Identification of the Plant Defensin (MsPDF) Gene Family in Medicago sativa and Analysis of Expression Patterns Under Abiotic Stress
Abstract
1. Introduction
2. Results
2.1. Identification and Characterization of PDF Gene Family Members in Alfalfa
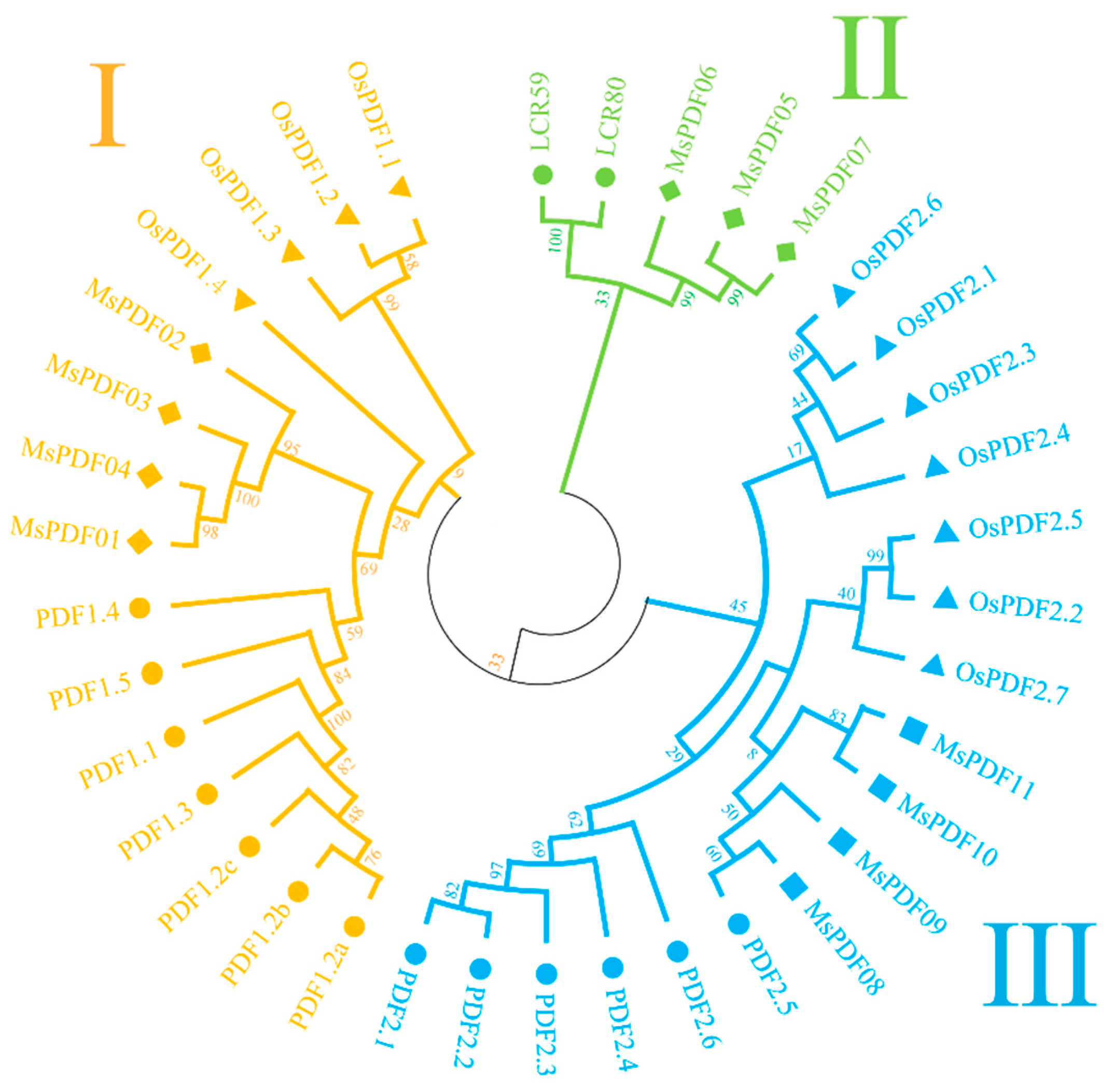
2.2. Phylogenetic Analysis of the PDF Gene Family in Alfalfa, Arabidopsis, and Rice
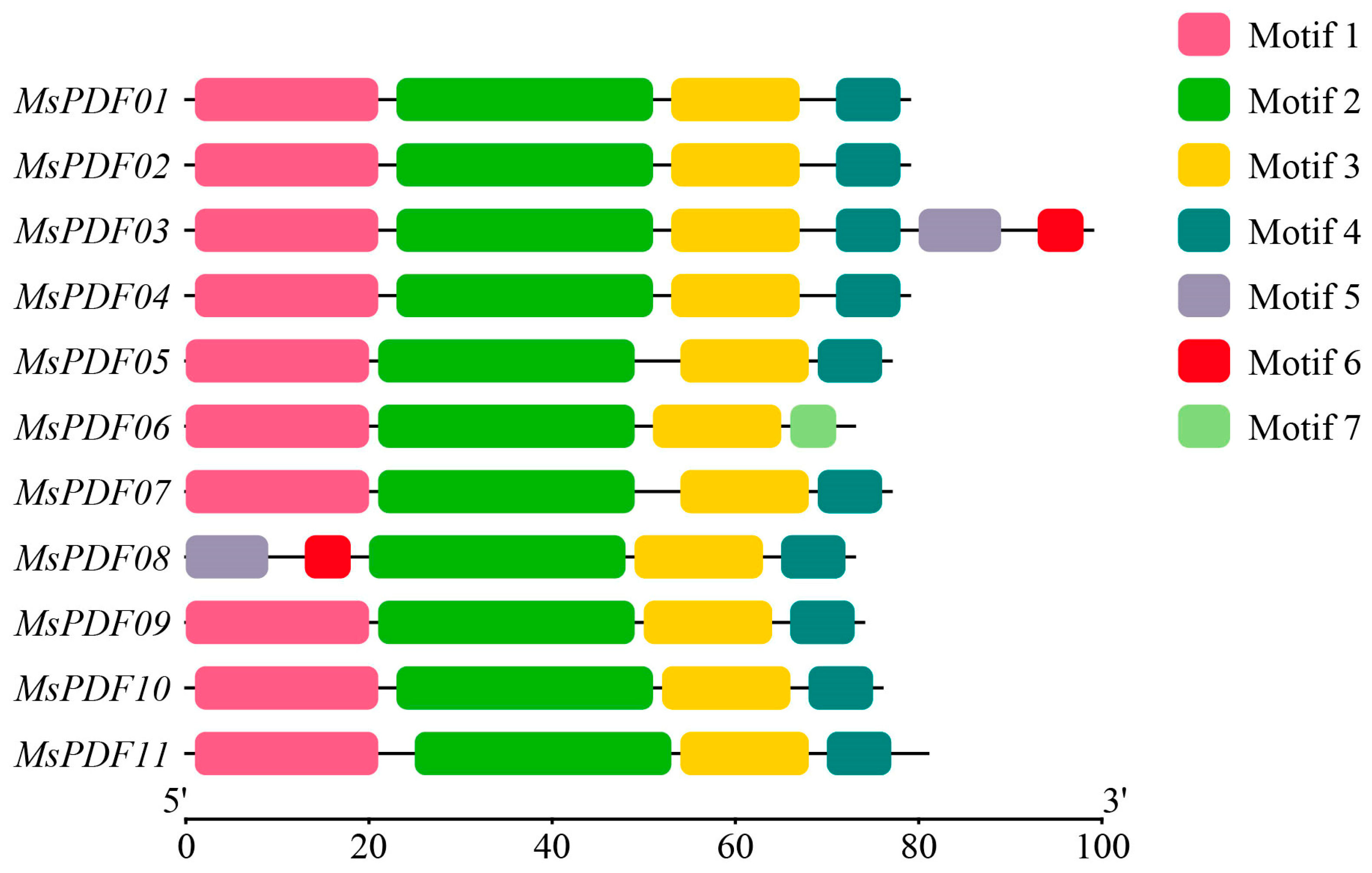
2.3. Analysis of Conserved Motifs in the PDF Gene Family in Alfalfa
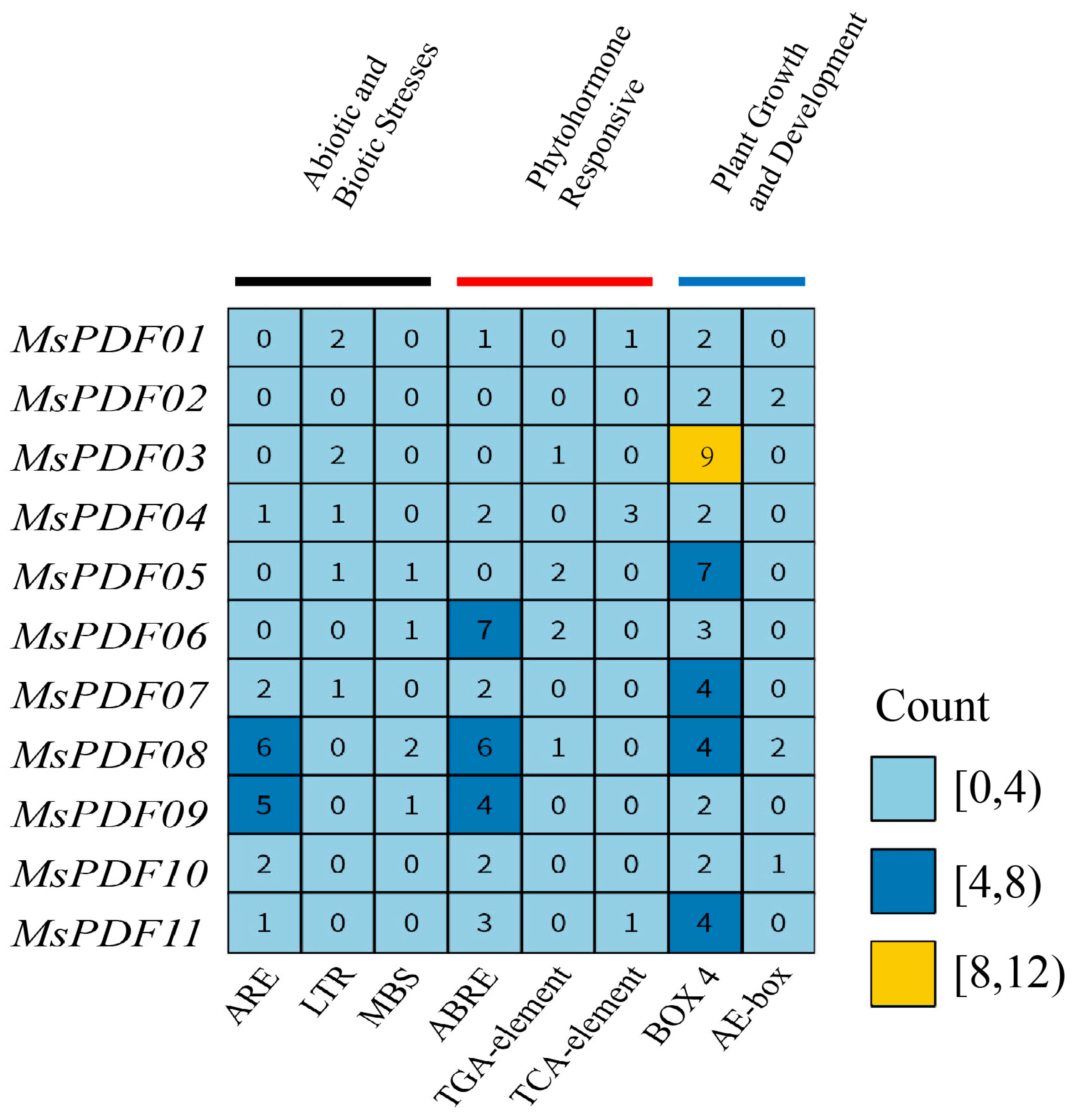
2.4. Analysis of Cis-Acting Elements in the Promoter Region of the PDF Gene Family in Alfalfa
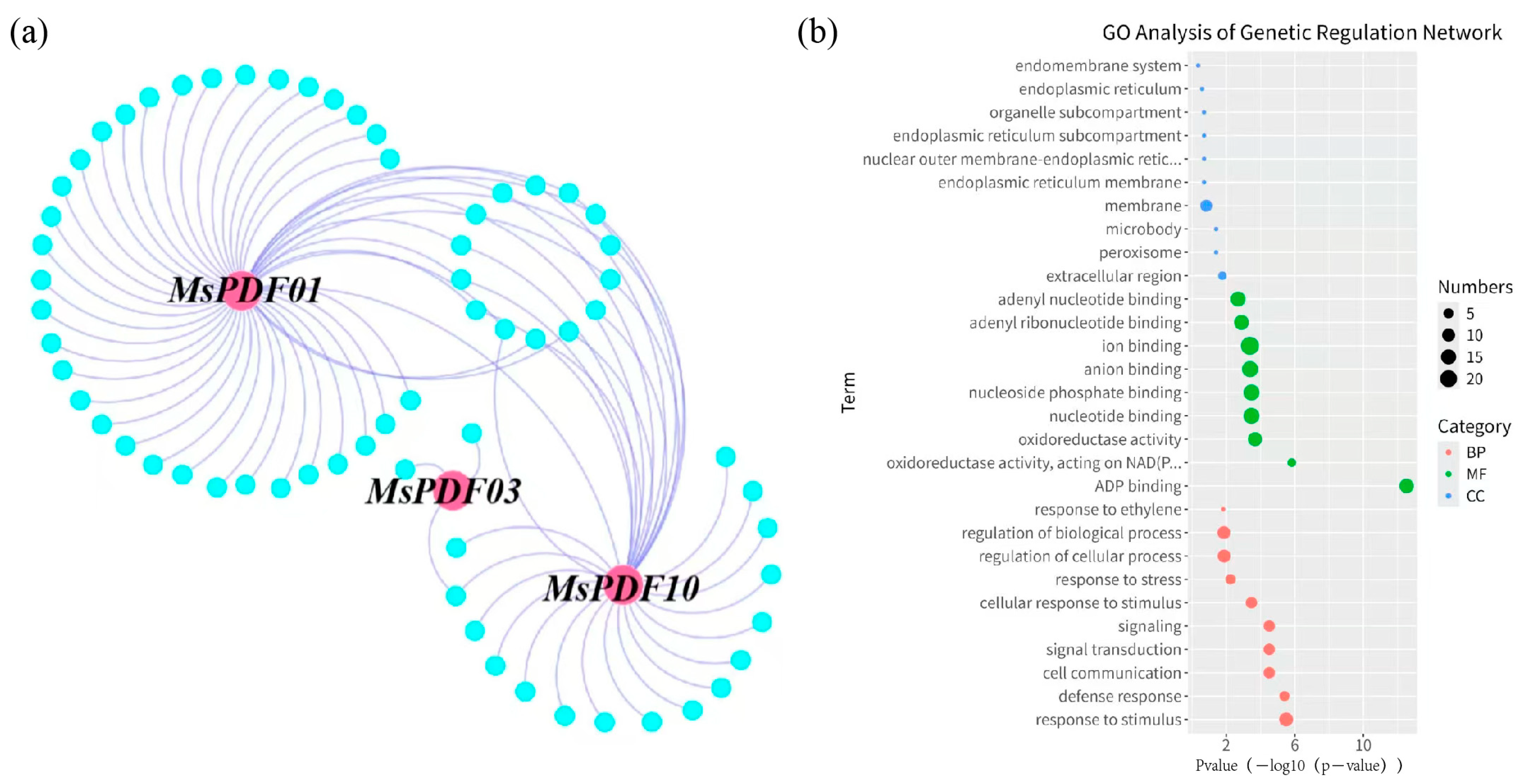
2.5. Regulatory Network and GO Analysis of MsPDFs Genes in Alfalfa
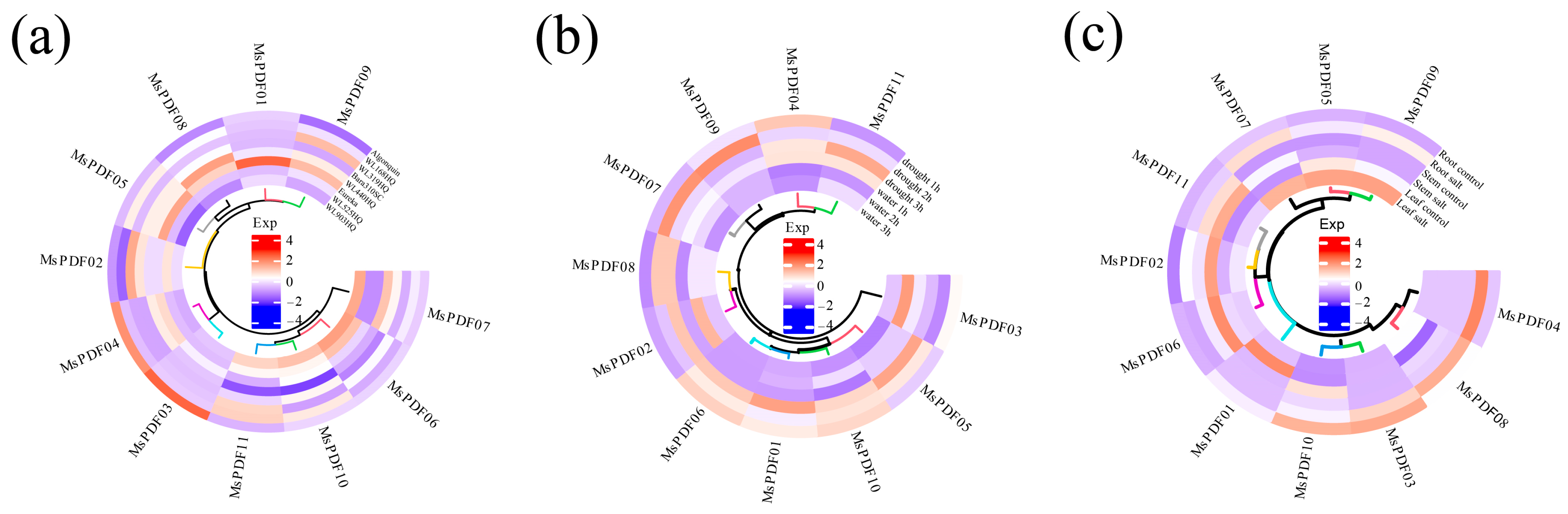
2.6. Expression Analysis of PDF Genes in Alfalfa Under Abiotic Stress
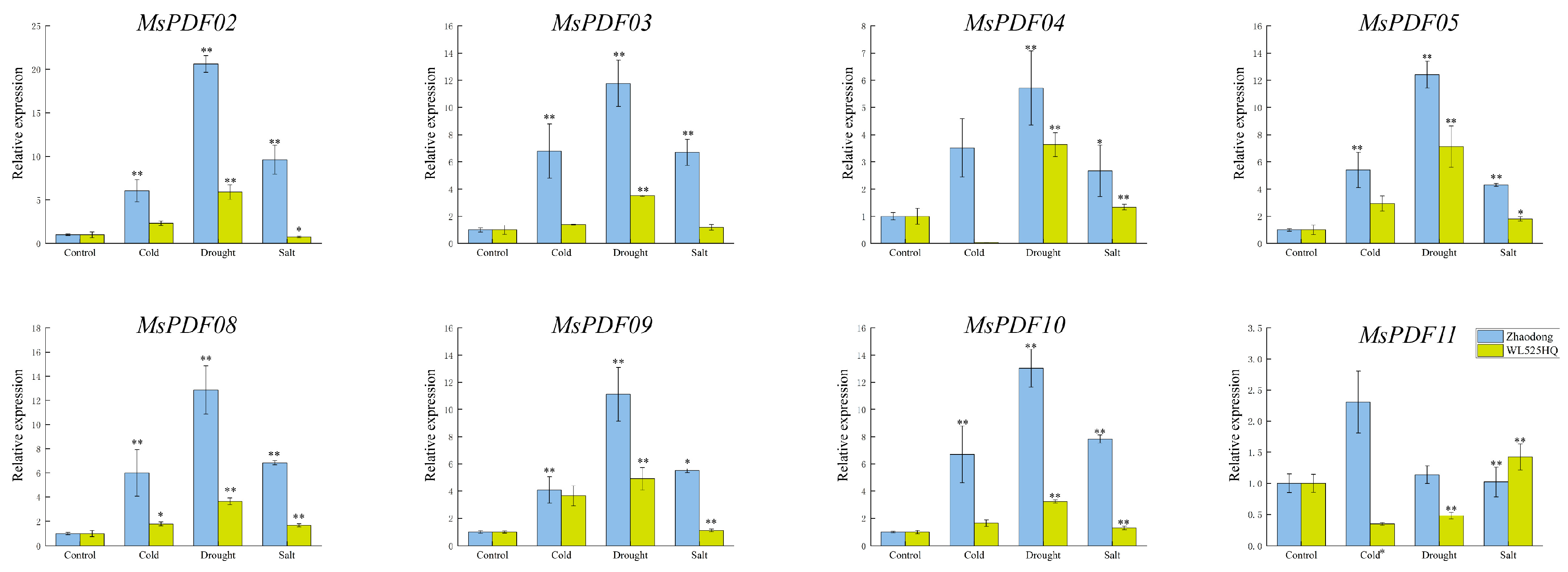
2.7. Expression Analysis of MsPDF Genes Under Cold, Drought, and Salinity Stress
2.8. Structure Prediction of Proteins Encoded by the MsPDF Genes
3. Discussion
4. Materials and Methods
4.1. Identification and Characterization of MsPDF Genes
4.2. Phylogenetic Analysis of the MsPDFs, AtPDFs and OsPDFs
4.3. Analysis of Conserved Motifs and Gene Structures of MsPDF Genes
4.4. Analysis of Cis-Acting Elements in the Promoters of MsPDF Genes
4.5. Gene Regulatory Network Analysis and Gene Ontology (GO) Annotation Analysis of MsPDF Genes
4.6. Expression Analysis of MsPDF Genes in Response to Abiotic Stresses
4.7. qRT-PCR Analysis
4.8. Prediction of the Protein Structures of MsPDF Genes
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Vriens, K.; Cammue, B.P.; Thevissen, K. Antifungal plant defensins: Mechanisms of action and production. Molecules 2014, 19, 12280–12303. [Google Scholar] [CrossRef] [PubMed]
- Lay, F.T.; Anderson, M.A. Defensins--components of the innate immune system in plants. Curr. Protein Pept. Sci. 2005, 6, 85–101. [Google Scholar] [CrossRef]
- Fant, F.; Vranken, W.; Broekaert, W.; Borremans, F. Determination of the three-dimensional solution structure of Raphanus sativus antifungal protein 1 by 1H NMR. J. Mol. Biol. 1998, 279, 257–270. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.J.; Cheng, C.S.; Lai, S.M.; Hsu, M.P.; Chen, C.S.; Lyu, P.C. Solution structure of the plant defensin VrD1 from mung bean and its possible role in insecticidal activity against bruchids. Proteins 2006, 63, 777–786. [Google Scholar] [CrossRef] [PubMed]
- Almeida, M.S.; Cabral, K.M.; Kurtenbach, E.; Almeida, F.C.; Valente, A.P. Solution structure of Pisum sativum defensin 1 by high resolution NMR: Plant defensins, identical backbone with different mechanisms of action. J. Mol. Biol. 2002, 315, 749–757. [Google Scholar] [CrossRef]
- García-Olmedo, F.; Molina, A.; Alamillo, J.M.; Rodríguez-Palenzuéla, P. Plant defense peptides. Biopolymers 1998, 47, 479–491. [Google Scholar] [CrossRef]
- Zeya, H.I.; Spitznagel, J.K. Cationic Proteins of Polymorphonuclear Leukocyte Lysosomes I. Resolution of Antibacterial and Enzymatic Activities. J. Bacteriol. 1966, 91, 750–754. [Google Scholar] [CrossRef]
- Ganz, T.; Selsted, M.E.; Szklarek, D.; Harwig, S.S.; Daher, K.; Bainton, D.F.; Lehrer, R.I. Defensins. Natural peptide antibiotics of human neutrophils. J. Clin. Investig. 1985, 76, 1427–1435. [Google Scholar] [CrossRef]
- Insect immunity: Isolation from immune blood of the dipteran Phormia terranovae of two insect antibacterial peptides with sequence homology to rabbit lung macrophage bactericidal peptides. Proc. Natl. Acad. Sci. USA 1989, 86, 3321. [CrossRef]
- Mendez, E.; Moreno, A.; Colilla, F.; Pelaez, F.; Limas, G.G.; Mendez, R.; Soriano, F.; Salinas, M.; de Haro, C. Primary structure and inhibition of protein synthesis in eukaryotic cell-free system of a novel thionin, gamma-hordothionin, from barley endosperm. Eur. J. Biochem. 1990, 194, 533–539. [Google Scholar] [CrossRef]
- Colilla, F.J.; Rocher, A.; Mendez, E. gamma-Purothionins: Amino acid sequence of two polypeptides of a new family of thionins from wheat endosperm. FEBS Lett. 1990, 270, 191–194. [Google Scholar] [CrossRef] [PubMed]
- Terras, F.R.; Schoofs, H.M.; De Bolle, M.F.; Van Leuven, F.; Rees, S.B.; Vanderleyden, J.; Cammue, B.P.; Broekaert, W.F. Analysis of two novel classes of plant antifungal proteins from radish (Raphanus sativus L.) seeds. J. Biol. Chem. 1992, 267, 15301–15309. [Google Scholar] [CrossRef]
- Carrasco, L.; Vázquez, D.; Hernández-Lucas, C.; Carbonero, P.; García-Olmedo, F. Thionins: Plant peptides that modify membrane permeability in cultured mammalian cells. Eur. J. Biochem. 1981, 116, 185–189. [Google Scholar] [CrossRef]
- Terras, F.R.; Eggermont, K.; Kovaleva, V.; Raikhel, N.V.; Osborn, R.W.; Kester, A.; Rees, S.B.; Torrekens, S.; Van Leuven, F.; Vanderleyden, J.; et al. Small cysteine-rich antifungal proteins from radish: Their role in host defense. Plant Cell 1995, 7, 573–588. [Google Scholar] [CrossRef] [PubMed]
- Penninckx, I.A.; Eggermont, K.; Terras, F.R.; Thomma, B.P.; De Samblanx, G.W.; Buchala, A.; Métraux, J.P.; Manners, J.M.; Broekaert, W.F. Pathogen-induced systemic activation of a plant defensin gene in Arabidopsis follows a salicylic acid-independent pathway. Plant Cell 1996, 8, 2309–2323. [Google Scholar] [CrossRef]
- Whitbred, J.M.; Schuler, M.A. Molecular Characterization of CYP73A9 andCYP82A1 P450 Genes Involved in Plant Defense in Pea. Plant Physiol. 2000, 124, 47–58. [Google Scholar] [CrossRef] [PubMed]
- Zhu-Salzman, K.; Salzman, R.A.; Ahn, J.-E.; Koiwa, H. Transcriptional Regulation of Sorghum Defense Determinants against a Phloem-Feeding Aphid. Plant Physiol. 2004, 134, 420–431. [Google Scholar] [CrossRef]
- Hanks, J.N.; Snyder, A.K.; Graham, M.A.; Shah, R.K.; Blaylock, L.A.; Harrison, M.J.; Shah, D.M. Defensin gene family in Medicago truncatula: Structure, expression and induction by signal molecules. Plant Mol. Biol. 2005, 58, 385–399. [Google Scholar] [CrossRef]
- Finkina, E.I.; Bogdanov, I.V.; Shevchenko, O.V.; Fateeva, S.I.; Ignatova, A.A.; Balandin, S.V.; Ovchinnikova, T.V. Immunomodulatory Effects of the Tobacco Defensin NaD1. Antibiotics 2024, 13, 1101. [Google Scholar] [CrossRef]
- Tantong, S.; Pringsulaka, O.; Weerawanich, K.; Meeprasert, A.; Rungrotmongkol, T.; Sarnthima, R.; Roytrakul, S.; Sirikantaramas, S. Two novel antimicrobial defensins from rice identified by gene coexpression network analyses. Peptides 2016, 84, 7–16. [Google Scholar] [CrossRef]
- Kerenga, B.K.; McKenna, J.A.; Harvey, P.J.; Quimbar, P.; Garcia-Ceron, D.; Lay, F.T.; Phan, T.K.; Veneer, P.K.; Vasa, S.; Parisi, K.; et al. Salt-Tolerant Antifungal and Antibacterial Activities of the Corn Defensin ZmD32. Front. Microbiol. 2019, 10, 795. [Google Scholar] [CrossRef]
- Bártová, V.; Bárta, J.; Jarošová, M. Antifungal and antimicrobial proteins and peptides of potato (Solanum tuberosum L.) tubers and their applications. Appl. Microbiol. Biotechnol. 2019, 103, 5533–5547. [Google Scholar] [CrossRef] [PubMed]
- Du, B.; Haensch, R.; Alfarraj, S.; Rennenberg, H. Strategies of plants to overcome abiotic and biotic stresses. Biol. Rev. Camb. Philos. Soc. 2024, 99, 1524–1536. [Google Scholar] [CrossRef]
- Tang, R.; Tan, H.; Dai, Y.; Li, L.; Huang, Y.; Yao, H.; Cai, Y.; Yu, G. Application of antimicrobial peptides in plant protection: Making use of the overlooked merits. Front. Plant Sci. 2023, 14, 1139539. [Google Scholar] [CrossRef] [PubMed]
- Manzanares, P.; Giner-Llorca, M.; Marcos, J.F.; Garrigues, S. Fighting pathogenic yeasts with plant defensins and anti-fungal proteins from fungi. Appl. Microbiol. Biotechnol. 2024, 108, 277. [Google Scholar] [CrossRef]
- Deepthi, V.; Mohanakumar, K.P.; Rajamma, U. Efficacy of defensins as neutralizing agents against the deadly SARS-CoV-2. J. Biomol. Struct. Dyn. 2023, 41, 2911–2925. [Google Scholar] [CrossRef] [PubMed]
- Kushmerick, C.; de Souza Castro, M.; Santos Cruz, J.; Bloch, C., Jr.; Beirão, P.S. Functional and structural features of gamma-zeathionins, a new class of sodium channel blockers. FEBS Lett. 1998, 440, 302–306. [Google Scholar] [CrossRef]
- Spelbrink, R.G.; Dilmac, N.; Allen, A.; Smith, T.J.; Shah, D.M.; Hockerman, G.H. Differential antifungal and calcium channel-blocking activity among structurally related plant defensins. Plant Physiol. 2004, 135, 2055–2067. [Google Scholar] [CrossRef]
- Vriens, K.; Peigneur, S.; De Coninck, B.; Tytgat, J.; Cammue, B.P.A.; Thevissen, K. The antifungal plant defensin AtPDF2.3 from Arabidopsis thaliana blocks potassium channels. Sci. Rep. 2016, 6, 32121. [Google Scholar] [CrossRef]
- Kovalchuk, N.; Li, M.; Wittek, F.; Reid, N.; Singh, R.; Shirley, N.; Ismagul, A.; Eliby, S.; Johnson, A.; Milligan, A.S.; et al. Defensin promoters as potential tools for engineering disease resistance in cereal grains. Plant Biotechnol. J. 2010, 8, 47–64. [Google Scholar] [CrossRef]
- Zhou, Q.; Luo, D.; Chai, X.; Wu, Y.; Wang, Y.; Nan, Z.; Yang, Q.; Liu, W.; Liu, Z. Multiple Regulatory Networks Are Activated during Cold Stress in Medicago sativa L. Int. J. Mol. Sci. 2018, 19, 3169. [Google Scholar] [CrossRef] [PubMed]
- Quan, W.; Liu, X. Tandem mass tag (TMT)-based quantitative proteomics analysis reveals the different responses of contrasting alfalfa varieties to drought stress. BMC Genom. 2024, 25, 806. [Google Scholar] [CrossRef]
- Xie, J.; Li, Y.; Jiang, G.; Sun, H.; Liu, X.; Han, L. Seed color represents salt resistance of alfalfa seeds (Medicago sativa L.): Based on the analysis of germination characteristics, seedling growth and seed traits. Front. Plant Sci. 2023, 14, 1104948. [Google Scholar] [CrossRef] [PubMed]
- Mäser, P.; Thomine, S.; Schroeder, J.I.; Ward, J.M.; Hirschi, K.; Sze, H.; Talke, I.N.; Amtmann, A.; Maathuis, F.J.; Sanders, D.; et al. Phylogenetic relationships within cation transporter families of Arabidopsis. Plant Physiol. 2001, 126, 1646–1667. [Google Scholar] [CrossRef]
- Dong, Y.; Wang, Y.; Tang, M.; Chen, W.; Chai, Y.; Wang, W. Bioinformatic analysis of wheat defensin gene family and function verification of candidate genes. Front. Plant Sci. 2023, 14, 1279502. [Google Scholar] [CrossRef]
- Giacomelli, L.; Nanni, V.; Lenzi, L.; Zhuang, J.; Dalla Serra, M.; Banfield, M.J.; Town, C.D.; Silverstein, K.A.; Baraldi, E.; Moser, C. Identification and characterization of the defensin-like gene family of grapevine. Mol. Plant-Microbe Interact. MPMI 2012, 25, 1118–1131. [Google Scholar] [CrossRef]
- Lee, S.C.; Luan, S. ABA signal transduction at the crossroad of biotic and abiotic stress responses. Plant Cell Environ. 2012, 35, 53–60. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.; Muhae-Ud-Din, G.; Liu, T.; Chen, W.; Liu, C.; Gao, L. Wheat Varietal Response to Tilletia controversa J. G. Kühn Using qRT-PCR and Laser Confocal Microscopy. Genes 2021, 12, 425. [Google Scholar] [CrossRef]
- Krishnaswamy, S.S.; Srivastava, S.; Mohammadi, M.; Rahman, M.H.; Deyholos, M.K.; Kav, N.N.V. Transcriptional profiling of pea ABR17 mediated changes in gene expression in Arabidopsis thaliana. BMC Plant Biol. 2008, 8, 91. [Google Scholar] [CrossRef]
- Dai, X.; Xu, Y.; Ma, Q.; Xu, W.; Wang, T.; Xue, Y.; Chong, K. Overexpression of an R1R2R3 MYB gene, OsMYB3R-2, increases tolerance to freezing, drought, and salt stress in transgenic Arabidopsis. Plant Physiol. 2007, 143, 1739–1751. [Google Scholar] [CrossRef]
- Cheng, M.C.; Liao, P.M.; Kuo, W.W.; Lin, T.P. The Arabidopsis ETHYLENE RESPONSE FACTOR1 regulates abiotic stress-responsive gene expression by binding to different cis-acting elements in response to different stress signals. Plant Physiol. 2013, 162, 1566–1582. [Google Scholar] [CrossRef] [PubMed]
- Sharma, A.; Sidhu, G.P.S.; Araniti, F.; Bali, A.S.; Shahzad, B.; Tripathi, D.K.; Brestic, M.; Skalicky, M.; Landi, M. The Role of Salicylic Acid in Plants Exposed to Heavy Metals. Molecules 2020, 25, 540. [Google Scholar] [CrossRef] [PubMed]
- Gu, T.; Qi, Z.; Wang, Y.; Chen, S.; Yan, J.; Qiu, H.; Yu, Y.; Fang, Z.; Wang, J.; Gong, J. An endophytic fungus interacts with the defensin-like protein OsCAL1 to regulate cadmium allocation in rice. Mol. Plant 2024, 17, 312–324. [Google Scholar] [CrossRef] [PubMed]
- Kaur, G.; Tak, Y.; Asthir, B. Salicylic acid: A key signal molecule ameliorating plant stresses. Cereal Res. Commun. 2022, 50, 617–626. [Google Scholar] [CrossRef]
- Luo, Y.; Song, Y. Mechanism of Antimicrobial Peptides: Antimicrobial, Anti-Inflammatory and Antibiofilm Activities. Int. J. Mol. Sci. 2021, 22, 11401. [Google Scholar] [CrossRef]
- Mostofa, M.G.; Rahman, M.M.; Ghosh, T.K.; Kabir, A.H.; Abdelrahman, M.; Rahman Khan, M.A.; Mochida, K.; Tran, L.P. Potassium in plant physiological adaptation to abiotic stresses. Plant Physiol. Biochem. PPB 2022, 186, 279–289. [Google Scholar] [CrossRef]
- Tong, T.; Li, Q.; Jiang, W.; Chen, G.; Xue, D.; Deng, F.; Zeng, F.; Chen, Z.H. Molecular Evolution of Calcium Signaling and Transport in Plant Adaptation to Abiotic Stress. Int. J. Mol. Sci. 2021, 22, 12308. [Google Scholar] [CrossRef]
- Yang, T.; Chaudhuri, S.; Yang, L.; Du, L.; Poovaiah, B.W. A calcium/calmodulin-regulated member of the receptor-like kinase family confers cold tolerance in plants. J. Biol. Chem. 2010, 285, 7119–7126. [Google Scholar] [CrossRef]
- Du, Y.Y.; Wang, P.C.; Chen, J.; Song, C.P. Comprehensive functional analysis of the catalase gene family in Arabidopsis thaliana. J. Integr. Plant Biol. 2008, 50, 1318–1326. [Google Scholar] [CrossRef]
- Yu, W.; Kong, G.; Ya, H.; He, L.; Wu, Y.; Zhang, H. Comprehensive Analysis of the Catalase (CAT) Gene Family and Expression Patterns in Rubber Tree (Hevea brasiliensis) under Various Abiotic Stresses and Multiple Hormone Treatments. Int. J. Mol. Sci. 2023, 25, 70. [Google Scholar] [CrossRef]
- Wei, Y.; Chen, H.; Wang, L.; Zhao, Q.; Wang, D.; Zhang, T. Cold acclimation alleviates cold stress-induced PSII inhibition and oxidative damage in tobacco leaves. Plant Signal. Behav. 2022, 17, 2013638. [Google Scholar] [CrossRef]
- Singh, V.; Hallan, V.; Pati, P.K. Withania somnifera osmotin (WsOsm) confers stress tolerance in tobacco and establishes novel interactions with the defensin protein (WsDF). Physiol. Plant. 2024, 176, e14513. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Liu, Y.; Hao, X.; Chen, Y.; Wang, Z.; Shen, Y. Enhancing plant defensins in a desert shrub: Exploring a regulatory pathway of AnWRKY29. Int. J. Biol. Macromol. 2024, 270, 132259. [Google Scholar] [CrossRef]
- Wu, Y.R.; Lin, Y.C.; Chuang, H.W. Laminarin modulates the chloroplast antioxidant system to enhance abiotic stress tolerance partially through the regulation of the defensin-like gene expression. Plant Sci. Int. J. Exp. Plant Biol. 2016, 247, 83–92. [Google Scholar] [CrossRef] [PubMed]
- Meng, L.; Feng, Y.; Zhao, M.; Jang, T.; Bi, H.; Ai, X. Hydrogen peroxide mediates melatonin-induced chilling tolerance in cucumber seedlings. Plant Cell Rep. 2024, 43, 279. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Zeng, Y.; Yang, Y.; Huang, L.; Tang, B.; Zhang, H.; Hao, F.; Liu, W.; Li, Y.; Liu, Y.; et al. Allele-aware chromosome-level genome assembly and efficient transgene-free genome editing for the autotetraploid cultivated alfalfa. Nat. Commun. 2020, 11, 2494. [Google Scholar] [CrossRef]
- Finn, R.D.; Coggill, P.; Eberhardt, R.Y.; Eddy, S.R.; Mistry, J.; Mitchell, A.L.; Potter, S.C.; Punta, M.; Qureshi, M.; Sangrador-Vegas, A.; et al. The Pfam protein families database: Towards a more sustainable future. Nucleic Acids Res. 2016, 44, D279–D285. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [CrossRef]
- Minh, B.Q.; Nguyen, M.A.; von Haeseler, A. Ultrafast approximation for phylogenetic bootstrap. Mol. Biol. Evol. 2013, 30, 1188–1195. [Google Scholar] [CrossRef]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Wu, Y.; Li, J.; Wang, X.; Zeng, Z.; Xu, J.; Liu, Y.; Feng, J.; Chen, H.; He, Y.; et al. TBtools-II: A “one for all, all for one” bioinformatics platform for biological big-data mining. Mol. Plant 2023, 16, 1733–1742. [Google Scholar] [CrossRef] [PubMed]
- Lee, T.; Yang, S.; Kim, E.; Ko, Y.; Hwang, S.; Shin, J.; Shim, J.E.; Shim, H.; Kim, H.; Kim, C.; et al. AraNet v2: An improved database of co-functional gene networks for the study of Arabidopsis thaliana and 27 other nonmodel plant species. Nucleic Acids Res. 2015, 43, D996–D1002. [Google Scholar] [CrossRef] [PubMed]
- Doncheva, N.T.; Morris, J.H.; Gorodkin, J.; Jensen, L.J. Cytoscape StringApp: Network Analysis and Visualization of Proteomics Data. J. Proteome Res. 2019, 18, 623–632. [Google Scholar] [CrossRef]
- Alexa, A.; Rahnenführer, J.; Lengauer, T. Improved scoring of functional groups from gene expression data by decorrelating GO graph structure. Bioinformatics 2006, 22, 1600–1607. [Google Scholar] [CrossRef]
- Zhang, X.; Yang, H.; Li, M.; Bai, Y.; Chen, C.; Guo, D.; Guo, C.; Shu, Y. A Pan-Transcriptome Analysis Indicates Efficient Downregulation of the FIB Genes Plays a Critical Role in the Response of Alfalfa to Cold Stress. Plants 2022, 11, 3148. [Google Scholar] [CrossRef]
- Medina, C.A.; Samac, D.A.; Yu, L.X. Pan-transcriptome identifying master genes and regulation network in response to drought and salt stresses in Alfalfa (Medicago sativa L.). Sci. Rep. 2021, 11, 17203. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Zhang, Y.; Skolnick, J. Scoring function for automated assessment of protein structure template quality. Proteins 2004, 57, 702–710. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Ma, P.; Li, D.W.; Brüschweiler, R. Predicting protein flexibility with AlphaFold. Proteins 2023, 91, 847–855. [Google Scholar] [CrossRef] [PubMed]
- Bryant, P.; Noé, F. Improved protein complex prediction with AlphaFold-multimer by denoising the MSA profile. PLoS Comput. Biol. 2024, 20, e1012253. [Google Scholar] [CrossRef] [PubMed]

| Name | Arabidopsis | Chromosomal Locations | Group | Intron | Length(aa) | |
|---|---|---|---|---|---|---|
| Homologous Gene | ||||||
| Gene | Gene | |||||
| Accession NO. | Name | |||||
| MsPDF01 | AT1G19610 | PDF1.4 | chr8.1:84174234-84174563 | I | 1 | 79 |
| MsPDF02 | AT1G55010 | PDF1.5 | chr8.1:83983356-83983809 | I | 1 | 79 |
| MsPDF03 | AT1G19610 | PDF1.4 | chr8.3:76391825-76392231 | I | 1 | 99 |
| MsPDF04 | AT1G19610 | PDF1.4 | chr8.3:76401833-76402163 | I | 1 | 79 |
| MsPDF05 | AT2G26010 | PDF1.3 | chr2.1:61335159-61335818 | II | 1 | 78 |
| MsPDF06 | AT1G61070 | PDF2.4 | chr2.3:18266357-18267529 | II | 1 | 73 |
| MsPDF07 | AT1G61070 | PDF2.4 | chr2.4:59797715-59798955 | II | 1 | 77 |
| MsPDF08 | AT5G63660 | PDF2.5 | chr3.2:81190189-81190502 | III | 1 | 73 |
| MsPDF09 | AT2G02100 | PDF2.2 | chr4.3:7552947-7553374 | III | 1 | 74 |
| MsPDF10 | AT2G02130 | PDF2.3 | chr8.4:32871559-32871973 | III | 1 | 76 |
| MsPDF11 | AT2G02100 | PDF2.2 | chr8.1:31454195-31454834 | III | 1 | 81 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guo, M.; Chen, X.; Li, S.; Tian, J.; Huang, W.; Shu, Y. Identification of the Plant Defensin (MsPDF) Gene Family in Medicago sativa and Analysis of Expression Patterns Under Abiotic Stress. Plants 2025, 14, 1312. https://doi.org/10.3390/plants14091312
Guo M, Chen X, Li S, Tian J, Huang W, Shu Y. Identification of the Plant Defensin (MsPDF) Gene Family in Medicago sativa and Analysis of Expression Patterns Under Abiotic Stress. Plants. 2025; 14(9):1312. https://doi.org/10.3390/plants14091312
Chicago/Turabian StyleGuo, Meiyan, Xiuhua Chen, Shuaixian Li, Jiang Tian, Wangqi Huang, and Yongjun Shu. 2025. "Identification of the Plant Defensin (MsPDF) Gene Family in Medicago sativa and Analysis of Expression Patterns Under Abiotic Stress" Plants 14, no. 9: 1312. https://doi.org/10.3390/plants14091312
APA StyleGuo, M., Chen, X., Li, S., Tian, J., Huang, W., & Shu, Y. (2025). Identification of the Plant Defensin (MsPDF) Gene Family in Medicago sativa and Analysis of Expression Patterns Under Abiotic Stress. Plants, 14(9), 1312. https://doi.org/10.3390/plants14091312







