Genome-Wide Analysis of GRETCHEN HAGEN3 Genes and Characterization of IAA-Amido Synthetase Gene CsGH3.1 in Rhizome Proliferation in Cymbidium sinense ‘Qijianbaimo’
Abstract
1. Introduction
2. Results
2.1. The Genome of CSQ Contains Seven GH3 Genes
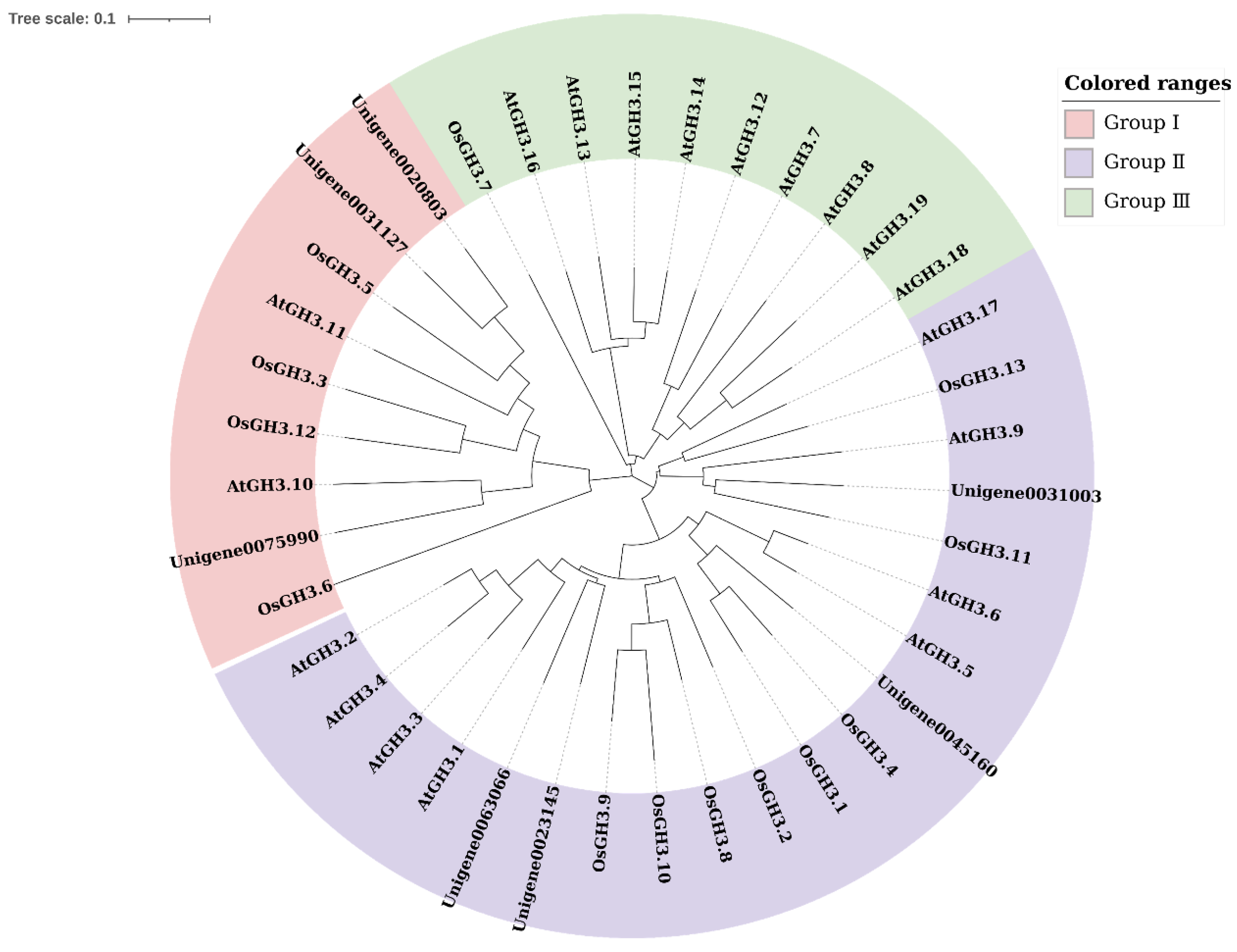
2.2. Phylogenetic Relationship of CsGH3s in CSQ
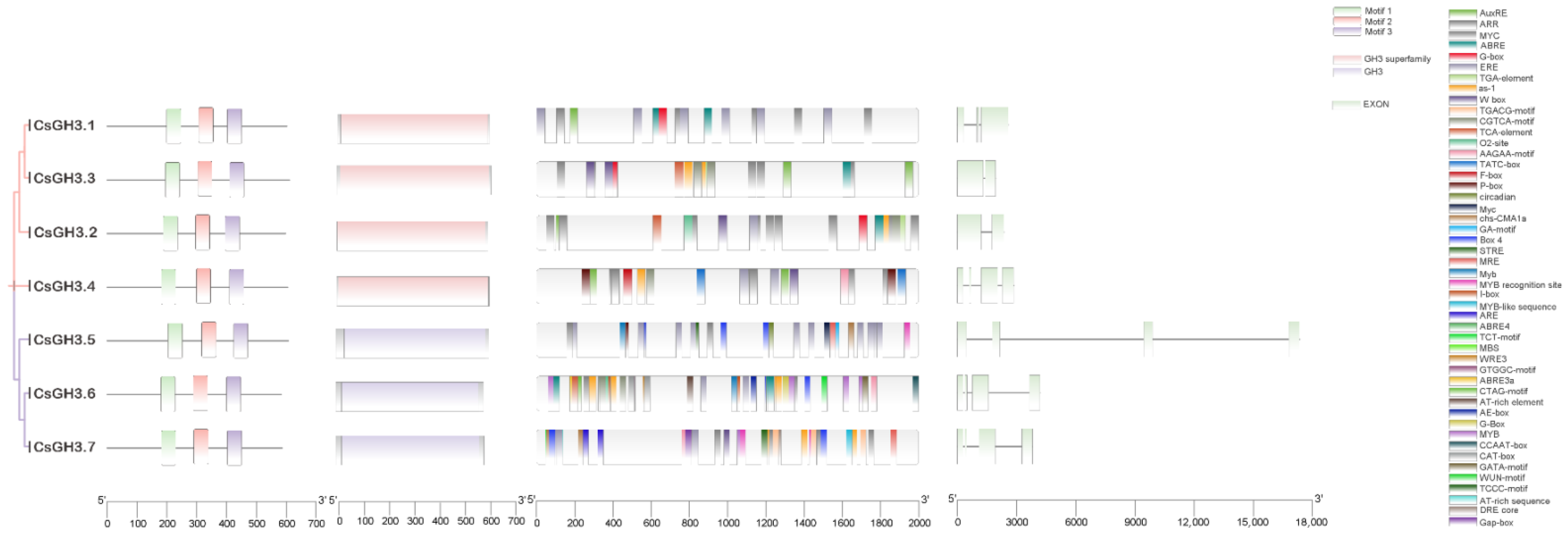
2.3. Analysis of CsGH3 Gene Structures and Cis Elements in the CsGH3 Gene Promoters
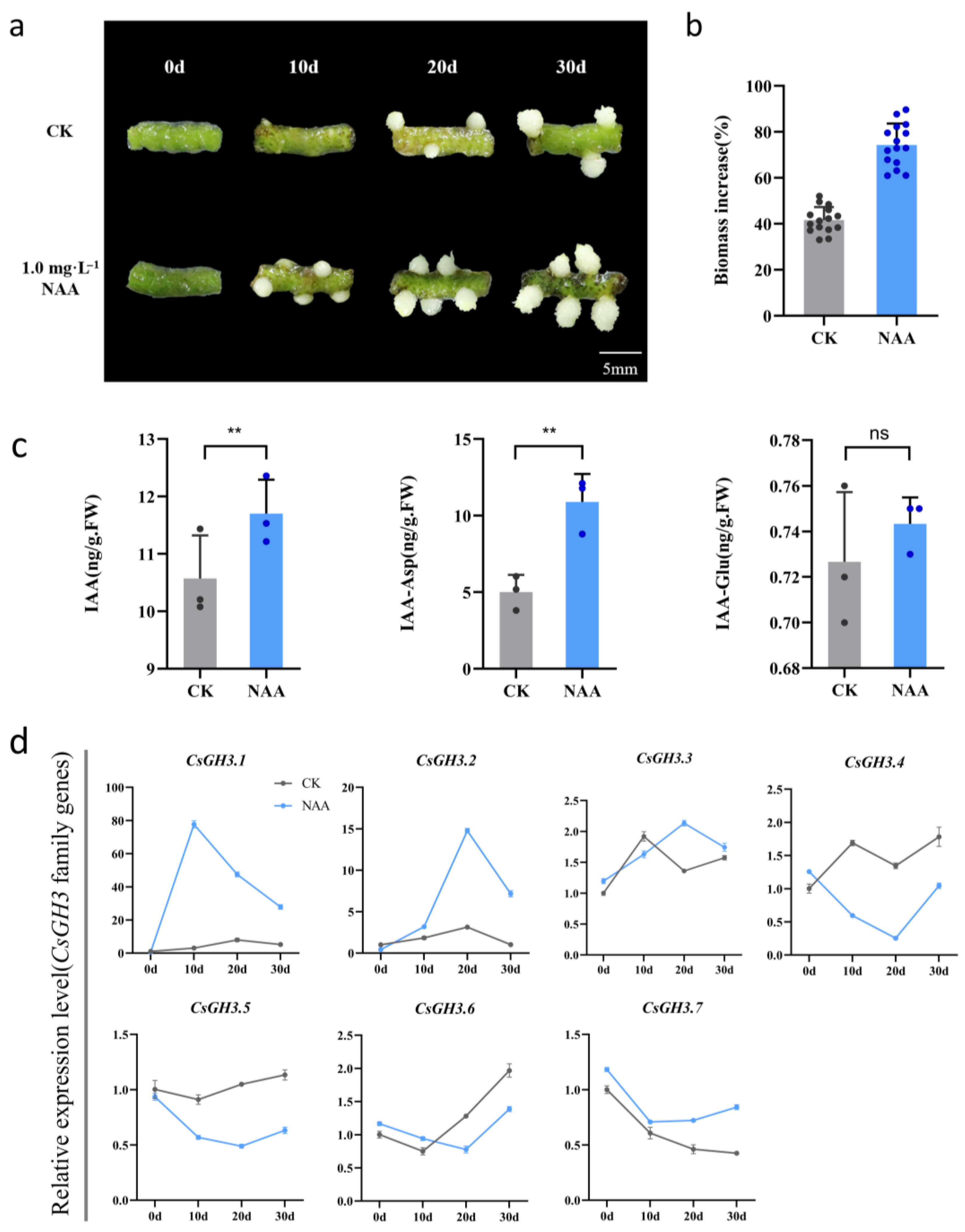
2.4. Function of CsGH3 Genes During Rhizome Proliferation in CSQ
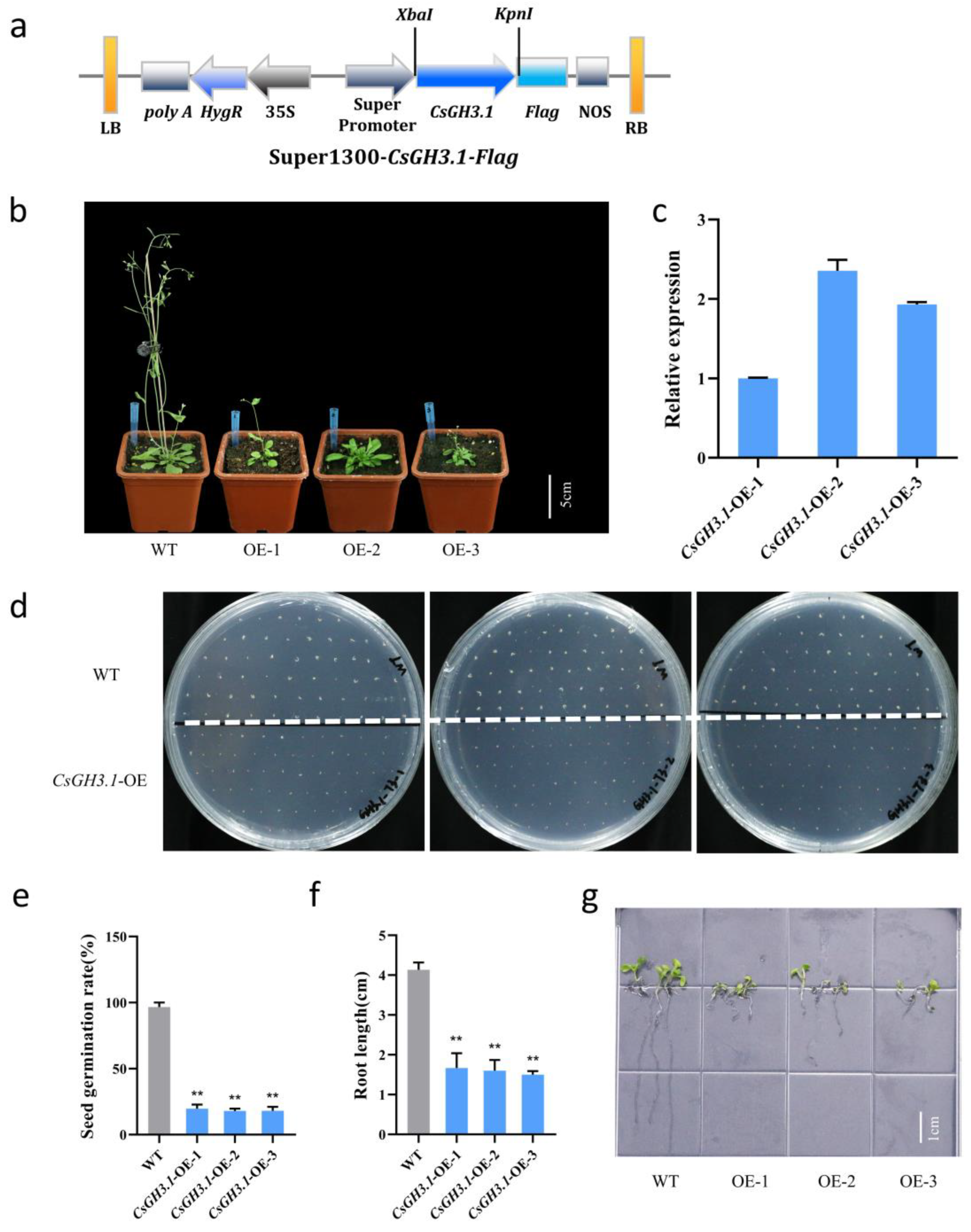
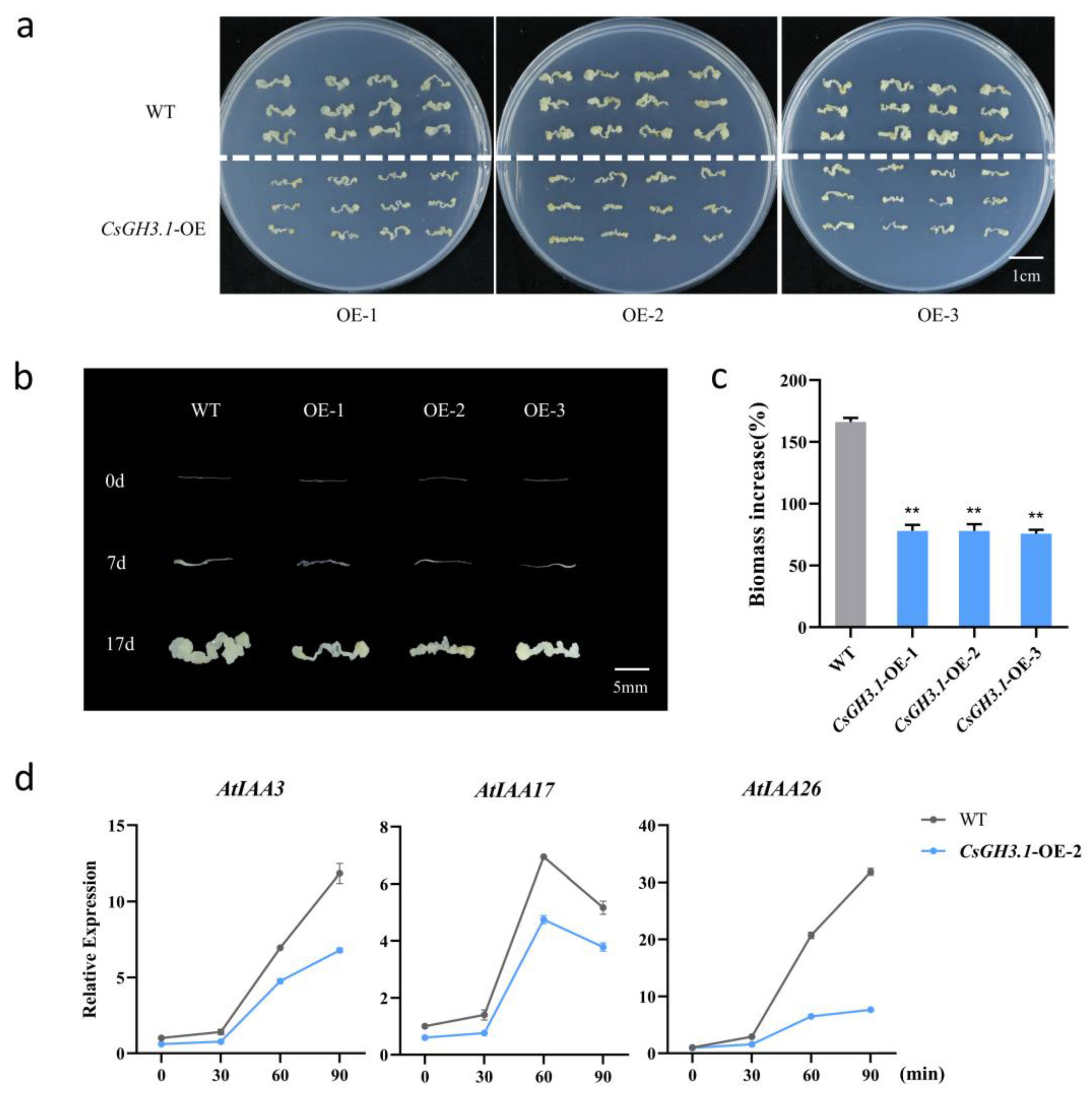
2.5. Overexpression of CsGH3.1 Leads to Decrease in Callus Proliferation Rate in Arabidopsis
3. Discussion
4. Materials and Methods
4.1. Identification and Bioinformatics Analysis of the GH3 Gene Family in CSQ
4.2. Rhizome Proliferation Culture of CSQ
4.3. Endogenous Hormone Measurement
4.4. Expression Analysis of CsGH3 Genes
4.5. Generation of Overexpressed-CsGH3.1 Arabidopsis Transgenic Materials
4.6. Phenotypic Analysis of CsGH3.1 Overexpression Arabidopsis Mutants
4.7. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Abbreviations
| CSQ | Cymbidium sinense ‘Qijianbaimo’ |
| IAA | indoleacetic acid |
| NAA | naphthaleneacetic acid |
| IBA | indolebutyric acid |
References
- Woodward, A.W.; Bartel, B. Auxin: Regulation, action, and interaction. Ann. Bot. 2005, 95, 707–735. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Z.; Guo, Y.; Novák, O.; Chen, W.; Ljung, K.; Noel, J.P.; Chory, J. Local auxin metabolism regulates environment-induced hypocotyl elongation. Nat. Plants 2016, 2, 16025. [Google Scholar] [CrossRef]
- Mateo-Bonmatí, E.; Casanova-Sáez, R.; Šimura, J.; Ljung, K. Broadening the roles of UDP-glycosyltransferases in auxin homeostasis and plant development. New Phytol. 2021, 232, 642–654. [Google Scholar] [CrossRef]
- Westfall, C.S.; Zubieta, C.; Herrmann, J.; Kapp, U.; Nanao, M.H.; Jez, J.M. Structural basis for prereceptor modulation of plant hormones by GH3 proteins. Science 2012, 336, 1708–1711. [Google Scholar] [CrossRef]
- Luo, P.; Li, T.-T.; Shi, W.-M.; Ma, Q.; Di, D.-W. The roles of GRETCHEN HAGEN3 (GH3)-dependent auxin conjugation in the regulation of plant development and stress adaptation. Plants 2023, 12, 4111. [Google Scholar] [CrossRef]
- Staswick, P.E.; Serban, B.; Rowe, M.; Tiryaki, I.; Maldonado, M.T.; Maldonado, M.C.; Suza, W. Characterization of an Arabidopsis enzyme family that conjugates amino acids to indole-3-acetic acid. Plant Cell 2005, 17, 616–627. [Google Scholar] [CrossRef]
- Hagen, G.; Guilfoyle, T. Auxin-responsive gene expression: Genes, promoters and regulatory factors. Plant Mol. Biol. 2002, 49, 373–385. [Google Scholar] [CrossRef]
- Fu, J.; Yu, H.; Li, X.; Xiao, J.; Wang, S. Rice GH3 gene family: Regulators of growth and development. Plant Signal. Behav. 2011, 6, 570–574. [Google Scholar] [CrossRef]
- Jain, M.; Kaur, N.; Tyagi, A.K.; Khurana, J.P. The auxin-responsive GH3 gene family in rice (Oryza sativa). Funct. Integr. Genom. 2006, 6, 36–46. [Google Scholar] [CrossRef]
- Kong, W.; Zhang, Y.; Deng, X.; Li, S.; Zhang, C.; Li, Y. Comparative genomic and transcriptomic analysis suggests the evolutionary dynamic of GH3 genes in Gramineae crops. Front. Plant Sci. 2019, 10, 1297. [Google Scholar] [CrossRef]
- Pinto, R.T.; Freitas, N.C.; Máximo, W.P.F.; Cardoso, T.B.; de Oliveira Prudente, D.; Paiva, L.V. Genome-wide analysis, transcription factor network approach and gene expression profile of GH3 genes over early somatic embryogenesis in Coffea spp. BMC Genom. 2019, 20, 812. [Google Scholar] [CrossRef] [PubMed]
- Monsalve, L.; Ayala-Raso, A.; Bernales, M.; Valdenegro, M.; Defilippi, B.; González-Agüero, M.; Cherian, S.; Fuentes, L. Dataset on quality and physiological changes of raspberry fruit during their development and under auxin in-vitro assay. Data Brief. 2018, 21, 1521–1525. [Google Scholar] [CrossRef] [PubMed]
- Terol, J.; Domingo, C.; Talón, M. The GH3 family in plants: Genome wide analysis in rice and evolutionary history based on EST analysis. Gene 2006, 371, 279–290. [Google Scholar] [CrossRef] [PubMed]
- Kumar, R.; Agarwal, P.; Tyagi, A.K.; Sharma, A.K. Genome-wide investigation and expression analysis suggest diverse roles of auxin-responsive GH3 genes during development and response to different stimuli in tomato (Solanum lycopersicum). Mol. Genet. Genom. 2012, 287, 221–235. [Google Scholar] [CrossRef]
- Nakazawa, M.; Yabe, N.; Ichikawa, T.; Yamamoto, Y.Y.; Yoshizumi, T.; Hasunuma, K.; Matsui, M. DFL1, an auxin-responsive GH3 gene homologue, negatively regulates shoot cell elongation and lateral root formation, and positively regulates the light response of hypocotyl length. Plant J. 2001, 25, 213–221. [Google Scholar]
- Park, J.-E.; Park, J.-Y.; Kim, Y.-S.; Staswick, P.E.; Jeon, J.; Yun, J.; Kim, S.-Y.; Kim, J.; Lee, Y.-H.; Park, C.-M. GH3-mediated auxin homeostasis links growth regulation with stress adaptation response in Arabidopsis. J. Biol. Chem. 2007, 282, 10036–10046. [Google Scholar] [CrossRef]
- Park, J.-E.; Seo, P.J.; Lee, A.-K.; Jung, J.-H.; Kim, Y.-S.; Park, C.-M. An Arabidopsis GH3 gene, encoding an auxin-conjugating enzyme, mediates phytochrome B-regulated light signals in hypocotyl growth. Plant Cell Physiol. 2007, 48, 1236–1241. [Google Scholar] [CrossRef]
- Takase, T.; Nakazawa, M.; Ishikawa, A.; Kawashima, M.; Ichikawa, T.; Takahashi, N.; Shimada, H.; Manabe, K.; Matsui, M. ydk1-D, an auxin-responsive GH3 mutant that is involved in hypocotyl and root elongation. Plant J. 2004, 37, 471–483. [Google Scholar] [CrossRef]
- Domingo, C.; Andrés, F.; Tharreau, D.; Iglesias, D.J.; Talón, M. Constitutive expression of OsGH3. 1 reduces auxin content and enhances defense response and resistance to a fungal pathogen in rice. Mol. Plant-Microbe Interact. 2009, 22, 201–210. [Google Scholar] [CrossRef]
- Du, H.; Wu, N.; Fu, J.; Wang, S.; Li, X.; Xiao, J.; Xiong, L. A GH3 family member, OsGH3-2, modulates auxin and abscisic acid levels and differentially affects drought and cold tolerance in rice. J. Exp. Bot. 2012, 63, 6467–6480. [Google Scholar] [CrossRef]
- Yuan, Z.; Fan, K.; Wang, Y.; Tian, L.; Zhang, C.; Sun, W.; He, H.; Yu, S. OsGRETCHENHAGEN3-2 modulates rice seed storability via accumulation of abscisic acid and protective substances. Plant Physiol. 2021, 186, 469–482. [Google Scholar] [CrossRef] [PubMed]
- Dai, Z.; Wang, J.; Yang, X.; Lu, H.; Miao, X.; Shi, Z. Modulation of plant architecture by the miR156f–OsSPL7–OsGH3. 8 pathway in rice. J. Exp. Bot. 2018, 69, 5117–5130. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.-W.; Li, C.-H.; Cao, J.; Zhang, Y.-C.; Zhang, S.-Q.; Xia, Y.-F.; Sun, D.-Y.; Sun, Y. Altered architecture and enhanced drought tolerance in rice via the down-regulation of indole-3-acetic acid by TLD1/OsGH3. 13 activation. Plant Physiol. 2009, 151, 1889–1901. [Google Scholar] [CrossRef] [PubMed]
- Yadav, S.R.; Khanday, I.; Majhi, B.B.; Veluthambi, K.; Vijayraghavan, U. Auxin-responsive OsMGH3, a common downstream target of OsMADS1 and OsMADS6, controls rice floret fertility. Plant Cell Physiol. 2011, 52, 2123–2135. [Google Scholar] [CrossRef]
- Li, X.; Yang, D.-L.; Sun, L.; Li, Q.; Mao, B.; He, Z. The systemic acquired resistance regulator OsNPR1 attenuates growth by repressing auxin signaling through promoting IAA-amido synthase expression. Plant Physiol. 2016, 172, 546–558. [Google Scholar] [CrossRef]
- Liu, Y.; Zhang, H.-L.; Guo, H.-R.; Xie, L.; Zeng, R.-Z.; Zhang, X.-Q.; Zhang, Z.-S. Transcriptomic and hormonal analyses reveal that YUC-mediated auxin biogenesis is involved in shoot regeneration from rhizome in Cymbidium. Front. Plant Sci. 2017, 8, 1866. [Google Scholar] [CrossRef]
- Xu, C.; Cao, H.; Zhang, Q.; Wang, H.; Xin, W.; Xu, E.; Zhang, S.; Yu, R.; Yu, D.; Hu, Y. Control of auxin-induced callus formation by bZIP59–LBD complex in Arabidopsis regeneration. Nat. Plants 2018, 4, 108–115. [Google Scholar] [CrossRef]
- Yang, F.-X.; Gao, J.; Wei, Y.-L.; Ren, R.; Zhang, G.-Q.; Lu, C.-Q.; Jin, J.-P.; Ai, Y.; Wang, Y.-Q.; Chen, L.-J.; et al. The genome of Cymbidium sinense revealed the evolution of orchid traits. Plant Biotechnol. J. 2021, 19, 2501–2516. [Google Scholar] [CrossRef]
- Hu, W.; Fagundez, S.; Katin-Grazzini, L.; Li, Y.; Li, W.; Chen, Y.; Wang, X.; Deng, Z.; Xie, S.; McAvoy, R.J. Endogenous auxin and its manipulation influence in vitro shoot organogenesis of citrus epicotyl explants. Hortic. Res. 2017, 4, 17071. [Google Scholar] [CrossRef]
- Wojtaczka, P.; Ciarkowska, A.; Starzynska, E.; Ostrowski, M. The GH3 amidosynthetases family and their role in metabolic crosstalk modulation of plant signaling compounds. Phytochemistry 2022, 194, 113039. [Google Scholar] [CrossRef]
- Bao, D.; Chang, S.; Li, X.; Qi, Y. Advances in the study of auxin early response genes: Aux/IAA, GH3, and SAUR. Crop J. 2024, 12, 964–978. [Google Scholar] [CrossRef]
- Jiang, W.; Yin, J.; Zhang, H.; He, Y.; Shuai, S.; Chen, S.; Cao, S.; Li, W.; Ma, D.; Chen, H. Genome-wide identification, characterization analysis and expression profiling of auxin-responsive GH3 family genes in wheat (Triticum aestivum L.). Mol. Biol. Rep. 2020, 47, 3885–3907. [Google Scholar] [CrossRef] [PubMed]
- Takase, T.; Nakazawa, M.; Ishikawa, A.; Manabe, K.; Matsui, M. DFL2, a new member of the Arabidopsis GH3 gene family, is involved in red light-specific hypocotyl elongation. Plant Cell Physiol. 2003, 44, 1071–1080. [Google Scholar] [CrossRef]
- Feng, S.; Yue, R.; Tao, S.; Yang, Y.; Zhang, L.; Xu, M.; Wang, H.; Shen, C. Genome-wide identification, expression analysis of auxin-responsive GH3 family genes in maize (Zea mays L.) under abiotic stresses. J. Integr. Plant Biol. 2015, 57, 783–795. [Google Scholar] [CrossRef]
- Khan, S.; Stone, J.M. Arabidopsis thaliana GH3. 9 influences primary root growth. Planta 2007, 226, 21–34. [Google Scholar] [CrossRef]
- Yan, W.; Jian, Y.; Duan, S.; Guo, X.; Hu, J.; Yang, X.; Li, G. Dissection of the plant hormone signal transduction network in late blight-resistant potato genotype SD20 and prediction of key resistance genes. Phytopathology® 2023, 113, 528–538. [Google Scholar] [CrossRef]
- Nobuta, K.; Okrent, R.A.; Stoutemyer, M.; Rodibaugh, N.; Kempema, L.; Wildermuth, M.C.; Innes, R.W. The GH3 acyl adenylase family member PBS3 regulates salicylic acid-dependent defense responses in Arabidopsis. Plant Physiol. 2007, 144, 1144–1156. [Google Scholar] [CrossRef]
- Liu, Z.-B.; Ulmasov, T.; Shi, X.; Hagen, G.; Guilfoyle, T.J. Soybean GH3 promoter contains multiple auxin-inducible elements. Plant Cell 1994, 6, 645–657. [Google Scholar]
- Ai, G.; Huang, R.; Zhang, D.; Li, M.; Li, G.; Li, W.; Ahiakpa, J.K.; Wang, Y.; Hong, Z.; Zhang, J. SlGH3. 15, a member of the GH3 gene family, regulates lateral root development and gravitropism response by modulating auxin homeostasis in tomato. Plant Sci. 2023, 330, 111638. [Google Scholar] [CrossRef]
- Zhao, D.; Wang, Y.; Feng, C.; Wei, Y.; Peng, X.; Guo, X.; Guo, X.; Zhai, Z.; Li, J.; Shen, X. Overexpression of MsGH3. 5 inhibits shoot and root development through the auxin and cytokinin pathways in apple plants. Plant J. 2020, 103, 166–183. [Google Scholar] [CrossRef]
- Hayashi, K.-i.; Arai, K.; Aoi, Y.; Tanaka, Y.; Hira, H.; Guo, R.; Hu, Y.; Ge, C.; Zhao, Y.; Kasahara, H. The main oxidative inactivation pathway of the plant hormone auxin. Nat. Commun. 2021, 12, 6752. [Google Scholar] [CrossRef] [PubMed]
- Yu, D.; Qanmber, G.; Lu, L.; Wang, L.; Li, J.; Yang, Z.; Liu, Z.; Li, Y.; Chen, Q.; Mendu, V. Genome-wide analysis of cotton GH3 subfamily II reveals functional divergence in fiber development, hormone response and plant architecture. BMC Plant Biol. 2018, 18, 350. [Google Scholar] [CrossRef] [PubMed]
- Ding, X.; Cao, Y.; Huang, L.; Zhao, J.; Xu, C.; Li, X.; Wang, S. Activation of the indole-3-acetic acid–amido synthetase GH3-8 suppresses expansin expression and promotes salicylate-and jasmonate-independent basal immunity in rice. Plant Cell 2008, 20, 228–240. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Bin, J.; Tan, Q.; Wen, S.; Huang, L.; Wang, H.; Imtiaz, M.; Zhang, Z.; Guo, H.; Xie, L.; Zeng, R.; et al. Comprehensive Analyses of Four PhNF-YC Genes from Petunia hybrida and Impacts on Flowering Time. Plants 2024, 13, 742. [Google Scholar] [CrossRef]
- Dai, X.; Wang, J.; Wang, L.; Liu, Z.; Li, Q.; Cai, Y.; Li, S.; Xiang, F. HY5 inhibits in vitro shoot stem cell niches initiation via directly repressing pluripotency and cytokinin pathways. Plant J. 2022, 110, 781–801. [Google Scholar] [CrossRef]
- Mallona, I.; Lischewski, S.; Weiss, J.; Hause, B.; Egea-Cortines, M. Validation of Reference Genes for Quantitative Real-Time PCR during Leaf and Flower Development in Petunia hybrida. BMC Plant Biol. 2010, 10, 4. [Google Scholar] [CrossRef]
| Gene Name | Gene Transcriptome ID | Gene Locus ID | Chr | Location | CDS (bp) | Protein (aa) | MW (Da) | pI | Formula |
|---|---|---|---|---|---|---|---|---|---|
| CsGH3.1 | Unigene0063066 | CYSIN007746 | 15 | 38,528,251–38,530,808 | 1806 | 602 | 67,940.94 | 5.79 | C3037H4761N815O893S30 |
| CsGH3.2 | Unigene0045160 | CYSIN015084 | 9 | 47,807,899–47,810,722 | 1791 | 597 | 66,682.16 | 5.61 | C2981H4642N800O881S28 |
| CsGH3.3 | Unigene0023145 | CYSIN028796 | 9 | 62,449,145–62,453,024 | 1833 | 611 | 68,633.54 | 5.59 | C3052H4805N829O910S30 |
| CsGH3.4 | Unigene0031003 | CYSIN005890 | 2 | 258,273,160–258,276,267 | 1815 | 605 | 67,626.52 | 5.68 | C3010H4723N807O895S34 |
| CsGH3.5 | Unigene0075990 | CYSIN028599 | 14 | 45,363,266–45,384,562 | 1821 | 607 | 68,287.87 | 6.27 | C3064H4774N826O902S21 |
| CsGH3.6 | Unigene0020803 | CYSIN006822 | 8 | 120,446,007–120,448,018 | 1758 | 586 | 65,523.81 | 6.2 | C2943H4587N779O871S22 |
| CsGH3.7 | Unigene0031127 | CYSIN027452 | 13 | 72,632,018–72,639,833 | 1761 | 587 | 65,895.12 | 6.22 | C2957H4604N782O880S22 |
| Hormone Name | Polarity | Parent Ion (m/z) | Daughter Ion (m/z) | Breakdown Voltage (V) | Collisional Energy (V) |
|---|---|---|---|---|---|
| IAA-ASP | + | 291.1 | 134/130.1 * | 110 | 8/35 |
| IAA-GLU | − | 303.0 | 146.0 */127.8/173.9 | −40 | −23/−25/−20 |
| IAA | + | 176.2 | 129.8 */102.9 | 65 | 12/42 |
| Primer Name | Primer Sequence (5′-3′) |
|---|---|
| RPS3-F | TGAAGCCAATGCAGAACAAACG |
| RPS3-R | ATGGATTTCACCACCGACAAGC |
| qP-BMGH3.1-F | TCTCCTTATGCCTGTTATG |
| qP-BMGH3.1-R | GAAATCGGAAACCAGAACT |
| qP-BMGH3.2-F | AGGCGACAATCACAACCAA |
| qP-BMGH3.2-R | CCGATACCGATACAAACCG |
| qP-BMGH3.3-F | GAACATCAGCGGGAGAGAG |
| qP-BMGH3.3-R | ACGAAGAGGAAGTAAAGGG |
| qP-BMGH3.4-F | TAGAGGCTGTGCTTACTGG |
| qP-BMGH3.4-R | GATGATGCATACATCGTGC |
| qP-BMGH3.5-F | CGTTGTATTGCCACCTTCT |
| qP-BMGH3.5-R | TTCTTCCCAAACATTCTCG |
| qP-BMGH3.6-F | ATCGAGCAGAGAGAGACAG |
| qP-BMGH3.6-R | AAACCTCCAAAAGTAGTGA |
| qP-BMGH3.7-F | TAGAGGCTGTGCTTACTGG |
| qP-BMGH3.7-R | GATGATGCATACATCGTGC |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jia, S.; Liu, C.; Liao, C.; Zeng, R.; Xie, L.; Wei, Q.; Zhang, Z. Genome-Wide Analysis of GRETCHEN HAGEN3 Genes and Characterization of IAA-Amido Synthetase Gene CsGH3.1 in Rhizome Proliferation in Cymbidium sinense ‘Qijianbaimo’. Plants 2025, 14, 1287. https://doi.org/10.3390/plants14091287
Jia S, Liu C, Liao C, Zeng R, Xie L, Wei Q, Zhang Z. Genome-Wide Analysis of GRETCHEN HAGEN3 Genes and Characterization of IAA-Amido Synthetase Gene CsGH3.1 in Rhizome Proliferation in Cymbidium sinense ‘Qijianbaimo’. Plants. 2025; 14(9):1287. https://doi.org/10.3390/plants14091287
Chicago/Turabian StyleJia, Sisi, Cuiling Liu, Chushu Liao, Ruizhen Zeng, Li Xie, Qian Wei, and Zhisheng Zhang. 2025. "Genome-Wide Analysis of GRETCHEN HAGEN3 Genes and Characterization of IAA-Amido Synthetase Gene CsGH3.1 in Rhizome Proliferation in Cymbidium sinense ‘Qijianbaimo’" Plants 14, no. 9: 1287. https://doi.org/10.3390/plants14091287
APA StyleJia, S., Liu, C., Liao, C., Zeng, R., Xie, L., Wei, Q., & Zhang, Z. (2025). Genome-Wide Analysis of GRETCHEN HAGEN3 Genes and Characterization of IAA-Amido Synthetase Gene CsGH3.1 in Rhizome Proliferation in Cymbidium sinense ‘Qijianbaimo’. Plants, 14(9), 1287. https://doi.org/10.3390/plants14091287






