The LBD Transcription Factor ZmLBD33 Confers Drought Tolerance in Transgenic Arabidopsis
Abstract
:1. Introduction
2. Results
2.1. Analysis of the Gene Structure and Expression of ZmLBD33
2.2. ZmLBD33 Is a Nucleus-Localized Protein and Could Form Dimers
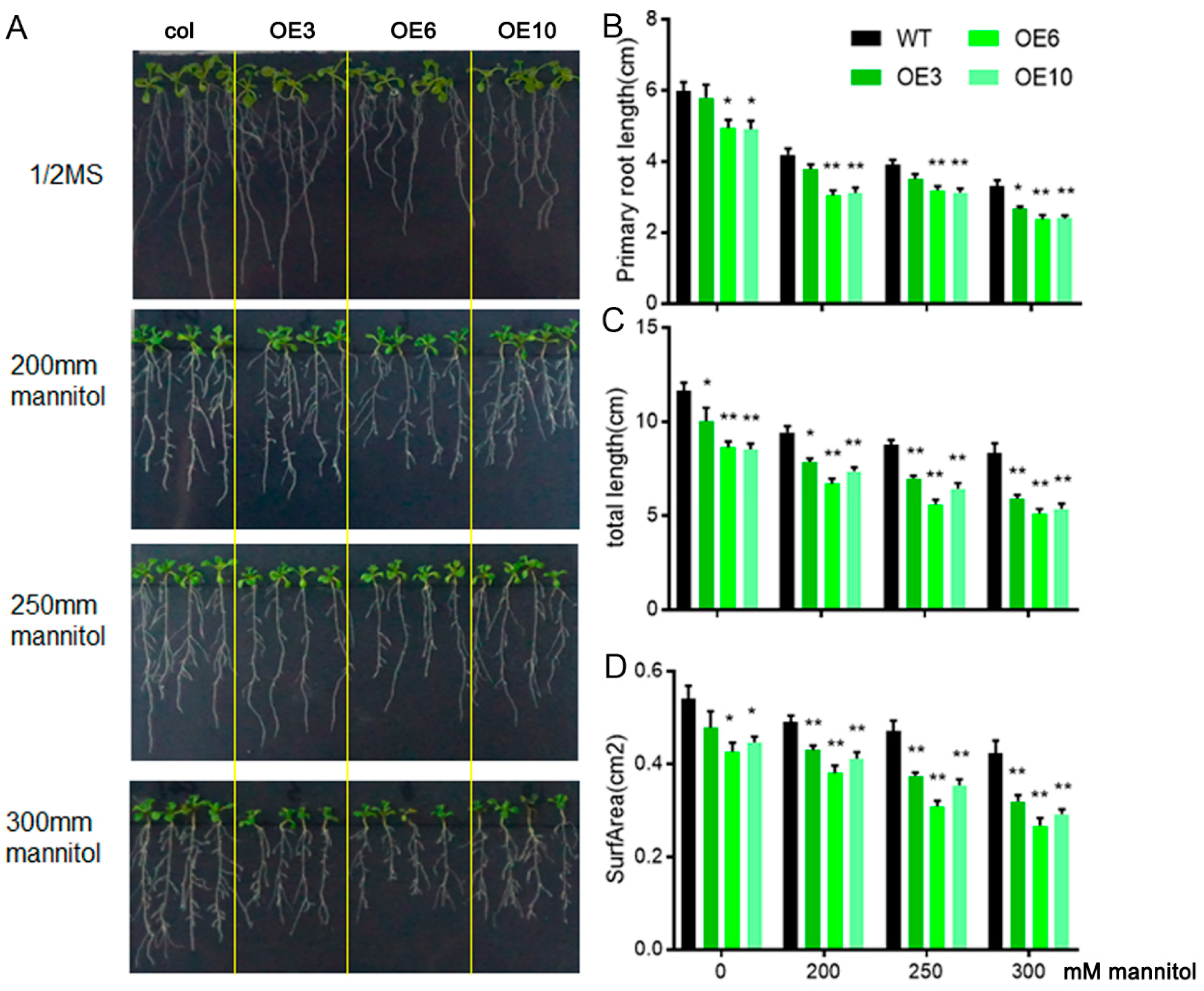
2.3. ZmLBD33 Overexpression Compromised Drought Tolerance in Arabidopsis
2.4. ZmLBD33 Promoted Water Loss Rate Through the Stomatal Density and Aperture
2.5. Overexpression of ZmLBD33 Augmented the Activity of Antioxidant Enzymes and Prevented the Accumulation of ROS in Arabidopsis
2.6. Drought-Inducible Genes Were Transcriptionally Suppressed in ZmLBD33-Overexpressing Arabidopsis
3. Discussion
4. Method and Materials
4.1. Plant Materials and Growth Conditions
4.2. Sequence Analysis and Phylogenetic Tree Construction
4.3. Subcellular Localization
4.4. RNA Extraction and Quantitative RT-qPCR Analysis
4.5. Transcriptional Activation Analysis and Y2H
4.6. Generation of Transgenic Plants and Phenotypic Analysis
4.7. Water Loss Measurement
4.8. Stomatal Density and Stomatal Aperture
4.9. ROS Measurements
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Meyer, E.; Aspinwall, M.J.; Lowry, D.B.; Palacio-Mejia, J.D.; Logan, T.L.; Fay, P.A.; Juenger, T.E. Integrating transcriptional, metabolomic, and physiological responses to drought stress and recovery in switchgrass (Panicum virgatum L.). BMC Genom. 2014, 15, 527. [Google Scholar] [CrossRef] [PubMed]
- Song, H.; Li, Y.; Zhou, L.; Xu, Z.; Zhou, G. Maize leaf functional responses to drought episode and rewatering. Agric. For. Meteorol. 2018, 249, 57–70. [Google Scholar] [CrossRef]
- Huang, X.; Bao, Y.N.; Wang, B.; Liu, L.J.; Chen, J.; Dai, L.J.; Peng, D.X. Identification and expression of Aux/IAA, ARF, and LBD family transcription factors in Boehmeria nivea. Biol. Plant. 2016, 60, 244–250. [Google Scholar] [CrossRef]
- FAO. The State of Food Security and Nutrition in the World; FAO: Rome, Italy, 2021; Available online: https://www.fao.org/faostat/en/#data (accessed on 1 July 2022).
- Colautti, A.; Mian, G.; Tomasi, D.; Bell, L.; Marcuzzo, P. Exploring the Influence of Soil Salinity on Microbiota Dynamics in Vitis vinifera cv. “Glera”: Insights into the Rhizosphere, Carposphere, and Yield Outcomes. Diversity 2024, 16, 247. [Google Scholar] [CrossRef]
- Shuai, B.; Reynaga-Peña, C.G.; Springer, P.S. The Lateral Organ Boundaries Gene Defines a Novel, Plant-Specific Gene Family. Plant Physiol. 2002, 129, 747–761. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.W.; Kim, M.J.; Park, M.Y.; Han, K.H.; Kim, J. The conserved proline residue in the LOB domain of LBD18 is critical for DNA-binding and biological function. Mol. Plant 2013, 6, 1722–1725. [Google Scholar] [CrossRef]
- Majer, C.; Hochholdinger, F. Defining the boundaries: Structure and function of LOB domain proteins. Trends Plant Sci. 2011, 16, 47–52. [Google Scholar] [CrossRef]
- Okushima, Y.; Fukaki, H.; Onoda, M.; Theologis, A.; Tasaka, M. ARF7 and ARF19 regulate lateral root formation via direct activation of LBD/ASL genes in Arabidopsis. Plant Cell 2007, 19, 118–130. [Google Scholar] [CrossRef] [PubMed]
- Rong, M.; Gao, S.-X.; Wen, D.; Xu, Y.-H.; Wei, J.-H. The LOB domain protein, a novel transcription factor with multiple functions: A review. Plant Physiol. Biochem. 2024, 214, 108922. [Google Scholar] [CrossRef]
- Xu, C.; Luo, F.; Hochholdinger, F. LOB Domain Proteins: Beyond Lateral Organ Boundaries. Trends Plant Sci. 2016, 21, 159–167. [Google Scholar] [CrossRef]
- Iwakawa, H.; Iwasaki, M.; Kojima, S.; Ueno, Y.; Soma, T.; Tanaka, H.; Semiarti, E.; Machida, Y.; Machida, C. Expression of the ASYMMETRIC LEAVES2 gene in the adaxial domain of Arabidopsis leaves represses cell proliferation in this domain and is critical for the development of properly expanded leaves. Plant J. 2007, 51, 173–184. [Google Scholar] [CrossRef] [PubMed]
- Semiarti, E.; Ueno, Y.; Tsukaya, H.; Iwakawa, H.; Machida, C.; Machida, Y. The ASYMMETRIC LEAVES2 gene of Arabidopsis thaliana regulates formation of a symmetric lamina, establishment of venation and repression of meristem-related homeobox genes in leaves. Development 2001, 128, 1771–1783. [Google Scholar] [CrossRef] [PubMed]
- Guo, M.; Thomas, J.; Collins, G.; Timmermans, M.C.P. Direct Repression of KNOX Loci by the ASYMMETRIC LEAVES1 Complex of Arabidopsis. Plant Cell 2008, 20, 48–58. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, T.-H.; Kim, M.J.; Kim, J. The transcription factor LBD10 sustains pollen tube growth and integrity by modulating reactive oxygen species homeostasis via the regulation of flavonol biosynthesis in Arabidopsis. New Phytol. 2024, 244, 131–146. [Google Scholar] [CrossRef]
- Timmermans, M.C.; Hudson, A.; Becraft, P.W.; Nelson, T. ROUGH SHEATH2: A Myb protein that represses knox homeobox genes in maize lateral organ primordia. Science 1999, 284, 151–153. [Google Scholar] [CrossRef]
- Evans, M.M. The indeterminate gametophyte1 gene of maize encodes a LOB Domain Protein Required for Embryo Sac and Leaf Development. Plant Cell 2007, 19, 46–62. [Google Scholar] [CrossRef]
- Bortiri, E.; Chuck, G.; Vollbrecht, E.; Rocheford, T.; Martienssen, R.; Hake, S. ramosa2 encodes a LATERAL ORGAN BOUNDARY domain protein that determines the fate of stem cells in branch meristems of maize. Plant Cell 2006, 18, 574–585. [Google Scholar] [CrossRef]
- Inukai, Y.; Sakamoto, T.; Ueguchi, M.; Shibata, Y.; Gomi, K.; Umemura, I.; Hasegawa, Y.; Ashikari, M.; Kitano, H.; Matsuoka, M. Crown rootless1, which is essential for crown root formation in rice, is a target of an AUXIN RESPONSE FACTOR in auxin signaling. Plant Cell 2005, 17, 1387–1396. [Google Scholar] [CrossRef]
- Taramino, G.; Sauer, M.; Stauffer, J.L., Jr.; Multani, D.; Niu, X.; Sakai, H.; Hochholdinger, F. The maize (Zea mays L.) RTCS gene encodes a LOB domain protein that is a key regulator of embryonic seminal and post-embryonic shoot-borne root initiation. Plant J. 2007, 50, 649–659. [Google Scholar] [CrossRef]
- Majer, C.; Xu, C.; Berendzen, K.W.; Hochholdinger, F. Molecular interactions of ROOTLESS CONCERNING CROWN AND SEMINAL ROOTS, a LOB domain protein regulating shoot-borne root initiation in maize (Zea mays L.). Philos. Trans. R. Soc. B Biol. Sci. 2012, 367, 1542–1551. [Google Scholar] [CrossRef]
- Yamauchi, T.; Tanaka, A.; Inahashi, H.; Nishizawa, N.K.; Tsutsumi, N.; Inukai, Y.; Nakazono, M. Fine control of aerenchyma and lateral root development through AUX/IAA- and ARF-dependent auxin signaling. Proc. Natl. Acad. Sci. USA 2019, 116, 20770–20775. [Google Scholar] [CrossRef] [PubMed]
- Goh, T.; Kasahara, H.; Mimura, T.; Kamiya, Y.; Fukaki, H. Multiple AUX/IAA-ARF modules regulate lateral root formation: The role of Arabidopsis SHY2/IAA3-mediated auxin signalling. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2012, 367, 1461–1468. [Google Scholar] [CrossRef]
- Han, Z.; Yang, T.; Guo, Y.; Cui, W.H.; Yao, L.J.; Li, G.; Wu, A.M.; Li, J.H.; Liu, L.J. The transcription factor PagLBD3 contributes to the regulation of secondary growth in Populus. J. Exp. Biol. 2021, 72, 7092–7106. [Google Scholar] [CrossRef]
- Kong, X.; Zhang, C.; Zheng, H.; Sun, M.; Zhang, F.; Zhang, M.; Cui, F.; Lv, D.; Liu, L.; Guo, S.; et al. Antagonistic Interaction between Auxin and SA Signaling Pathways Regulates Bacterial Infection through Lateral Root in Arabidopsis. Cell Rep. 2020, 32, 108060. [Google Scholar] [CrossRef]
- Jeon, B.W.; Kim, J. Role of LBD14 during ABA-mediated control of root system architecture in Arabidopsis. Plant Signal. Behav. 2018, 13, e1507405. [Google Scholar] [CrossRef]
- Jeon, E.; Young Kang, N.; Cho, C.; Joon Seo, P.; Chung Suh, M.; Kim, J. LBD14/ASL17 Positively Regulates Lateral Root Formation and is Involved in ABA Response for Root Architecture in Arabidopsis. Plant Cell Physiol. 2017, 58, 2190–2201. [Google Scholar] [CrossRef] [PubMed]
- Guo, Z.; Xu, H.; Lei, Q.; Du, J.; Li, C.; Wang, C.; Yang, Y.; Yang, Y.; Sun, X. The Arabidopsis transcription factor LBD15 mediates ABA signaling and tolerance of water-deficit stress by regulating ABI4 expression. Plant J. 2020, 104, 510–521. [Google Scholar] [CrossRef] [PubMed]
- Ma, W.; Wu, F.; Sheng, P.; Wang, X.; Zhang, Z.; Zhou, K.; Zhang, H.; Hu, J.; Lin, Q.; Cheng, Z.; et al. The LBD12-1 Transcription Factor Suppresses Apical Meristem Size by Repressing Argonaute 10 Expression. Plant Physiol. 2017, 173, 801–811. [Google Scholar] [CrossRef]
- Feng, X.; Xiong, J.; Zhang, W.; Guan, H.; Zheng, D.; Xiong, H.; Jia, L.; Hu, Y.; Zhou, H.; Wen, Y.; et al. ZmLBD5, a class-II LBD gene, negatively regulates drought tolerance by impairing abscisic acid synthesis. Plant J. 2022, 112, 1364–1376. [Google Scholar] [CrossRef]
- Xu, J.; Hu, P.; Tao, Y.; Song, P.; Gao, H.; Guan, Y. Genome-wide identification and characterization of the Lateral Organ Boundaries Domain (LBD) gene family in polyploid wheat and related species. PeerJ 2021, 9, e11811. [Google Scholar] [CrossRef]
- Yang, Y.; Yu, X.; Wu, P. Comparison and evolution analysis of two rice subspecies LATERAL ORGAN BOUNDARIES domain gene family and their evolutionary characterization from Arabidopsis. Mol. Phylogenet. Evol. 2006, 39, 248–262. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; He, Y.; He, W.; Su, H.; Wang, Y.; Hong, G.; Xu, P. Structural and functional insights into the LBD family involved in abiotic stress and flavonoid synthases in Camellia sinensis. Sci. Rep. 2019, 9, 15651. [Google Scholar] [CrossRef] [PubMed]
- Rubin, G.; Tohge, T.; Matsuda, F.; Saito, K.; Scheible, W.R. Members of the LBD family of transcription factors repress anthocyanin synthesis and affect additional nitrogen responses in Arabidopsis. Plant Cell 2009, 21, 3567–3584. [Google Scholar] [CrossRef] [PubMed]
- Zentella, R.; Zhang, Z.L.; Park, M.; Thomas, S.G.; Endo, A.; Murase, K.; Fleet, C.M.; Jikumaru, Y.; Nambara, E.; Kamiya, Y.; et al. Global analysis of della direct targets in early gibberellin signaling in Arabidopsis. Plant Cell 2007, 19, 3037–3057. [Google Scholar] [CrossRef]
- Albinsky, D.; Kusano, M.; Higuchi, M.; Hayashi, N.; Kobayashi, M.; Fukushima, A.; Mori, M.; Ichikawa, T.; Matsui, K.; Kuroda, H.; et al. Metabolomic screening applied to rice FOX Arabidopsis lines leads to the identification of a gene-changing nitrogen metabolism. Mol. Plant 2010, 3, 125–142. [Google Scholar] [CrossRef]
- Ariel, F.; Diet, A.; Verdenaud, M.; Gruber, V.; Frugier, F.; Chan, R.; Crespi, M. Environmental regulation of lateral root emergence in Medicago truncatula requires the HD-Zip I transcription factor HB1. Plant Cell 2010, 22, 2171–2183. [Google Scholar] [CrossRef]
- Thatcher, L.F.; Kazan, K.; Manners, J.M. Lateral organ boundaries domain transcription factors: New roles in plant defense. Plant Signal. Behav. 2012, 7, 1702–1704. [Google Scholar] [CrossRef]
- Lee, H.W.; Kim, N.Y.; Lee, D.J.; Kim, J. LBD18/ASL20 Regulates Lateral Root Formation in Combination with LBD16/ASL18 Downstream of ARF7 and ARF19 in Arabidopsis. Plant Physiol. 2009, 151, 1377–1389. [Google Scholar] [CrossRef]
- Xiong, J.; Zhang, W.; Zheng, D.; Xiong, H.; Feng, X.; Zhang, X.; Wang, Q.; Wu, F.; Xu, J.; Lu, Y. ZmLBD5 Increases Drought Sensitivity by Suppressing ROS Accumulation in Arabidopsis. Plants 2022, 11, 1382. [Google Scholar] [CrossRef]
- Zhu, X.; Wang, D.; Xie, L.; Zhou, T.; Zhao, J.; Zhang, Q.; Yang, M.; Wu, W.; Lian, X. Rice transcription factors OsLBD37/38/39 regulate nitrate uptake by repressing OsNRT2.1/2.2/2.3 under high-nitrogen conditions. Crop J. 2022, 10, 1623–1632. [Google Scholar] [CrossRef]
- Jiang, X.; Cui, H.; Wang, Z.; Kang, J.; Yang, Q.; Guo, C. Genome-Wide Analysis of the LATERAL ORGAN BOUNDARIES Domain (LBD) Members in Alfalfa and the Involvement of MsLBD48 in Nitrogen Assimilation. Int. J. Mol. Sci. 2023, 24, 4644. [Google Scholar] [CrossRef] [PubMed]
- Li, H.-H.; Liu, X.; An, J.-P.; Hao, Y.-J.; Wang, X.-F.; You, C.-X. Cloning and elucidation of the functional role of apple MdLBD13 in anthocyanin biosynthesis and nitrate assimilation. Plant Cell Tissue Organ Cult. (PCTOC) 2017, 130, 47–59. [Google Scholar] [CrossRef]
- Herath, V. Small family, big impact: In silico analysis of DREB2 transcription factor family in rice. Comput. Biol. Chem. 2016, 65, 128–139. [Google Scholar] [CrossRef]
- Matsukura, S.; Mizoi, J.; Yoshida, T.; Todaka, D.; Ito, Y.; Maruyama, K.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Comprehensive analysis of rice DREB2-type genes that encode transcription factors involved in the expression of abiotic stress-responsive genes. Mol. Genet. Genom. 2010, 283, 185–196. [Google Scholar] [CrossRef] [PubMed]
- Qin, F.; Kakimoto, M.; Sakuma, Y.; Maruyama, K.; Osakabe, Y.; Tran, L.-S.P.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Regulation and functional analysis of ZmDREB2A in response to drought and heat stresses in Zea mays L. Plant J. 2007, 50, 54–69. [Google Scholar] [CrossRef]
- Xiang, Y.; Sun, X.; Gao, S.; Qin, F.; Dai, M. Deletion of an Endoplasmic Reticulum Stress Response Element in a ZmPP2C-A Gene Facilitates Drought Tolerance of Maize Seedlings. Mol. Plant 2017, 10, 456–469. [Google Scholar] [CrossRef]







| Site Name | Sequence | Position | Strand | Function |
|---|---|---|---|---|
| ABRE | ACGTG | 682 | + | abscisic acid responsiveness |
| ABRE | GCCGCGTGGC | 761 | − | abscisic acid responsiveness |
| ABRE | ACGTG | 624 | − | abscisic acid responsiveness |
| ABRE | CACGTG | 681 | − | abscisic acid responsiveness |
| ARE | AAACCA | 129 | + | anaerobic induction |
| ARE | AAACCA | 806 | + | anaerobic induction |
| ARE | AAACCA | 590 | − | anaerobic induction |
| G-Box | CACGTT | 624 | + | light responsiveness |
| G-Box | CACGTG | 681 | − | light responsiveness |
| G-box | CACGTG | 681 | − | light responsiveness |
| Sp1 | GGGCGG | 492 | − | light responsiveness |
| Sp1 | GGGCGG | 744 | + | light responsiveness |
| Sp1 | GGGCGG | 511 | − | light responsiveness |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xiong, J.; Mi, X.; Du, L.; Wang, X. The LBD Transcription Factor ZmLBD33 Confers Drought Tolerance in Transgenic Arabidopsis. Plants 2025, 14, 1305. https://doi.org/10.3390/plants14091305
Xiong J, Mi X, Du L, Wang X. The LBD Transcription Factor ZmLBD33 Confers Drought Tolerance in Transgenic Arabidopsis. Plants. 2025; 14(9):1305. https://doi.org/10.3390/plants14091305
Chicago/Turabian StyleXiong, Jing, Xin Mi, Lijuan Du, and Xianqiu Wang. 2025. "The LBD Transcription Factor ZmLBD33 Confers Drought Tolerance in Transgenic Arabidopsis" Plants 14, no. 9: 1305. https://doi.org/10.3390/plants14091305
APA StyleXiong, J., Mi, X., Du, L., & Wang, X. (2025). The LBD Transcription Factor ZmLBD33 Confers Drought Tolerance in Transgenic Arabidopsis. Plants, 14(9), 1305. https://doi.org/10.3390/plants14091305





