An AP2-Family Gene Correlates with the Double-Flower Trait in Petunia × hybrida
Abstract
1. Introduction
2. Results
2.1. Inheritance of Double Flower Trait
2.2. Morphology of Floral Organs and Their Surface Cell
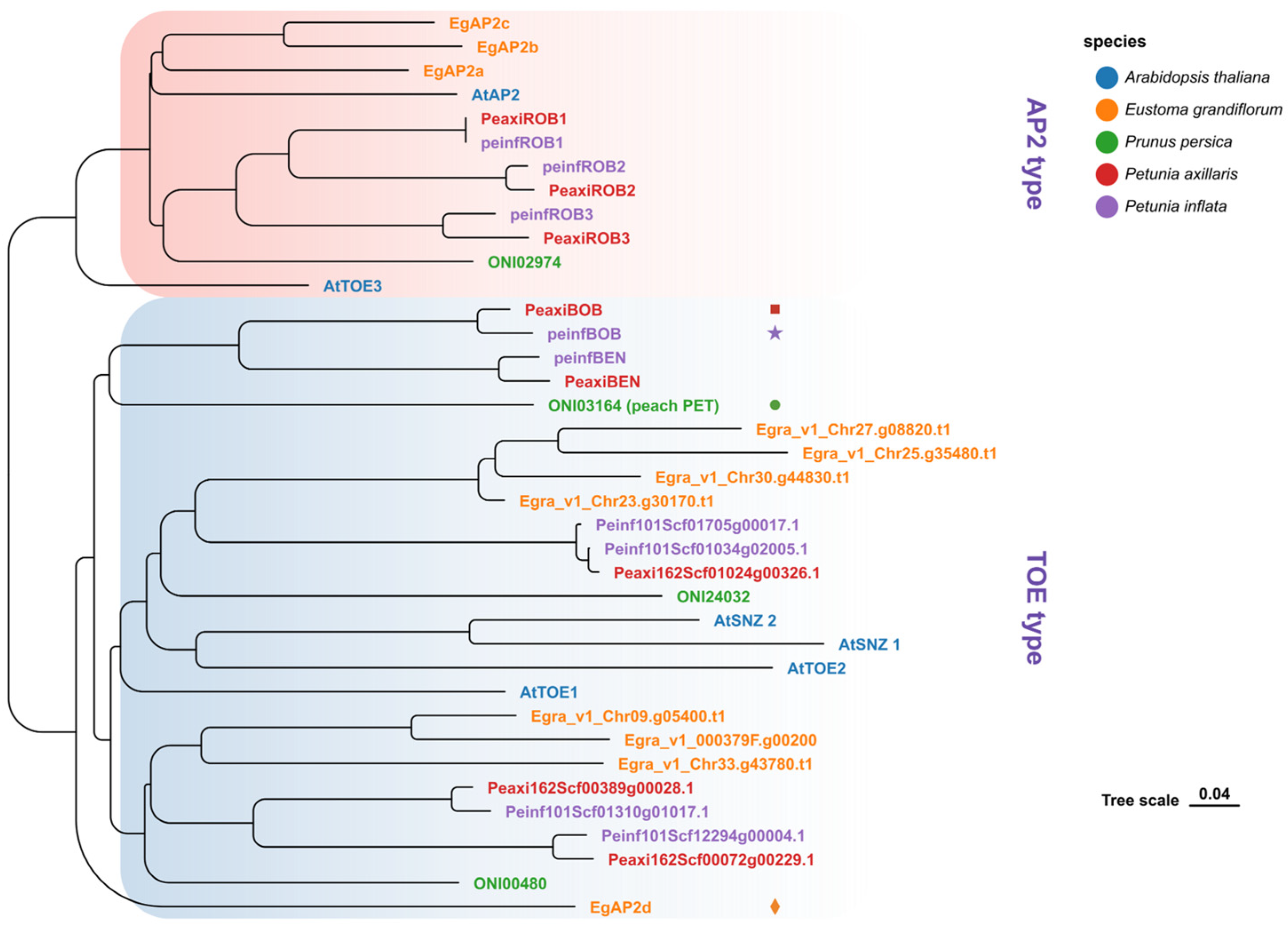
2.3. Sequence Variation of Double-Flower-Related Genes in Petunia × hybrida
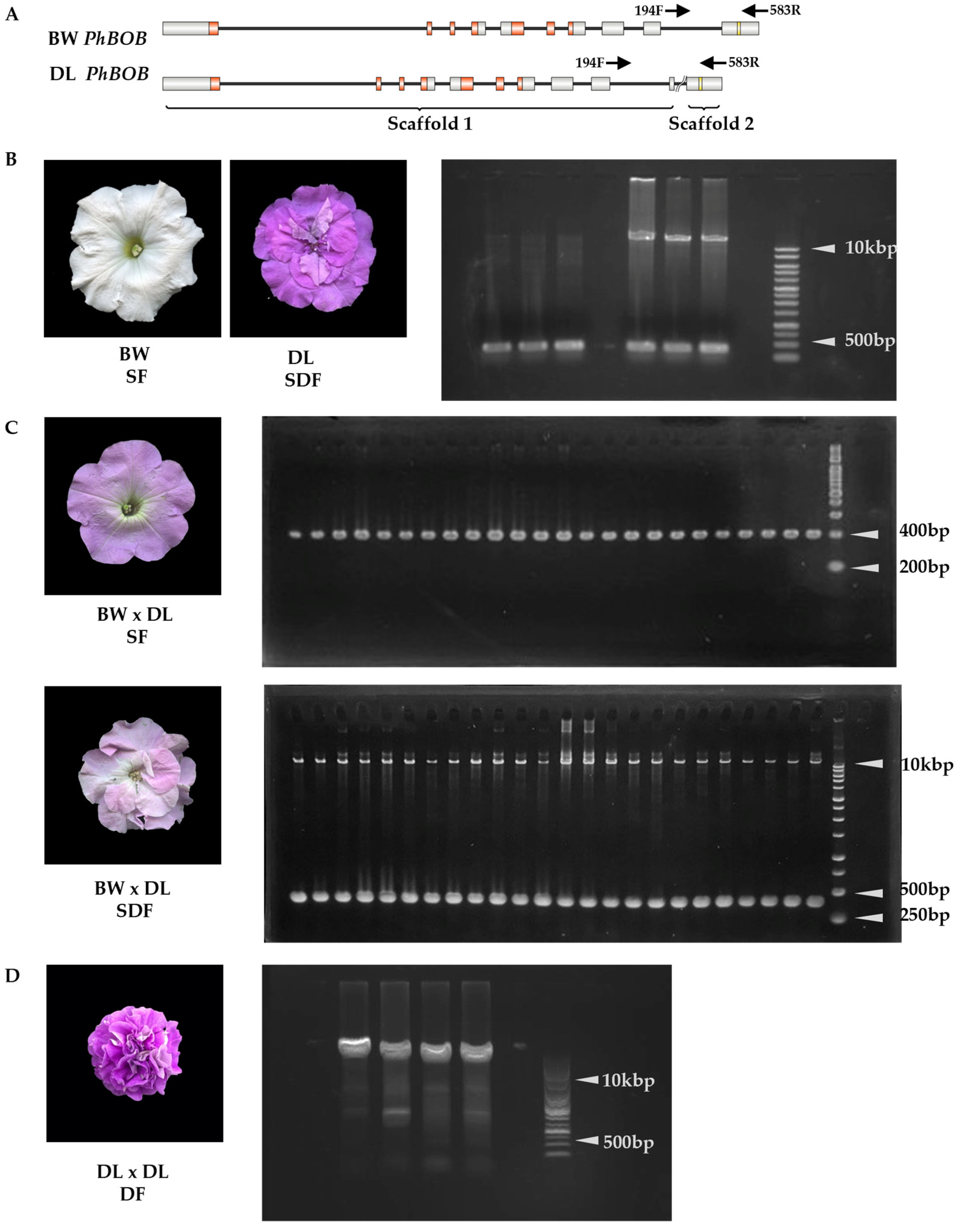
2.4. The 10 kbp Insertion in PhBOB Is Highly Linked to Double-Flowering in Petunias
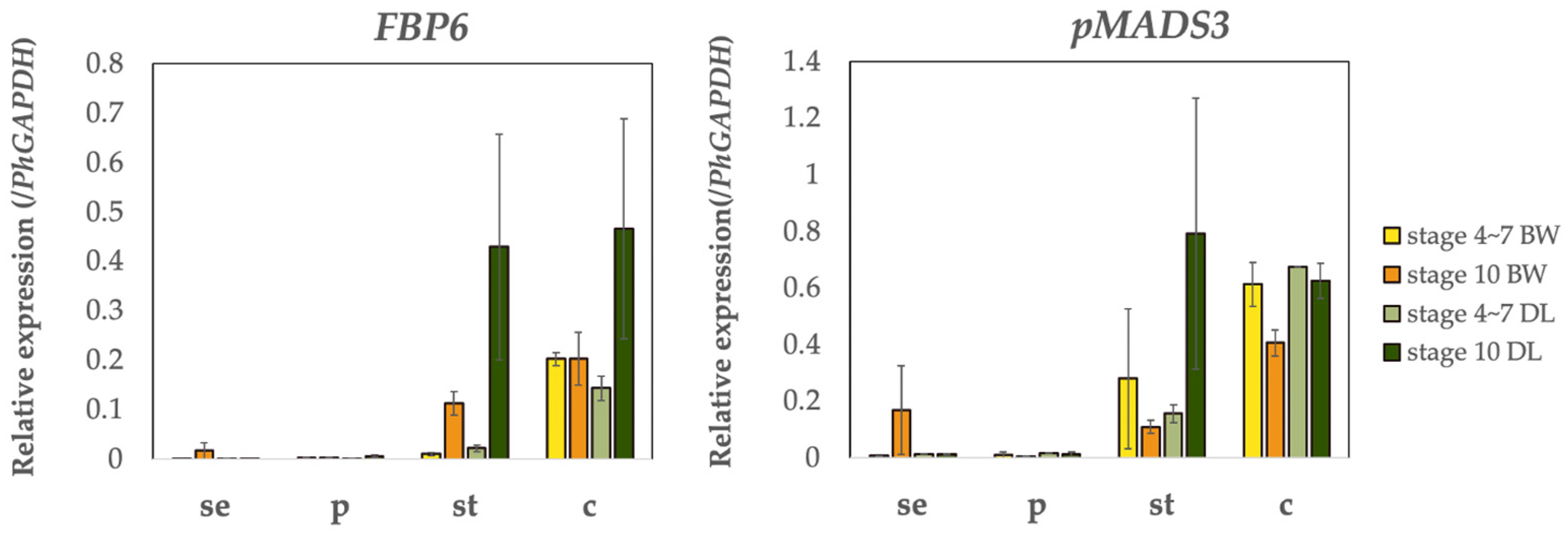
2.5. Expression of MADS-Box and AP2 Genes in BW and DL
3. Discussion
3.1. PhBOB 10 kb Insertion Is Responsible for the Double-Flower Trait in Petunia
3.2. PhBOB 10 kb Insertion Is Highly Conserved in Solanaceae and Contains CMC-EnSpm Transposon Sequences
3.3. The Expression of C-Class Genes (FBP6 and pMADS3) Is Controlled by PhBOB
3.4. AP2 Genes in the TOE-Type Clade Are Associated with the Double-Flower Trait
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. Paraffin Sectioning
4.3. Scanning Electron Microscopy (SEM) Observation
4.4. Whole-Genome Sequencing
4.5. Identification and Screening of AP2 Family Genes in Petunia and Other Ornamental Plants
4.6. Sequence Alignment and Homolog Identification of ABC-Class Genes in Petunia × hybrida
4.7. Linkage Analysis
4.8. Gene Expression Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Souer, E.; Rebocho, A.B.; Bliek, M.; Kusters, E.; De Bruin, R.A.M.; Koes, R. Patterning of Inflorescences and Flowers by the F-Box Protein DOUBLE TOP and the LEAFY Homolog ABERRANT LEAF AND FLOWER of Petunia. Plant Cell 2008, 20, 2033–2048. [Google Scholar] [CrossRef] [PubMed]
- Bombarely, A.; Moser, M.; Amrad, A.; Bao, M.; Bapaume, L.; Barry, C.S.; Bliek, M.; Boersma, M.R.; Borghi, L.; Bruggmann, R.; et al. Insight into the Evolution of the Solanaceae from the Parental Genomes of Petunia Hybrida. Nat. Plants 2016, 2, 16074. [Google Scholar] [CrossRef] [PubMed]
- Gautam, P.; Terfa, M.T.; Olsen, J.E.; Torre, S. Red and Blue Light Effects on Morphology and Flowering of Petunia×hybrida. Sci. Hortic. 2015, 184, 171–178. [Google Scholar] [CrossRef]
- Chopy, M.; Cavallini-Speisser, Q.; Chambrier, P.; Morel, P.; Just, J.; Hugouvieux, V.; Rodrigues Bento, S.; Zubieta, C.; Vandenbussche, M.; Monniaux, M. Cell layer–specific expression of the homeotic MADS-box transcription factor PhDEF contributes to modular petal morphogenesis in petunia. Plant Cell 2024, 36, 324–345. [Google Scholar] [CrossRef]
- Geitmann, A. Petunia. Evolutionary, Developmental and Physiological Genetics. Ann. Bot. 2011, 107, vi. [Google Scholar] [CrossRef]
- Krizek, B.A.; Fletcher, J.C. Molecular Mechanisms of Flower Development: An Armchair Guide. Nat. Rev. Genet. 2005, 6, 688–698. [Google Scholar] [CrossRef]
- Schwarz-Sommer, Z.; Huijser, P.; Nacken, W.; Saedler, H.; Sommer, H. Genetic Control of Flower Development by Homeotic Genes in Antirrhinum majus. Science 1990, 250, 931–936. [Google Scholar] [CrossRef]
- Pelaz, S.; Ditta, G.S.; Baumann, E.; Wisman, E.; Yanofsky, M.F. B and C floral Organ Identity Functions Require SEPALLATA MADS-Box Genes. Nature 2000, 405, 200–203. [Google Scholar] [CrossRef]
- Pinyopich, A.; Ditta, G.S.; Savidge, B.; Liljegren, S.J.; Baumann, E.; Wisman, E.; Yanofsky, M.F. Assessing the Redundancy of MADS-Box Genes during Carpel and Ovule Development. Nature 2003, 424, 85–88. [Google Scholar] [CrossRef]
- Ehlers, K.; Bhide, A.S.; Tekleyohans, D.G.; Wittkop, B.; Snowdon, R.J.; Becker, A. The MADS Box Genes ABS, SHP1, and SHP2 Are Essential for the Coordination of Cell Divisions in Ovule and Seed Coat Development and for Endosperm Formation in Arabidopsis thaliana. PLoS ONE 2016, 11, e0165075. [Google Scholar] [CrossRef]
- Mizzotti, C.; Ezquer, I.; Paolo, D.; Rueda-Romero, P.; Guerra, R.F.; Battaglia, R.; Rogachev, I.; Aharoni, A.; Kater, M.M.; Caporali, E.; et al. SEEDSTICK Is a Master Regulator of Development and Metabolism in the Arabidopsis Seed Coat. PLoS Genet. 2014, 10, e1004856. [Google Scholar] [CrossRef] [PubMed]
- Bowman, J.L.; Smyth, D.R.; Meyerowitz, E.M. Genes Directing Flower Development in Arabidopsis. Plant Cell 1989, 1, 37–52. [Google Scholar] [CrossRef] [PubMed]
- Yao, J.; Xu, J.; Tomes, S.; Cui, W.; Luo, Z.; Deng, C.; Ireland, H.S.; Schaffer, R.J.; Gleave, A.P. Ectopic Expression of the PISTILLATAHomologous Md PI Inhibits Fruit Tissue Growth and Changes Fruit Shape in Apple. Plant Direct 2018, 2, e00051. [Google Scholar] [CrossRef] [PubMed]
- Ishimori, M. Genome Analysis of Eustoma Grandiflorum and Identification of the Causative Gene for the Double-Flower Trait. Ph.D. Thesis, The University of Tokyo, Tokyo, Japan, 2015. [Google Scholar]
- Gattolin, S.; Cirilli, M.; Pacheco, I.; Ciacciulli, A.; Da Silva Linge, C.; Mauroux, J.-B.; Lambert, P.; Cammarata, E.; Bassi, D.; Pascal, T.; et al. Deletion of the miR172 Target Site in a TOE-Type Gene Is a Strong Candidate Variant for Dominant Double-Flower Trait in Rosaceae. Plant J. 2018, 96, 358–371. [Google Scholar] [CrossRef]
- François, L.; Verdenaud, M.; Fu, X.; Ruleman, D.; Dubois, A.; Vandenbussche, M.; Bendahmane, A.; Raymond, O.; Just, J.; Bendahmane, M. A miR172 Target-Deficient AP2-like Gene Correlates with the Double Flower Phenotype in Roses. Sci. Rep. 2018, 8, 12912. [Google Scholar] [CrossRef]
- Gattolin, S.; Cirilli, M.; Chessa, S.; Stella, A.; Bassi, D.; Rossini, L. Mutations in Orthologous PETALOSA TOE-Type Genes Cause a Dominant Double-Flower Phenotype in Phylogenetically Distant Eudicots. J. Exp. Bot. 2020, 71, 2585–2595. [Google Scholar] [CrossRef]
- Liu, W.; Zheng, T.; Qiu, L.; Guo, X.; Li, P.; Yong, X.; Li, L.; Ahmad, S.; Wang, J.; Cheng, T.; et al. A 49-Bp Deletion of PmAP2L Results in a Double Flower Phenotype in Prunus Mume. Hortic. Res. 2024, 11, uhad278. [Google Scholar] [CrossRef]
- Magnani, E.; Sjölander, K.; Hake, S. From Endonucleases to Transcription Factors: Evolution of the AP2 DNA Binding Domain in Plants[W]. Plant Cell 2004, 16, 2265–2277. [Google Scholar] [CrossRef]
- Allen, M.D.; Yamasaki, K.; Ohme-Takagi, M.; Tateno, M.; Suzuki, M. A Novel Mode of DNA Recognition by a β-Sheet Revealed by the Solution Structure of the GCC-Box Binding Domain in Complex with DNA. EMBO J. 1998, 17, 5484–5496. [Google Scholar] [CrossRef]
- Cao, D.; Lin, Z.; Huang, L.; Damaris, R.N.; Yang, P. Genome-Wide Analysis of AP2/ERF Superfamily in Lotus (Nelumbo nucifera) and the Association between NnADAP and Rhizome Morphology. BMC Genomics 2021, 22, 171. [Google Scholar] [CrossRef]
- Nakano, T.; Suzuki, K.; Fujimura, T.; Shinshi, H. Genome-Wide Analysis of the ERF Gene Family in Arabidopsis and Rice. Plant Physiol. 2006, 140, 411–432. [Google Scholar] [CrossRef] [PubMed]
- Magar, M.M.; Liu, H.; Yan, G. Genome-Wide Analysis of AP2/ERF Superfamily Genes in Contrasting Wheat Genotypes Reveals Heat Stress-Related Candidate Genes. Front. Plant Sci. 2022, 13, 853086. [Google Scholar] [CrossRef]
- Licausi, F.; Ohme-Takagi, M.; Perata, P. APETALA 2/Ethylene Responsive Factor (AP 2/ERF) Transcription Factors: Mediators of Stress Responses and Developmental Programs. New Phytol. 2013, 199, 639–649. [Google Scholar] [CrossRef] [PubMed]
- Kagaya, Y.; Ohmiya, K.; Hattori, T. RAV1, a Novel DNA-Binding Protein, Binds to Bipartite Recognition Sequence through Two Distinct DNA-Binding Domains Uniquely Found in Higher Plants. Nucleic Acids Res. 1999, 27, 470–478. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Chen, X. Secrets of the MIR172 Family in Plant Development and Flowering Unveiled. PLOS Biol. 2021, 19, e3001099. [Google Scholar] [CrossRef]
- Díaz-Manzano, F.E.; Cabrera, J.; Ripoll, J.-J.; del Olmo, I.; Andrés, M.F.; Silva, A.C.; Barcala, M.; Sánchez, M.; Ruíz-Ferrer, V.; de Almeida-Engler, J.; et al. A Role for the Gene Regulatory Module microRNA172/TARGET OF EARLY ACTIVATION TAGGED 1/FLOWERING LOCUS T (miRNA172/TOE1/FT) in the Feeding Sites Induced by Meloidogyne Javanica in Arabidopsis thaliana. New Phytol. 2018, 217, 813–827. [Google Scholar] [CrossRef]
- Cirilli, M.; Rossini, L.; Chiozzotto, R.; Baccichet, I.; Florio, F.E.; Mazzaglia, A.; Turco, S.; Bassi, D.; Gattolin, S. Less Is More: Natural Variation Disrupting a miR172 Gene at the Di Locus Underlies the Recessive Double-Flower Trait in Peach (P. persica L. Batsch). BMC Plant Biol. 2022, 22, 318. [Google Scholar] [CrossRef]
- Wang, L.; Ma, H.; Lin, J. Angiosperm-Wide and Family-Level Analyses of AP2/ERF Genes Reveal Differential Retention and Sequence Divergence After Whole-Genome Duplication. Front. Plant Sci. 2019, 10, 196. [Google Scholar] [CrossRef]
- Yin, F.; Zeng, Y.; Ji, J.; Wang, P.; Zhang, Y.; Li, W. The Halophyte Halostachys Caspica AP2/ERF Transcription Factor HcTOE3 Positively Regulates Freezing Tolerance in Arabidopsis. Front. Plant Sci. 2021, 12, 638788. [Google Scholar] [CrossRef]
- Zhang, B.; Wang, L.; Zeng, L.; Zhang, C.; Ma, H. Arabidopsis TOE Proteins Convey a Photoperiodic Signal to Antagonize CONSTANS and Regulate Flowering Time. Genes Dev. 2015, 29, 975–987. [Google Scholar] [CrossRef]
- Morel, P.; Heijmans, K.; Ament, K.; Chopy, M.; Trehin, C.; Chambrier, P.; Rodrigues Bento, S.; Bimbo, A.; Vandenbussche, M. The Floral C-Lineage Genes Trigger Nectary Development in Petunia and Arabidopsis. Plant Cell 2018, 30, 2020–2037. [Google Scholar] [CrossRef] [PubMed]
- Morel, P.; Heijmans, K.; Rozier, F.; Zethof, J.; Chamot, S.; Bento, S.R.; Vialette-Guiraud, A.; Chambrier, P.; Trehin, C.; Vandenbussche, M. Divergence of the Floral A-Function between an Asterid and a Rosid Species. Plant Cell 2017, 29, 1605–1621. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Kim, Y.J.; Müller, R.; Yumul, R.E.; Liu, C.; Pan, Y.; Cao, X.; Goodrich, J.; Chen, X. AGAMOUS Terminates Floral Stem Cell Maintenance in Arabidopsis by Directly Repressing WUSCHEL through Recruitment of Polycomb Group Proteins. Plant Cell 2011, 23, 3654–3670. [Google Scholar] [CrossRef] [PubMed]
- Nagahara, U. The Inheritance of the Double-Flower Trait in Petunia. J-STAGE 1930, 6, 205. [Google Scholar]
- Cheng, C.; Krishnakumar, V.; Chan, A.P.; Thibaud-Nissen, F.; Schobel, S.; Town, C.D. Araport11: A Complete Reannotation of the Arabidopsis Thaliana Reference Genome. Plant J. 2017, 89, 789–804. [Google Scholar] [CrossRef]
- Swarbreck, D.; Wilks, C.; Lamesch, P.; Berardini, T.Z.; Garcia-Hernandez, M.; Foerster, H.; Li, D.; Meyer, T.; Muller, R.; Ploetz, L.; et al. The Arabidopsis Information Resource (TAIR): Gene Structure and Function Annotation. Nucleic Acids Res. 2007, 36, D1009–D1014. [Google Scholar] [CrossRef]
- Xie, Y.; Li, H.; Luo, X.; Li, H.; Gao, Q.; Zhang, L.; Teng, Y.; Zhao, Q.; Zuo, Z.; Ren, J. IBS 2.0: An Upgraded Illustrator for the Visualization of Biological Sequences. Nucleic Acids Res. 2022, 50, W420–W426. [Google Scholar] [CrossRef]
- Aukerman, M.J.; Sakai, H. Regulation of Flowering Time and Floral Organ Identity by a MicroRNA and Its APETALA2-Like Target Genes. Plant Cell 2003, 15, 2730–2741. [Google Scholar] [CrossRef]
- Wang, T.; Ping, X.; Cao, Y.; Jian, H.; Gao, Y.; Wang, J.; Tan, Y.; Xu, X.; Lu, K.; Li, J.; et al. Genome-Wide Exploration and Characterization of miR172/euAP2 Genes in Brassica napus L. for Likely Role in Flower Organ Development. BMC Plant Biol. 2019, 19, 336. [Google Scholar] [CrossRef]
- Zhao, Q.; Sun, C.; Liu, D.-D.; Hao, Y.-J.; You, C.-X. Ectopic Expression of the Apple Md-miR172e Gene Alters Flowering Time and Floral Organ Identity in Arabidopsis. Plant Cell Tissue Organ Cult. PCTOC 2015, 123, 535–546. [Google Scholar] [CrossRef]
- Chung, M.-Y.; Nath, U.K.; Vrebalov, J.; Gapper, N.; Lee, J.M.; Lee, D.-J.; Kim, C.K.; Giovannoni, J. Ectopic Expression of miRNA172 in Tomato (Solanum lycopersicum) Reveals Novel Function in Fruit Development through Regulation of an AP2 Transcription Factor. BMC Plant Biol. 2020, 20, 283. [Google Scholar] [CrossRef] [PubMed]
- Pei, M.-S. Comparative Transcriptome Analyses of Fruit Development among Pears, Peaches, and Strawberries Provide New Insights into Single Sigmoid Patterns. BMC Plant Biol. 2020, 20, 108. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Li, F.; Qian, M.; Han, M.; Liu, H.; Zhang, D.; Ma, J.; Zhao, C. Characteristics and Regulatory Pathway of the PrupeSEP1 SEPALLATA Gene during Ripening and Softening in Peach Fruits. Plant Sci. 2017, 257, 63–73. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Kim, Y.; Dinh, T.T.; Chen, X. miR172 Regulates Stem Cell Fate and Defines the Inner Boundary of APETALA3 and PISTILLATA Expression Domain in Arabidopsis Floral Meristems: miR172 in Flower Development. Plant J. 2007, 51, 840–849. [Google Scholar] [CrossRef]
- Wei, B.; Liu, H.; Liu, X.; Xiao, Q.; Wang, Y.; Zhang, J.; Hu, Y.; Liu, Y.; Yu, G.; Huang, Y. Genome-Wide Characterization of Non-Reference Transposons in Crops Suggests Non-Random Insertion. BMC Genom. 2016, 17, 536. [Google Scholar] [CrossRef]
- Liang, L.; Chang, Y.; Lu, J.; Wu, X.; Liu, Q.; Zhang, W.; Su, X.; Zhang, B. Global Methylomic and Transcriptomic Analyses Reveal the Broad Participation of DNA Methylation in Daily Gene Expression Regulation of Populus Trichocarpa. Front. Plant Sci. 2019, 10, 243. [Google Scholar] [CrossRef]
- Panhwar, S.A.; Wang, D.; Lin, F.; Wang, Y.; Liu, M.; Chen, R.; Huang, Y.; Wu, W.; Huang, D.; Xiao, Y.; et al. Characterization of Active Transposable Elements and Their New Insertions in Tuber Propagated Greater Yam (Dioscorea alata). BMC Genom. 2024, 25, 864. [Google Scholar] [CrossRef]
- Dreni, L.; Kater, M.M. MADSReloaded: Evolution of the AGAMOUS Subfamily Genes. New Phytol. 2014, 201, 717–732. [Google Scholar] [CrossRef]
- Dreni, L.; Zhang, D. Flower Development: The Evolutionary History and Functions of the AGL6 Subfamily MADS-Box Genes. J. Exp. Bot. 2016, 67, 1625–1638. [Google Scholar] [CrossRef]
- Ishimori, M.; Kawabata, S. Conservation and Diversification of Floral Homeotic MADS-Box Genes in Eustoma Grandiflorum. J. Jpn. Soc. Hortic. Sci. 2014, 83, 172–180. [Google Scholar] [CrossRef]
- Simpson, J.T.; Wong, K.; Jackman, S.D.; Schein, J.E.; Jones, S.J.; Birol, I. ABySS: A Parallel Assembler for Short Read Sequence Data. Genome Res. 2009, 19, 1117–1123. [Google Scholar] [CrossRef] [PubMed]
- Xie, J.; Chen, Y.; Cai, G.; Cai, R.; Hu, Z.; Wang, H. Tree Visualization By One Table (tvBOT): A Web Application for Visualizing, Modifying and Annotating Phylogenetic Trees. Nucleic Acids Res. 2023, 51, W587–W592. [Google Scholar] [CrossRef]
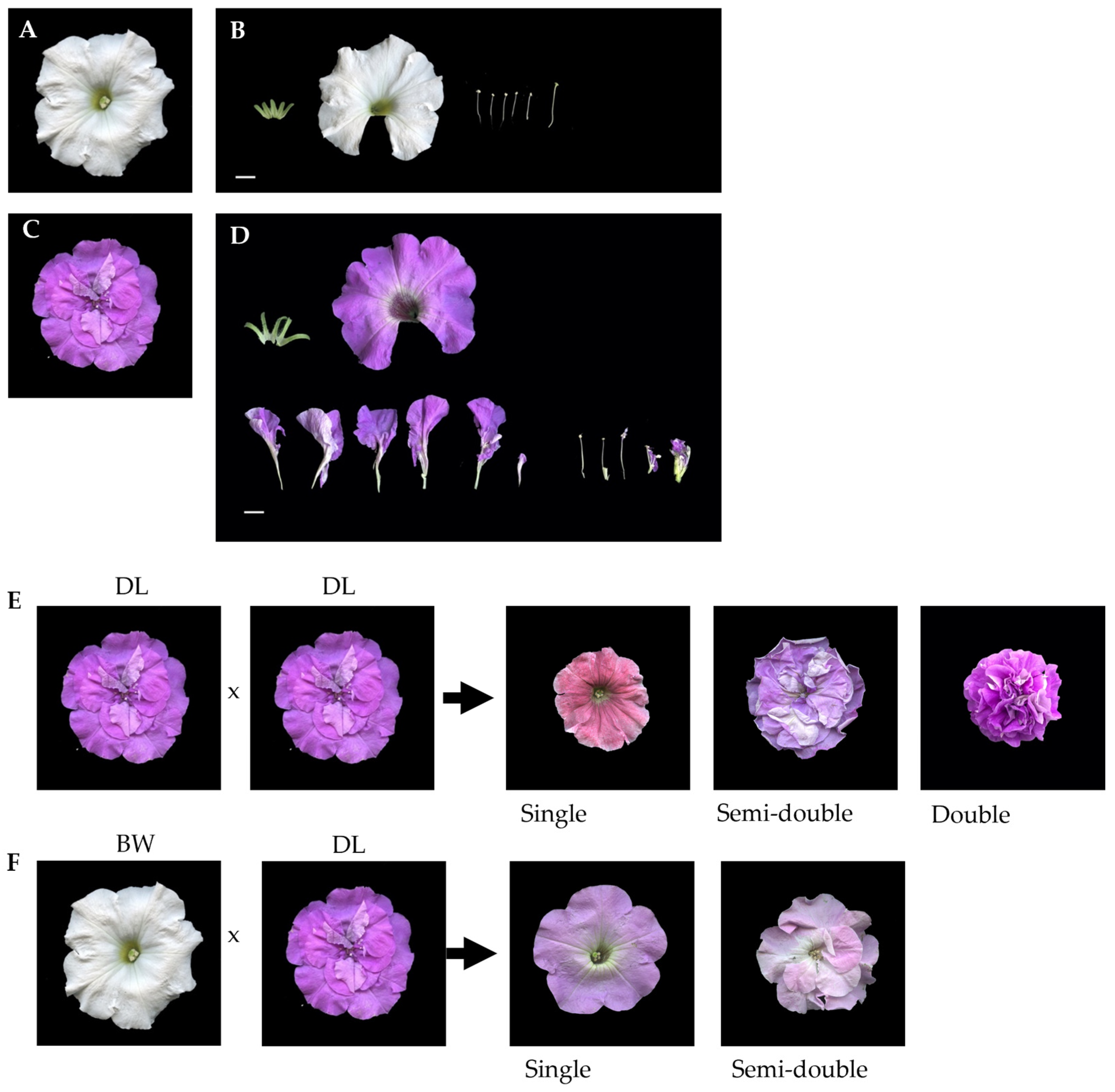



| Cross | Total | Flower Type | Exp. Ratio | X2 | ||
|---|---|---|---|---|---|---|
| Single | Semi-Double | Double | ||||
| BW | 8 | 8 | 0 | 0 | - | - |
| DL | 8 | 0 | 8 | 0 | - | - |
| BW × BW | 8 | 8 | 0 | 0 | 1:0:0 | - |
| DL × DL | 17 | 5 | 8 | 4 | 1:2:1 | 0.916 |
| BW × DL rep1 | 196 | 94 | 102 | 0 | 1:1:0 | 0.567 |
| BW × DL rep2 | 142 | 75 | 67 | 0 | 1:1:0 | 0.502 |
| Flower Phenotype | Genotype (−/−) (No Insertion) | Genotype (+/−) Heterozygous for Insertion | Total (n) |
|---|---|---|---|
| Single | 96 | 0 | 96 |
| Semi-double | 0 | 96 | 96 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xie, T.; Kawabata, S. An AP2-Family Gene Correlates with the Double-Flower Trait in Petunia × hybrida. Plants 2025, 14, 1314. https://doi.org/10.3390/plants14091314
Xie T, Kawabata S. An AP2-Family Gene Correlates with the Double-Flower Trait in Petunia × hybrida. Plants. 2025; 14(9):1314. https://doi.org/10.3390/plants14091314
Chicago/Turabian StyleXie, Tong, and Saneyuki Kawabata. 2025. "An AP2-Family Gene Correlates with the Double-Flower Trait in Petunia × hybrida" Plants 14, no. 9: 1314. https://doi.org/10.3390/plants14091314
APA StyleXie, T., & Kawabata, S. (2025). An AP2-Family Gene Correlates with the Double-Flower Trait in Petunia × hybrida. Plants, 14(9), 1314. https://doi.org/10.3390/plants14091314





