. 2018, 8, 9363. [Google Scholar] [CrossRef]
- Singroha, G.; Sharma, P.; Sunkur, R. Current Status of microRNA-Mediated Regulation of Drought Stress Responses in Cereals. Physiol. Plant. 2021, 172, 1808–1821. [Google Scholar] [CrossRef]
- Ferdous, J.; Hussain, S.S.; Shi, B.-J. Role of microRNAs in Plant Drought Tolerance. Plant Biotechnol. J. 2015, 13, 293–305. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: Multiple Sequence Alignment with High Accuracy and High Throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef]
- Nguyen, L.T.; Schmidt, H.A.; Von Haeseler, A.; Minh, B.Q. IQ-TREE: A Fast and Effective Stochastic Algorithm for Estimating Maximum-Likelihood Phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef]
- Hu, B.; Jin, J.; Guo, A.Y.; Zhang, H.; Luo, J.; Gao, G. GSDS 2.0: An Upgraded Gene Feature Visualization Server. Bioinformatics 2015, 31, 1296–1297. [Google Scholar] [CrossRef]
- Rzywinski, M.; Schein, J.; Birol, I.; Connors, J.; Gascoyne, R.; Horsman, D.; Jones, S.J.; Marra, M.A. Circos: An Information Aesthetic for Comparative Genomics. Genome Res. 2009, 19, 1639–1645. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Tang, H.; Debarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.H.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A Toolkit for Detection and Evolutionary Analysis of Gene Synteny and Collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef]
- Wang, D.; Zhang, Y.; Zhang, Z.; Zhu, J.; Yu, J. KaKs_Calculator 2.0: A Toolkit Incorporating Gamma-Series Methods and Sliding Window Strategies. Genom. Proteom. Bioinform. 2010, 8, 77–80. [Google Scholar] [CrossRef] [PubMed]
- Shevliakova, E.; Sarmiento, J.; Gloor, M.; Lynch, M.; Conery, J.S. The Evolutionary Fate and Consequences of Duplicate Genes. Science 2000, 290, 1151. [Google Scholar]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van De Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a Database of Plant Cis-Acting Regulatory Elements and a Portal to Tools for in Silico Analysis of Promoter Sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Li, J.; Feng, J.; Liu, B.; Feng, L.; Yu, X.; Li, G.; Zhai, J.; Meyers, B.C.; Xia, R. sRNAanno—A Database Repository of Uniformly Annotated Small RNAs in Plants. Hortic. Res. 2021, 8, 45. [Google Scholar] [CrossRef]
- Dai, X.; Zhuang, Z.; Zhao, P.X. psRNATarget: A Plant Small RNA Target Analysis Server (2017 Release). Nucleic Acids Res. 2018, 46, W49–W54. [Google Scholar] [CrossRef]
- Gasteiger, E.; Hoogland, C.; Gattiker, A.; Duvaud, S.; Wilkins, M.R.; Appel, R.D.; Bairoch, A. Protein Identification and Analysis Tools on the ExPASy Server. In The Proteomics Protocols Handbook; Springer: Berlin/Heidelberg, Germany, 2005; pp. 571–607. [Google Scholar] [CrossRef]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME Suite: Tools for Motif Discovery and Searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef]
- Hsiao, T.C. Plant Responses to Water Stress. Annu. Rev. Plant Physiol. 1973, 24, 519–570. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Schmittgen, T.D.; Livak, K.J. Analyzing Real-Time PCR Data by the Comparative CT Method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
de Souza-Vieira, Y.; Felix-Mendes, E.; Valente-Almeida, G.; Felix-Cordeiro, T.; Corrêa, R.L.; Jardim-Messeder, D.; Sachetto-Martins, G. Analysis of the Genes from Gibberellin, Jasmonate, and Auxin Signaling Under Drought Stress: A Genome-Wide Approach in Castor Bean (Ricinus communis L.). Plants 2025, 14, 1256. https://doi.org/10.3390/plants14081256
de Souza-Vieira Y, Felix-Mendes E, Valente-Almeida G, Felix-Cordeiro T, Corrêa RL, Jardim-Messeder D, Sachetto-Martins G. Analysis of the Genes from Gibberellin, Jasmonate, and Auxin Signaling Under Drought Stress: A Genome-Wide Approach in Castor Bean (Ricinus communis L.). Plants. 2025; 14(8):1256. https://doi.org/10.3390/plants14081256
Chicago/Turabian Stylede Souza-Vieira, Ygor, Esther Felix-Mendes, Gabriela Valente-Almeida, Thais Felix-Cordeiro, Régis L. Corrêa, Douglas Jardim-Messeder, and Gilberto Sachetto-Martins. 2025. "Analysis of the Genes from Gibberellin, Jasmonate, and Auxin Signaling Under Drought Stress: A Genome-Wide Approach in Castor Bean (Ricinus communis L.)" Plants 14, no. 8: 1256. https://doi.org/10.3390/plants14081256
APA Stylede Souza-Vieira, Y., Felix-Mendes, E., Valente-Almeida, G., Felix-Cordeiro, T., Corrêa, R. L., Jardim-Messeder, D., & Sachetto-Martins, G. (2025). Analysis of the Genes from Gibberellin, Jasmonate, and Auxin Signaling Under Drought Stress: A Genome-Wide Approach in Castor Bean (Ricinus communis L.). Plants, 14(8), 1256. https://doi.org/10.3390/plants14081256







