Abstract
(1) Background: The study analyzed wheat morphological traits to assess the role of roots structure in the tolerance of drought and to recognize the mechanisms of root structure adjustment to dry soil environment. (2) Methods: Root-box and root-basket methods were applied to maintain an intact root system for analysis. (3) Results: Phenotypic differences among six genotypes with variable drought susceptibility index were found. Under drought, the resistant genotypes lowered their shoot-to-root ratio. Dry matter, number, length, and diameter of nodal and lateral roots were higher in drought-tolerant genotypes than in sensitive ones. The differences in the surface area of the roots were greater in the upper parts of the root system (in the soil layer between 0 and 15 cm) and resulted from the growth of roots of the tolerant plant at an angle of 0–30° and 30–60°. (4) Conclusions: Regulation of root bending in a more downward direction can be important but is not a priority in avoiding drought effects by tolerant plants. If this trait is reduced and accompanied by restricted root development in the upper part of the soil, it becomes a critical factor promoting plant sensitivity to water-limiting conditions.
1. Introduction
Roots, as one of the primary organs, play an important role in vascular plants. They are a hidden part of a plant, responsible for growth, development and productivity, anchorage, and supplying stem and leaves with water and nutrients [1,2,3,4,5,6,7,8]. Roots constitute a source of organic matter, they influence the soil structure, aeration, and its biological activity [9,10,11]. Our knowledge on root interaction with soil environment is relatively poor; however, recent advances in imaging and sensor technologies are making root phenomic studies more possible and efficient [2,3].
Root system structure (RSS) is defined as a spatial configuration of roots in the soil profile (refers to the shape and physical space of the roots) and is controlled genetically [12,13,14]. The RSS consists of several components, such as seminal, seminal adventitious, nodal, and lateral roots. The nodal roots build up a framework, while lateral roots provide a network of the root system [15,16,17,18,19]. In cereals, two types of lateral roots can be distinguished according to their length, diameter, and histological structure. The L-type lateral roots are long, thick, and branch into the higher-order lateral roots, while S-type lateral roots are short, slender, and non-branching. Individual components of one root system can differ in origin, age, phenotypic plasticity, anatomical and morphological features, and physiological function [5,16,19,20]. Based on a “root box—pin board” method (also known as “rhizobox”), “concentrated” or “scattered” RSS types were described in cereals [15,18]. “Concentrated” type of the root system develops a greater number of densely distributed nodal roots with a relatively small rooting angle. The second type, designated as “scattered”, develops fewer but longer nodal roots, many of which grow obliquely in the soil profile (larger rooting angle). In “root-basket” method, the RSS is classified as “monomorphic-shallow”, “monomorphic-deep”, and “dimorphic” root systems in cereals [5]. The deep root system effectively absorbs water and nutrients from deeper soil layers [21], while shallow root systems ensure better nutrient uptake from fertilized upper soil profile [4,5]. The dimorphic root system comprises both deep and shallow roots, and thus it can adapt to multiple adverse factors such as water and nutrient shortages [22,23,24].
Soil drought is a common environmental phenomenon affecting plant growth, development, and yield. Strong differences in susceptibility to drought exist among genotypes, varieties, and species, as demonstrated by in various studies in, e.g., wheat [25,26], maize [23,27,28,29], oat [30,31], triticale [24,32,33], grasses [34], Arabidopsis, and poplar [35]. Acquisition of water (and mineral nutrients) is primarily determined by the dimension of the entire root zone, root density, their differentiation and elongation, and is closely related to soil water availability [18,33,36,37]. In water-limited environments, plants develop small root systems and the reduction in RSS size is proportional to the changes in water shortage [18]. In cereals exposed to long and severe drought, the rate of root elongation slows down due to reduced nutrient supply to the cortical cell layer. Since soil drought significantly affects the morphology and anatomy of root system components, a symbiosis between the roots and mycorrhizal fungi can be disrupted. As a result, both the flow of carbon from the root system to rhizosphere microorganisms and the flow of oxidized NH4 from the microorganisms to the plant could be inhibited [6,15,33]. Further, in the atmosphere, changes of the climatic factors (relative humidity, air temperature) occur more frequently and faster than in the soil, where they are mitigated by greater inertia [5,8,17,28]. Therefore, the relation between the size of the above-ground part and roots (S/R ratio) is of vital importance for the plant water balance. Lower S/R is more advantageous for plant exposed to water-limited environment and is an important mechanism of plant drought tolerance.
The changes in water status of the soil–root–leaf continuum negatively influence various physiological processes, such as intensity of leaf gas exchange and photosynthesis, membrane injury, or chlorophyll content [33,38,39,40]. Plants have developed different mechanisms that can reduce or repair the harmful effects of drought [9,10,25,34,41], including RSS plasticity, which allows plants to adapt to different soil water content [21,41]. Studies published so far have focused mostly on the above-ground organs, and physiological changes in the roots and the relationships between root system architecture and soil water availability are much less understood [29,42,43]. This greatly limits the development of new drought-resistant genotypes in breeding programs. Therefore, the objective of this study was to evaluate changes in root morphological traits in three drought-resistant and three drought-sensitive wheat genotypes grown under long soil drought. The study also aimed at improving methods that would help to determine the root features and their ecological and physiological role. The results of the experiments may help the breeders in determination of genetic diversity of wheat and in the development of new cultivars with improved water use efficiency and water economy.
2. Results
2.1. Plant Traits Under Control (C) Conditions
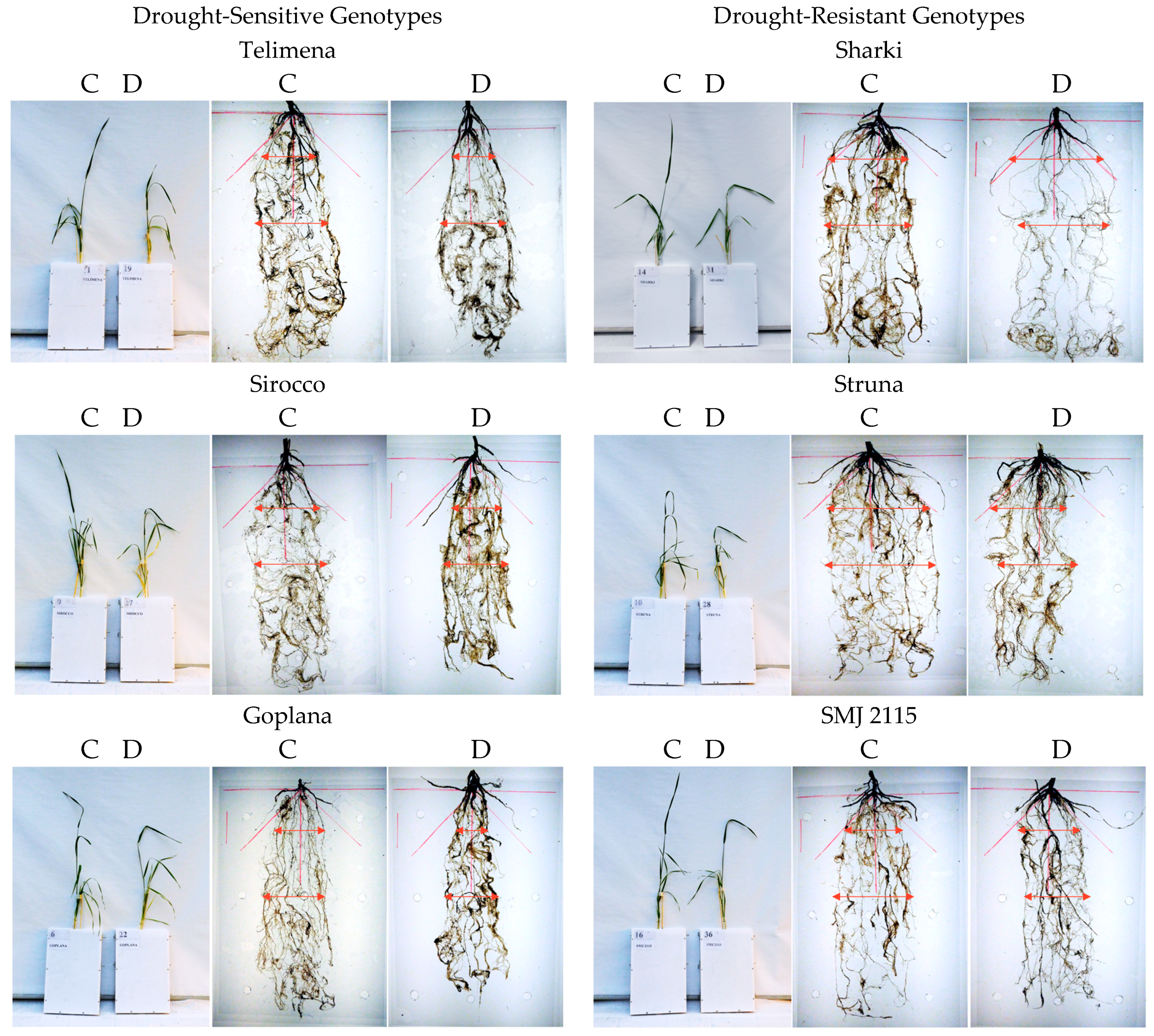
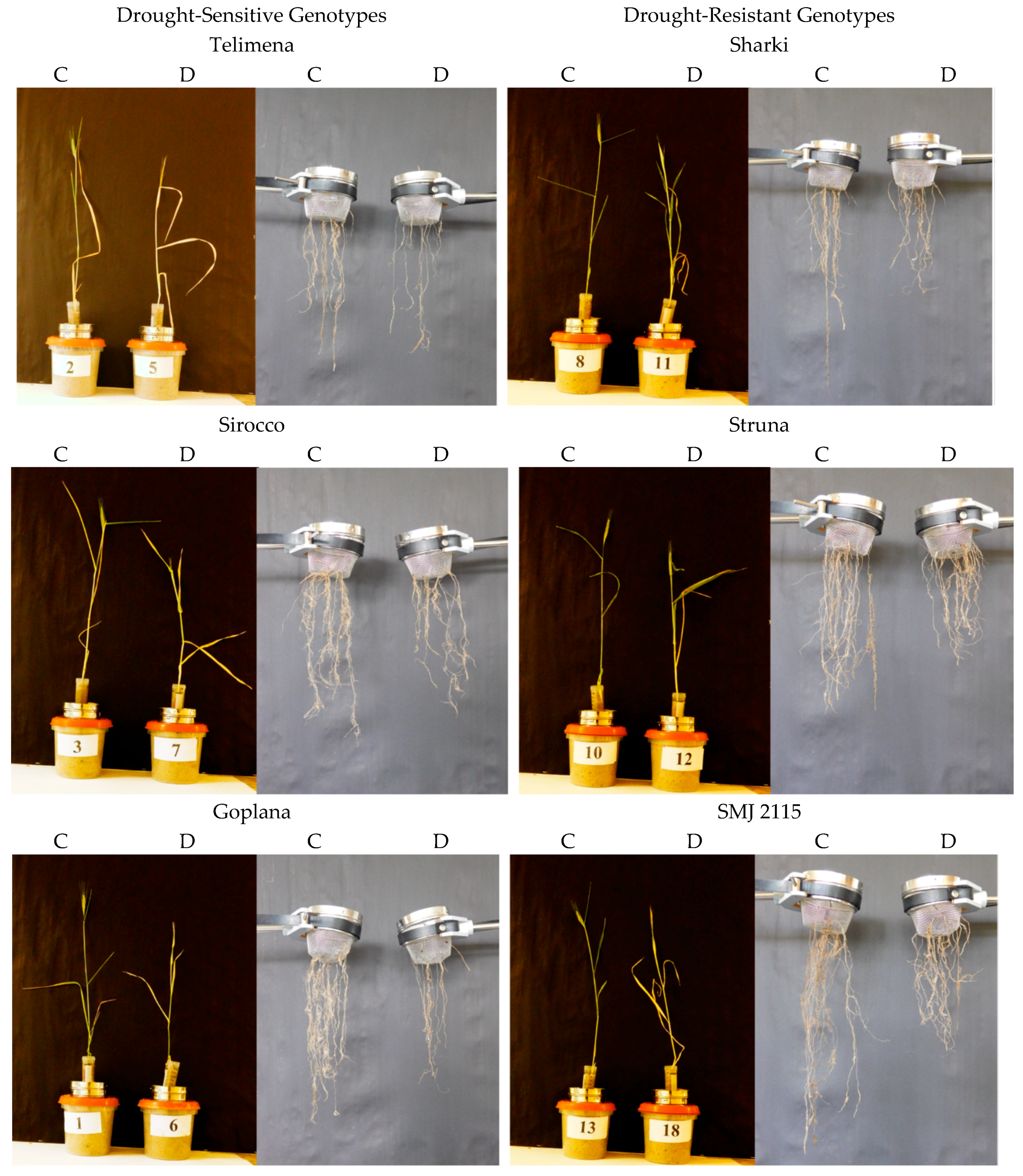
Wheat genotypes, both sensitive and resistant to drought, developed the “concentrated” and “deep” type of the root system, as most of the roots were located in the range up to 45 from the main axis of the root system (Figure 1 and Figure 2). In control conditions, all genotypes developed one main seminal, two seminal adventitious roots, and a similar number of nodal roots. Root development followed different patterns in the sensitive and resistant genotypes. The drought-resistant genotypes had more extensive and branched root system even at optimal water content.
Figure 1.
Representative examples of 8-week-old wheat plants and their roots in control (C) and drought (D) conditions during the root-box experiment. Cvs. “Telimena”, “Sirocco”, and “Goplana” were selected as drought-sensitive genotypes, while cvs. “Sharki”, “Struna”, and “SMJ 2115” as drought-resistant genotypes. The angle of 45° from main root axis is marked in red, and arrows indicate the width of the root system.
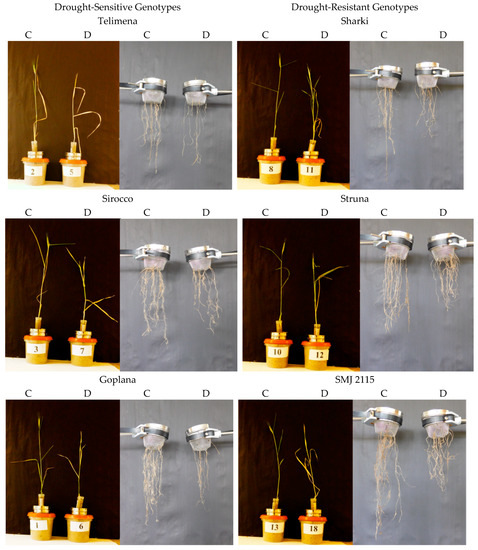
Figure 2.
Representative examples of 8-week-old wheat plants and their roots in control (C) and drought (D) conditions during the root-basket experiment. Cvs. “Telimena”, “Sirocco”, and “Goplana” were selected as drought-sensitive genotypes, while cvs. “Sharki”, “Struna”, and “SMJ 2115” as drought-resistant genotypes.
In root-box experiment under control conditions, most genotypes included in the sensitive group were higher (H) and developed a larger number of leaves (LNo), when compared to drought-resistant genotypes (Table 1). Dry mass of the above-ground parts (S), the roots (R), and the S/R ratio were also greater. Contrary, total root length (RL) and mean root length (RML) were lower in the sensitive than resistant plants (Table 2). The root-basket experiment confirmed higher growth (H) of sensitive plants (Table 3). Dry mass of above-ground parts (S), S + R, and S/R ratio were also greater (Table 3), whereas dry matter (R) and length of nodal roots (RL) growing at an angle of 0–30° and 30–60° were smaller in sensitive genotypes than in resistant ones (Table 4).

Table 1.
Effect of soil drought on wheat genotypes differing in their drought susceptibility index (DSI) during the root-box experiment. Plant height (H); leaf number (LNo); dry matter of shoot (S); dry matter of roots growing in two soil levels (RU, upper from 0 to 15 cm; RB, bottom from 15 to 40 cm); total root dry matter (R); total plant dry matter (S + R); shoot-to-root ratio (S/R); control treatment (C); drought treatment (D); relative trait change index (RTC); the mean of plant group is provided together with the standard error (SE); (n = 3).

Table 2.
Effect of soil drought on wheat genotypes differing in their drought susceptibility index (DSI) during the root-box experiment. Length of one seminal root (R1SL); total length of two seminal adventitious roots (R2SAL); number of nodal roots (RNo); length of nodal roots (RL); mean root length (RML); control treatment (C); drought treatment (D); relative trait change index (RTC); the mean of plant group is provided together with the standard error (SE); (n = 3).

Table 3.
Effect of soil drought on wheat genotypes differing in their drought susceptibility index (DSI) during the root-basket experiment. Plant height (H); leaf number (LNo); dry matter of shoot (S); total root dry matter (R); total plant dry matter (S + R ); shoot-to-root ratio (S/R); control treatment (C); drought treatment (D); relative trait change index (RTC); the mean of plant group is provided together with the standard error (SE); (n = 4).

Table 4.
Effect of soil drought on wheat genotypes differing in their drought susceptibility index (DSI) during the root-basket experiment. Total root dry matter (R); number of nodal roots (RNo); length of nodal roots (RL); control treatment (C); drought treatment (D); relative trait change index (RTC); the mean of plant group is provided together with the standard error (SE); (n = 4).
2.2. Plant Traits Under Drought (D vs. C)
The differences between drought-resistant and drought-sensitive genotypes under drought treatment were observed in density of the roots in the upper parts of the root system structure. In the resistant genotypes exposed to drought, a tendency to increase the number of roots in the upper part and occupy space above the angle of 45° was observed (Figure 1 and Figure 2). In root-box containers, drought-sensitive genotypes exhibited significantly decreased height (H), the strongest reduction of the root size, and smaller dry matter of shoot and roots (S, S + R, RU, RB, R, respectively) when drought conditions were applied (Figure 1, Table 1). The number of sensitive leaves (LNo) and S/R ratio did not differ significantly in drought-treated and control plants. The sensitive genotypes showed also a significant decrease in nodal root length (RL) and mean length of nodal roots (RML) under drought (Figure 1, Table 2). Drought slightly diminished the majority of traits in the drought-resistant genotypes, with an exception of total root dry matter (Figure 1, Table 1), nodal root length (RL), and mean length of nodal roots (RML) (Figure 1, Table 2), which were similar in control and drought-treated tolerant genotypes. A total length of two seminal adventitious roots differed among genotypes and it did not depend on plant tolerance to water-limited environment. Lack of statistically significant differences in the length of seminal root were probably due to the duration of the experiment, which was long enough for the roots to finish their growth and reach maximum length. In the root-basket experiment, drought diminished the height of all plants and dry matter of their shoots (Figure 2, Table 3). Importantly, the number of leaves significantly decreased in resistant plants but not in the sensitive ones, while dry mass of roots decreased significantly in sensitive plants but not in resistant ones. As a result, the shoot-to-root ratio dropped significantly only for the resistant genotypes. Furthermore, in drought-sensitive plants growing in root-basket containers, all measured root traits, i.e., root dry matter, number of nodal roots, and length of nodal roots growing at an angle 60–90°, significantly decreased under drought in comparison with control conditions (Table 4). Contrary trends occurred in resistant plants, in which drought increased dry matter of the roots growing at an angle 0–30°, as well as the number of nodal roots and the length of nodal roots growing at an angle 0–60°.
The relative trait change index (RTC) showed that in both root-box and root-basket experiments, the water-limiting conditions caused a greater decrease in H, S, R, and S + R, but not in LNo in the sensitive genotypes, when compared to resistant ones (Table 1 and Table 3). Drought-resistant genotypes accumulated greater dry biomass of roots in the upper layer of the soil (at a depth of 0–15 cm) and slightly smaller in the lower layer (at a depth of 15–40 cm) than the sensitive genotypes (Table 1 and Table 3). Roots of the tolerant plants that grew at the angle of 0–60° surpassed the sensitive plant roots in most of the parameters (Table 4). Sensitive genotypes exhibited the highest decrease of nodal root length (Table 2), and diminished all traits of the roots growing at the angle of 60–90°.
2.3. Correlation Between Drought Susceptibility Index (DSI) and Relative Trait Changes (RTC)
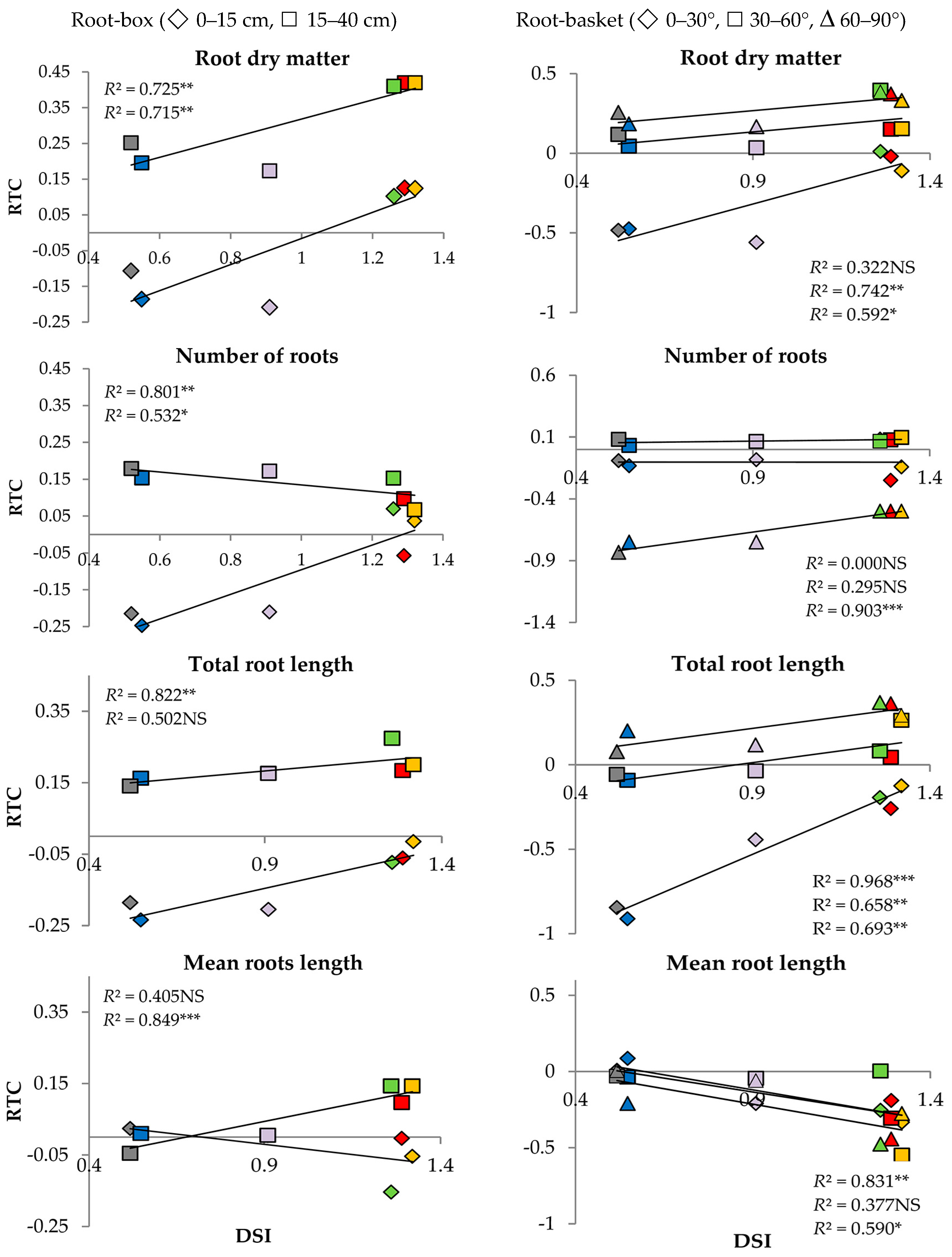
Linear correlation coefficients “r” between DSI ([26] and Materials and Methods section) and RTC of the measured traits in the plants growing in root-box and root-basket containers were high and statistically significant (Table 5). Contrary to that, insignificant correlation coefficients “r” were calculated for the length of seminal and seminal adventitious roots (0.605 and 0.104) and the number of roots growing at the angle of 0–30° and 30–60° (−0.004 and 0.543).

Table 5.
Correlation coefficient “r” between selected traits and relative trait change index (RTC) for wheat genotypes grown in root-box and root-basket containers. Plant height (H); leaf number (LNo); dry matter of shoot (S); total root dry matter (R); total plant dry matter (S+R); shoot-to-root ratio (S/R); number of nodal roots (RNo); length of nodal roots (RL); mean root length (RML); (df = 4).
The relationship between DSI and RTC calculated for the roots is presented in Figure 3. The root-box experiment revealed significant correlations between DSI and root dry matter and between DSI and the number of developed roots in both soil profiles: from 0 to 15 cm and from 15 to 40 cm. Another significant correlation was found for the length of roots growing at 0–15 cm below soil level and mean root length at the depth of 15–40 cm (Figure 3a). The root-basket experiment indicated a significant correlation between DSI and total root length when the roots grew at the angles of 0–30°, 30–60°, and 60–90° (Figure 3b). On the other hand, there was no statistically significant correlation between DSI and root dry matter (R) for the angle of 0–30°, the number of roots for the angle of 0°–30° and 30–60°, and mean root length (RML) for the angle of 30–60°.
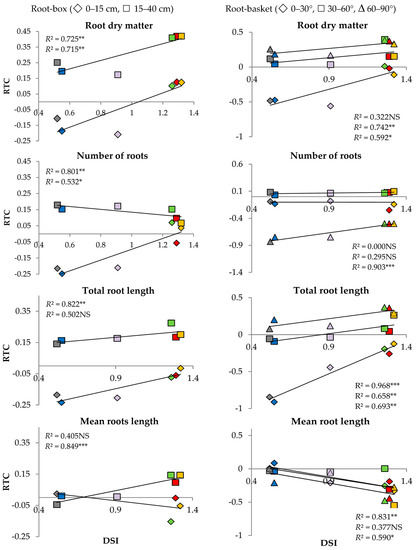
Figure 3.
Relationship between drought susceptibility index (DSI) and relative trait change index (RTC) for six wheat genotypes: Telimena (red), Sirocco (yellow), Goplana (green), Struna (blue), Sharki (violet), and SMJ 2115 (grey) in the root-box experiment and the root-basket experiment. NS—regression coefficient (R2) statistically not significant; *, **, *** R2 statistically significant at p < 0.1, 0.05, and 0.01, respectively.
2.4. Transcriptional Responses to Drought in Root Tissue
A number of transcripts can show overlapping and specific response under the experimental conditions in drought (Table 6, Supplementary Figures S1–S6). The transcripts up-regulated in roots under drought stress conditions were annotated to genes functional in pathways dependent on the transcription factors (TFs), auxin, cytokinin, mitogen-activated protein kinase, abscisic acid, ethylene, and antioxidative enzymes. Specifically, few transcription factors that make up the RNA polymerase II preinitiation complex (TFIID), a basic leucine zipper (BZIP) TFs, ethylene-responsive TFs, drought-responsive factor-like TFs (DRFL), wax production-like (WXPL) TFs, and further sequence-specific DNA binding TFs like LIM, MYB, MIKC-type MADS-box, NAC, and WRKY were identified to be the main players of drought tolerance via regulation of root traits. Additionally, several genes were found to be regulators of drought responses in wheat via signaling pathways involving an actin filament bundle assembly, transmembrane transport, cell differentiation, cell division, priming of cellular response to stress, and detoxification of reactive oxygen species.

Table 6.
Selected transcripts up-regulated in response of the roots to water deprivation. Analysis of multiple RNAseq experiments with meta-analytical approach of Genevestigator (details in Supplementary Figures S1–S6).
3. Discussion
The turn of the 21st century witnessed a drastic decrease in the yield of crops, including rice, wheat, and maize, despite increasing concentrations of CO2 that could benefit photosynthesis [44]. Drought turned out to be the most detrimental abiotic stress that limited crop yield by as much as 30–80% [45]. Scientists and breeders are gradually improving crop plant genotypes in terms of drought tolerance using both traditional selection methods and genetic engineering [26,34]. However, tolerance to drought stress is a complex feature, and finding suitable phenotypes requires time-consuming exposure of plants to simultaneously or sequentially occurring stress factors in natural conditions. Further, the progress in breeding is slower than expected and some of the reasons are that the genotypic variation in drought tolerance in crops has not yet been fully comprehended [9,46,47,48], and the genotype itself is not the only factor determining survival in suboptimal conditions [13,14,17,23,33,34,49,50,51]. Our research focused on both aspects in an attempt to recognize: (1) The role of root structure in soil drought tolerance, and (2) the mechanisms of root structure adjustment to the surrounding soil environment in order to increase competitiveness of water uptake by plant roots in dry conditions.
Wheat genotypes used in our study showed a wide range of drought tolerance traits in previous experiments [26]. We found difference and correlation between the physiological traits (photosynthesis, growth of aerial parts, and yield) and drought susceptibility index (DSI), proving the effect of genetic variation on the degree of drought tolerance, and grounds for the selection of suitable genotypes for water-limited environments. In the present study, the traits of the above-ground plant parts (height, leaf number, shoot dry matter) correspond to those for roots (dry matter, number, length), confirming that drought conditions were correctly optimized and applied in our experimental system. Shoot height and dry matter diminished severely in sensitive plants and moderately in resistant ones. Surprisingly, leaf number remained similar in sensitive genotypes but lowered significantly in resistant ones. Some leaves of sensitive plants showed slight chlorotic spots. Visible symptoms of drought resulted from a sequence of physiological processes, i.e., stomatal closure, reduced tissue turgor, leaf gas exchange, limited supply of CO2 to the mesophyll cells, and, subsequently, a decrease in photosynthesis rate [20,26,34,49,50]. The ability to maintain the structure and function of photosynthetic membranes under water deficit is one of the most important tolerance mechanisms. Efficient mechanisms that protect the function and structure of the membranes by stronger binding of molecules and maintaining intact lipid–protein complexes can prevent, i.e., disruption of thylakoid membranes and chlorophyll antennas [26,34,38,41,50,52]. Although smaller, the resistant leaves did not show the visible symptoms of damaged photosynthetic apparatus, thus confirming better acclimation to drought stress.
Root distribution in individual genotypes depended also on water availability and individual tolerance to drought. Water-limiting conditions exacerbated the decrease of the root system dry matter and reduced the number, length, and diameter of nodal and lateral roots in drought-sensitive genotypes, similarly as in many other studies [4,13,17,18,21,38,53,54]. Drought-resistant genotypes featured improved RSS (more extensive and branched root system), even at optimal water content. The most interesting and important differences between sensitive and tolerant plants included: (1) Better adaptation to reduced soil water content; and (2) greater plasticity of morphological changes in the root architecture. In the first case, the number of leaves decreased and the root system showed more intense development in the resistant plants exposed to drought. In the sensitive plants, number of leaves did not differ in control and water-limiting conditions, but dry mass of roots declined significantly. As a result, resistant genotypes had a lower S/R ratio. Lowering the number of leaves in dry conditions is a physiological priority that enables reduction of transpiration and improvement of plant water management and water use efficiency. Consequently, grain yield is greatly reduced; however, it remains sufficient to produce succeeding generations (Darwinian fitness). On the other hand, plants maintain more favorable distribution and supply of photosynthesis products to below-ground parts that promoted root growth and development [10,12,14,36,46,55]. Increased root surface area can improve nutrient and water absorption and contribute to the final yield and development of drought tolerance-related traits [44]. Higher number of roots can also protect against lodging and breaking roots in drying soil. In the second case, an intermediate number of roots seems to provide superior plant performance, as too extensive root system development may result in competition for soil and metabolic resources [11,53,56].
In our research, the differences in the root development between tolerant and sensitive genotypes were particularly visible in the upper part of the root system during soil drought. This resulted from a more vigorous growth of tolerant roots in the soil layer at the depth of 0–15 cm. More dry matter accumulated close to the ground level as a consequence of the highest number of nodal roots and their higher length. In the tolerant genotypes exposed to drought, growth and development of the root system were executed mainly by the roots growing at the angle of 0–30° and 30–60°. In the sensitive genotypes and in control conditions, dry matter, number of nodal roots, and their length close to the ground level were much smaller. In the plants sensitive to drought, all measured traits for the roots growing at the angle of 60–90° also decreased significantly under drought. These differences suggest that the roots of resistant genotypes grew through the soil more extensively than the roots of sensitive genotypes. The mechanism of plant root biomass allocation in the soil space translates into specific physiological functioning. Fast growth results in a deeper root system, formation of a denser root network in the upper soil layer, and in turn increases exploitation of soil resources. Higher root density in the active root zone area (up to 30 cm) improves water and nutrient uptake and enhances yield of the plants grown under water deficit conditions [57]. Wider root angles could also reduce the energy inputs to penetrate deeper soil layers to access water during limited rainfall [37] and reduce surface soil evaporative losses. Contrary to that, a more vertical angle and a greater number of seminal roots in wheat plants have been associated with more compact and deeper roots. As sufficient moisture remains available in deeper soil layers under drought, one can expect that deep roots are essential for small-statured crops, such as wheat, to extract water. However, wheat genotypes with deeper roots, higher root density in deeper soil layers, and smaller root density at the surface produce higher yield under rain-fed conditions [58].
Drought stress tolerance as a polygenic trait involves the expression of many sets of genes governing morpho-physiological traits, including root architecture [59,60,61]. The meta-analytical approach and analysis of multiple transcriptomic data have uncovered many transcription regulators and key genes. They can work via stress signaling, stress transduction, biosynthesis of stress-related proteins, enzymes and metabolites to coordinate the drought stress responses of wheat roots. We identified transcription factors such as TFIID, BZIP TFs, ethylene-responsive TFs, DRFL, WXPL TFs, LIM, MYB, MIKC-type MADS-box, NAC, and WRKY, which can be the enhancers and positive regulators of root traits under drought. For example, TFs of the MYB can be associated with the majority of biochemical and physiological processes in plants, via regulation of primary and secondary metabolism, hormone synthesis (ABA, ethylene, auxin), and signaling [62,63,64]. In our multi-transcriptome analysis of roots exposed to water deficit, numerous up-regulated MYB-encoding genes were found. It was reported that MYB can be involved in lateral root formation and in the biosynthesis of phenylpropanoid compounds, such as lignin [64]. The lower lignin content in roots can promote cell elongation of roots growing under drought; on the other hand, a higher rate of lignin accumulation can be required under prolonged drought to increase the mechanical strength of developed root tissues. Therefore, they can play a key role in root development and root structure arrangement. Similar to MYB TFs, abundant NAC TFs were up-regulated in drought-exposed roots. They can regulate development of thicker roots with enlarged stele, cortex, and epidermis, control a higher number of cells in stele and aerenchyma tissues, and induce lateral roots development and greater root biomass [64,65,66]. In our meta-analysis of multiple transcriptomes, NAC expression was not limited to the root; although, some of the NACs were found specifically or preferentially in this organ. Further, a direct role of ABA-dependent WRKY transcription factors was evident from their up-regulation in response to drought stress. The role of WRKY as a regulator of reactive oxygen species and antioxidant accumulation, cellular membrane stability, and altered osmotic adjustment was suggested [59,62,66]. We found, that the important mechanisms of drought sensing by roots were also regulated by hormones. The predominant role was attributed to abscisic acid (ABA), ethylene, auxins, and cytokinins. They are key regulators of complex heat–drought stress genetic networks in wheat [59,61,62,66]. ABA is important for early response to drought stress, but its high content in roots is not beneficial under prolonged and severe stress. Although the elevated expression of the zeaxanthin epoxidase (ZEP) encoding genes was not found in our transcriptomic analysis in roots, the other ABA-related genes such as genes encoding HVA22-like proteins were up-regulated. ABA biosynthesis is correlated with auxin transport to the root tips, which, in turn, enhances proton secretion in this zone [64]; therefore, their coexpression would be beneficial for development and morphology of the roots, as suggested by meta-analysis. Our analysis also indicates that universal stress players like antioxidative enzymes or mitogen-activated protein kinase play an important role in the roots during drought. We showed that their expression in wheat can be organ-specific. Particularly, two mitogen-activated protein kinases, as well as superoxide dismutase, catalase, glutathione reductase, and flavin-containing monooxygenase, can play an important role in acclimation of the root system to drought conditions. The above study of transcriptome-wide variation suggests the genes, which provide a promising example of multiple roles in drought stress, can be targets of molecular breeding strategies for improving drought tolerance, or at least candidate genes and putative molecular markers.
Concluding, a wide-spreading and much-branched root system is a specific feature of tolerant plants. Such a type of root system is required for stress avoidance or reduction of drought effects in a more efficient way than the deep-type root system. Extended root architecture and root size, also in the upper soil layer, can provide plants with a competitive edge regarding water uptake. Therefore, wide distribution of the roots in soil greatly affects the final crop yield. In breeding programs, selection of plants with both branched and deep roots should be taken into account to enhance soil water capture by new varieties and to help in yield stabilization under water stress conditions. Root traits such as specific root length, specific root area, and root density should be considered with the wide root angle, as they comprise the key traits for improving plant productivity under drought. Regulation of cell elongation at the root tip to bending of the root in a more downward direction can be important, but is not a priority in avoiding drought effects by tolerant wheat plants. However, if this trait is reduced and accompanied by restricted root development in the upper part of the soil, it becomes a critical factor promoting plant sensitivity to water-limiting conditions.
4. Materials and Methods
Plant material. Six commercial and breeding forms of wheat from DANKO Plant Breeders Ltd and Plant Breeding Centre Smolice Ltd., Co, were choosen based on our previous study. We selected three drought-sensitive (Sirocco, Telimena, Goplana) and three drought-resistant (Sharki, Struna, SMJ 2115) genotypes. Their drought susceptibility index (DSI = (1 − D/C)/(1 − xD/xC); where C, D—dry matter of aboveground part of the plant in control (C) and drought (D) conditions, xD, xC—average values for all examined genotypes) was 1.32, 1.29, 1.26, 0.91, 0.55, and 0.52, respectively [26], which gives clustering of sensitive and tolerant groups, but also more or less linear genotypic dependency on tolerance. The selected genotypes had also different origin, thus different genotypic diversity.
Growth conditions. The experiment was carried out in an air-conditioned glasshouse (Kraków, Poland), under the following day/night conditions: Temperature 23/18 ± 2.5 °C photoperiod 14/10, and relative humidity 70% ± 5%. Photosynthetically active radiation (PAR) was set to 350 ± 20 μmol photons m−2 s−1. Plants were grown in custom-made root-box or root-basket containers (Figure 4, Supplementary Figures S7 and S8) filled with a mixture of universal potting soil (Holas, Poland) and quartz-sand (Chemoform AG, Germany) in proportion 1:2 (v/v). Soil was mixed with compound fertilizer at the rate N: 2.8 mg, P: 1.8 mg, K: 1.4 mg per 1 kg of the soil substrate. Air-dried soil substrate was sieved through a 0.25 cm mesh and soil substrate compaction in root containers was set to 1.3 g cm−3. Field soil water capacity (FWC) for the soil mixture was determined according to the Kopecky method. Briefly, air-dried soil samples (100 cm3) were placed inside metal cylinders with 1 mm holes at the bottom. The cylinders were then placed inside a container filled with water for 30 min. The maximal soil water content in the samples was 0.43 g cm−3 after 8 h, and it decreased to 0.21 g cm−3 after 48 h. This final value was assumed as 100% FWC [67].

Figure 4.
Wheat plants in root-box (A) and root-basket (B) containers.
The “root box–pin board” and “root-basket“ methods were applied to enable non-destructive cleaning and analysis of all intact compartments of the root system [15]. The “root box–pin board method” utilized a plexiglass root-box (height 40 cm, width 20 cm, thickness 2 cm), a pin-board for sampling the roots, and a perforated polyethylene sheet (envelope) for handling and preserving the roots. The “root-basket” included two baskets with a horizontal mesh (diameter 9.0 cm, holes 0.25 × 0.25 cm, HTL-Trading B.V., Nederland), the upper one 2.0 cm high, and the lower one 6.0 cm high to maintain a constant distance between the grain placed in the upper basket and the mesh of the lower basket. The set of root baskets was placed in a pot (diameter 12.0 cm, height 13.0 cm). The root-box and root-basket containers were weighed three times per week and the amount of water lost through evapotranspiration was refilled to keep the constant mass.
In each root-box or root-basket container, a single pregerminated grain was planted at a depth of 2 cm (March 30, 2019). At the harvest (27 May 2019), the shoots and roots were separated. Roots were sampled in an envelope placed on the pin-board by removing the soil substrate with a gentle water stream. Root samples were preserved in formalin–acetic acid–ethanol (FAA) solution fixative (Fisher Scientific) and scanned.
Drought stress. Soil drought was induced by limiting watering. Drought treatment (D) at the level of 30–35% FWC was maintained for 6 weeks (from 14 to 56 days of plant growth). For control (C) treatment, soil water content was maintained at 65–70% FWC from sowing to harvest. The water content in pots was controlled during whole experiment by weighting of the containers. The loss of water caused by transpiration and evaporation was refilled every 2–3 days.
Plant traits. At the harvest, the above-ground parameters, such as plant height (H), shoot dry matter (S), and the number of leaves (LNo), were assessed. The below-ground traits were recorded as a number (RNo) and length (RL) of particular components of the root system (seminal root, seminal adventitious root, nodal root). In the root-box experiment, dry matter of roots (R) was analyzed for two levels of soil profile, i.e., roots collected from the layer of 0–15 cm below ground (RU), and from 15 to 40 cm (RB). The root-basket experiment assessed the number and length of roots growing at the angles of 0–30°, 30–60°, and 60–90° (horizontal ground line = 0°; vertical main root axis = 90°). Dry matter of the above-ground plant parts (S) and roots (R) was measured after drying at 65 °C for 72 h. For each genotype and treatment, three plants from root-boxes and four plants from root-basket containers were collected.
The results are presented as relative trait change index RTC = (C−D)/C, where C represents control, and D refers to drought treatment. Positive values of RTC indicate a decrease of the trait value in comparison with control plants, and negative values of RTC indicate an increase of the trait value vs. control plants.
Meta-analysis of transcripts. The meta-analytical approach of Genevestigator (Genevestigator, Plant Biology, https://www.genevestigator.ethz.ch/, accessed on 27 October 2019) was used to study transcriptomes related to transcription factors, auxin, cytokinin, gibberellin, mitogen-activated protein kinase, abscisic acid, ethylene, and antioxidative enzymes. Multiple RNAseq experiments were chosen on the mRNA-Seq Gene Level Triticum aestivum platform. The experimental conditions were defined by the selection of drought and dehydration perturbations from the experiments with accession number TA-00015, TA-00025, TA-00028, TA-00030, TA-00042, TA-00045, TA-00059, TA-00073, TA-00128, TA-00138, and TA-00139. A filter “Anatomy” was used to analyze the transcriptomic data in order to gene expression in different plant organs, including roots. The Similarity Search Tool was used to define the Euclidean distance with the optimal leaf-ordering for both transcripts and wheat anatomy. All data are presented as heatmaps, which represent the maximum level of expression for a given probe across all measurements available in the database for this probe (Supplementary Figures S1–S6). The transcripts with the highest expression were selected and described (Table 6). A functional information and gene ontology were assigned using the EnsemblPlants (EnsemblPlants, https://plants.ensembl.org/index.html, accessed on 27 October 2019) and UniProt (UniProt, https://www.uniprot.org/, accessed on 27 October 2019) databases.
Statistical analysis. Statistical significance of data in the ANOVA analysis was evaluated in STATISTICA 12.0 (Statsoft, Tulsa, OK, USA). Three biological replicates were used in each root-box experiment, and four biological replicates were used in each root-basket experiment per plant and treatment. The means were compared using Duncan’s multiple range test at 0.05 level of probability. We also calculated the F-values, linear correlation coefficients, and probability levels.
5. Conclusions
Our study contributed to the understanding of RSS responses to soil drought and determining root morphological traits of wheat genotypes with different drought susceptibility. The results suggest that drought triggers specific modifications of the wheat root system structure that involve changes in the roots biomass, size, and distribution at different levels of the soil profile and changes in the angle of root growth in relation to the main root axis. The methods being developed are useful tools to capture and analyze the complex root systems in laboratory conditions under a limited space, but on the other hand, the results correspond to a high degree to (previously published) field phenotyping.
Supplementary Materials
The following are available online at https://www.mdpi.com/2223-7747/8/12/584/s1, Figure S1: Transcription factors (187 genes), Figure S2: MYB (A, 38 genes), NAC (B, 15 genes), and WRKY (C, 18 genes) transcription factors, Figure S3: Auxins-related genes (195 genes), Figure S4: Cytokinin (A, 40 genes), gibberellin (B, 3 genes), zeatin epoxidase (C, 3 genes), mitogen-activated protein kinase (D, 40 genes) related genes, Figure S5: Abscisic acid (A, 50 genes), ethylene (B, 68 genes) related genes, Figure S6: Antioxidative enzyme (156 genes) related genes, Figure S7: Root box-pin board set, Figure S8: Root-basket set.
Author Contributions
Conceptualization, M.T.G., S.G., and M.S.-H.; methodology, S.G; investigation, M.T.G., N.H., A.M., and S.G.; resources, M.S.-H.; writing—original draft preparation, M.T.G. and S.G.; writing—review and editing, M.T.G., S.G., and M.S.-H.; visualization, M.T.G., S.G., and M.S.-H.; project administration, M.S.-H.; funding acquisition, M.S.-H.
Funding
This research was funded by the MINISTRY OF AGRICULTURE AND RURAL DEVELOPMENT, grant number HOR.hn.802.1.2019/103.
Acknowledgments
The authors are thankful to Zofia Banaszak, Małgorzata Niewinska, and Boguslawa Lugowska from DANCO Plant Breeders Ltd. and Jarosław Bojarczuk from Smolice Plant Breeders Ltd. Smolice for providing wheat genotypes. The authors also would like to thank Marek Trybała and Piotr Calik from the technical section of Cracow “Phytotron” Lab for construction of root-box and root-basket containers.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Atkinson, A.; Pound, M.P.; Bennett, M.J.; Wells, D.M. Uncovering the hidden half of plants using new advances in root phenotyping. Curr. Opin. Biotech. 2019, 55, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Lu, W.; Wang, X.; Wang, F. Adaptive minirhizotron for pepper roots observation and its installation based on root system architecture traits. Plant Methods 2019, 15, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Fromm, H. Root plasticity in the pursuit of water. Plants (Basel) 2019, 8, 236. [Google Scholar] [CrossRef] [PubMed]
- Lynch, J.P. Steep, cheap and deep: An ideotype to optimize water and N acquisition by maize root systems. Ann. Bot. 2013, 112, 347–357. [Google Scholar] [CrossRef]
- Uga, Y.; Ebana, K.; Abe, J.; Morita, S.; Okuno, K.; Yano, M. Variation in root morphology and anatomy among accessions of cultivated rice (Oryza sativa L.) with different genetic backgrounds. Breed Sci. 2009, 59, 87–93. [Google Scholar] [CrossRef]
- Ogawa, S.; Valencia, M.O.; Ishitani, M.; Selvaraj, M.G. Root system architecture variation in response to different NH4 concentrations and its association with Nitrogen-deficient tolerance traits in rice. Acta Physiol. Plant. 2014, 36, 2361–2372. [Google Scholar] [CrossRef]
- Colombi, T.; Walter, A. Root responses of triticale and soybean to soil compaction in the field are reproducible under controlled conditions. Funct. Plant Biol. 2015, 43, 114–128. [Google Scholar] [CrossRef]
- Szechyńska-Hebda, M.; Marczyk, J.; Ziejewska, C.; Hordyńska, N.; Mikuła, J.; Hebda, M. Optimal design of pH-neutral geopolymer foams for their use in ecological plant cultivation systems. Materials 2019, 12, 2999. [Google Scholar] [CrossRef]
- Ashraf, M. Inducing drought tolerance in plants: Recent advances. Biotech. Adv. 2010, 28, 199–238. [Google Scholar] [CrossRef]
- King, J. Reaching for The Sun: How Plant Work; Cambridge University Press: Cambridge, UK, 2011; pp. 185–244. [Google Scholar]
- Colombi, T.; Walder, F.; Büchi, L.; Sommer, M.; Liu, K.; Six, J.; van der Heijden, M.G.A.; Charles, R.; Keller, T. On-farm study reveals positive relationship between gas transport capacity and organic carbon content in arable soil. Soil 2019, 5, 91–105. [Google Scholar] [CrossRef]
- Chang, T.T.; Armenta-Soto, J.L.; Mao, C.X.; Peiris, R.; Loresto, G.C. Genetic studies on the components of drought resistance in rice (Oryza sativa L.). In Rice Genetics; IRRI Los Banos: Manila, Philippines, 1986; pp. 389–398. [Google Scholar] [CrossRef]
- Wu, L.; McGechan, M.B.; Watson, C.A.; Baddeley, J.A. Developing existing plant root system architecture models to meet future agricultural challenges. Adv. Agron. 2005, 85, 181–219. [Google Scholar] [CrossRef]
- Fleury, D.; Jefferies, S.; Kuchel, H.; Langridge, P. Genetic and genomic tools to improve drought tolerance in wheat. J. Exp. Bot. 2010, 61, 3211–3222. [Google Scholar] [CrossRef] [PubMed]
- Kono, Y.; Yamauchi, A.; Nonoyama, T.; Tatsumi, T.; Kawamura, N. A revised system of root-soil interaction for laboratory work. Environ. Control Biol. 1987, 25, 141–151. [Google Scholar] [CrossRef]
- Iijima, M.; Kono, Y.; Yamauchi, A.; Pardales, P.J. Effects of soil compaction on the development of rice and maize root system. Environ. Exp. Bot. 1991, 30, 333–342. [Google Scholar] [CrossRef]
- Tardieu, F.; Katerji, N. Plant response to soil water reserve: Consequences of the root system environment. Irrig. Sci. 1991, 12, 145–152. [Google Scholar] [CrossRef]
- Yamauchi, A. Significance of root system structure in relation to stress tolerance in cereal crop. Low-Input Sustain. Crop Product. Syst. Asia 1993, 347–360. Available online: https://ci.nii.ac.jp/naid/10008236424/ (accessed on 7 December 2019).
- Bengough, A.G.; McKenzie, B.M.; Hallett, P.D.; Valentine, T.A. Root elongation, water stress, and mechanical impedance: A review of limiting stresses and beneficial root tip traits. J. Exp. Bot. 2011, 62, 59–68. [Google Scholar] [CrossRef]
- Grzesiak, M.T. Impact of soil compaction on root architecture, leaf water status, gas exchange and growth of maize and triticale seedlings. Plant Root 2009, 3, 10–16. [Google Scholar] [CrossRef]
- Yoshida, S.; Bhattacharjee, D.P.; Cabuslay, G.S. Relationship between plant type and root growth in rice. Soil Sci. Plant Nutr. 1982, 28, 473–482. [Google Scholar] [CrossRef]
- Ho, M.D.; Rosas, J.C.; Brown, K.M.; Lynch, J.P. Root architectural tradeoffs for water and phosphorus acquisition. Funct. Plant Biol. 2005, 32, 737–748. [Google Scholar] [CrossRef]
- Grzesiak, M.T.; Marcińska, I.; Janowiak, F.; Rzepka, A.; Hura, T. The relationship between seedling growth and grain yield under drought conditions in maize and triticale genotypes. Acta Physiol. Plant 2012, 34, 1757–1764. [Google Scholar] [CrossRef]
- Grzesiak, M.T.; Waligórski, P.; Janowiak, F.; Marcińska, I.; Hura, K.; Szczyrek, P.; Głąb, T. The relations between drought susceptibility index based on grain yield (DSIGY) and key physiological seedling traits in maize and triticale genotypes. Acta Physiol. Plant 2013, 35, 549–565. [Google Scholar] [CrossRef]
- Gietler, M.; Nykiel, M.; Zagdanska, B.M. Changes in the reduction state of ascorbate and glutathione, protein oxidation and hydrolysis leading to the development of dehydration intolerance in Triticum aestivum L. seedlings. Plant Growth Reg. 2016, 79, 287–297. [Google Scholar] [CrossRef]
- Grzesiak, S.; Hordynska, N.; Szczyrek, P.; Grzesiak, M.T.; Noga, A.; Szechynska-Hebda, M. Variation among wheat (Triticum easativum L.) genotypes in response to the drought stress: I- selection approaches. J. Plant Interact. 2019, 14, 30–44. [Google Scholar] [CrossRef]
- Lorens, G.F.; Bennett, J.M.; Loggale, L.B. Differences in drought resistance between two corn hybrids. I. Water relations and root length density. Agron. J. 1987, 79, 802–807. [Google Scholar] [CrossRef]
- Grzesiak, S. Reaction to drought of inbreds and hybrids of maize (Zea mays L.) as evaluated in field and greenhouse experiments. Maydica 1990, 35, 303–331. [Google Scholar]
- Golbashy, M.; Ebrahimi, M.; Khavari-Khorasani, S.; Choukan, R. Evaluation of drought tolerance of same corn (Zea mays L.) hybrids in Iran. Afr. J. Agr. Res. 2010, 5, 2714–2719. [Google Scholar]
- Larsson, S.; Górny, A.G. Grain yield and drought resistance indices of oat cultivars in field rain shelter and laboratory experiments. J. Agron. Crop Sci. 1988, 161, 277–286. [Google Scholar] [CrossRef]
- Marcinska, I.; Czyczyło-Mysza, I.; Skrzypek, E.; Grzesiak, M.T.; Popielarska-Konieczna, M.; Warchoł, M.; Grzesiak, S. Application of photochemical parameters and several indices based on phenotypical traits to assess intraspecific variation of oat (Avena sativa L.) tolerance to drought. Acta Physiol. Plant 2017, 39, 153–165. [Google Scholar] [CrossRef]
- Royo, C.; Abaza, M.; Bianco, R.; del Moral, L.F.G. Triticale grain growth and morphometry as affected by drought stress, late sowing and simulated drought stress. Aust. J. Physiol. 2000, 27, 1051–1059. [Google Scholar] [CrossRef]
- Grzesiak, M.T.; Hura, K.; Jurczyk, B.; Hura, T.; Rut, G.; Szczyrek, P.; Grzesiak, S. Physiological markers of stress susceptibility in maize and triticale under different soil compaction and/or soil water contents. J. Plant Inter. 2017, 12, 355–372. [Google Scholar] [CrossRef]
- Bandurska, H.; Jóźwiak, W. A comparison of the effects of drought on proline accumulation and peroxidases activity in leaves of Festuca rubra L. and Lolium perenne L. Acta Soc. Bot. Pol. 2010, 79, 111–116. [Google Scholar] [CrossRef]
- Szechyńska-Hebda, M.; Czarnocka, W.; Hebda, M.; Bernacki, M.J.; Karpiński, S. PAD4, LSD1 and EDS1 regulate drought tolerance, plant biomass production, and cell wall properties. Plant Cell Rep. 2016, 35, 527–539. [Google Scholar] [CrossRef] [PubMed]
- Crush, J.R.; Easton, H.S.; Waller, J.E.; Hume, D.E.; Faville, M.J. Genotypic variation in patterns of root distribution, nitrate interception and response to moisture stress of a perennial ryegrass (Lolium perenne L.) mapping population. Grass Forage Sci. 2007, 62, 265–273. [Google Scholar] [CrossRef]
- Colombi, T.; Kirchgessner, N.; Walter, A.; Keller, T. Root tip shape governs root elongation rate under increased soil strength. Plant Physiol. 2017, 174, 2289–2301. [Google Scholar] [CrossRef]
- Geravandi, M.; Farshadfar, E.; Kahrizi, D. Evaluation of some physiological traits as indicators of drought tolerance in bread wheat genotypes. Russ. J. Plant Physiol. 2011, 58, 69–75. [Google Scholar] [CrossRef]
- Bandurska, H. Does proline accumulated in leaves of water deficit stressed barley plants confine cell membrane injury? I. Free proline accumulation and membrane injury index in drought and osmotically stressed plants. Acta Physiol. Plant 2012, 22, 409–415. [Google Scholar] [CrossRef]
- Pieczynski, M.; Wyrzykowska, A.; Milanowska, K.; Boguszewska-Mankowska, D.; Zagdanska, B.; Karlowski, W.; Jarmolowski, A.; Szweykowska-Kulinska, Z. Genomewide identification of genes involved in thepotato response to drought indicates functional evolutionary conservation with Arabidopsis plants. Plant Biotechnol. J. 2017, 16, 603–614. [Google Scholar] [CrossRef]
- Walter, A.; Silk, W.K.; Schurr, U. Environmental effects on spatial and temporal patterns of leaf and root growth. Annu. Rev. Plant Biol. 2009, 60, 279–304. [Google Scholar] [CrossRef]
- Batey, T. Soil compaction and soil management—A review. Soil Use Manag. 2009, 25, 335–345. [Google Scholar] [CrossRef]
- Drikvand, R.; Doosty, B.; Hosseinpour, T. Response of rainfed wheat genotypes to drought stress using drought tolerance indices. J. Agric. Sci. 2012, 4, 126–131. [Google Scholar] [CrossRef][Green Version]
- Ye, H.; Roorkiwal, M.; Valliyodan, B.; Zhou, L.; Chen, P.; Varshney, R.K.; Nguyen, T.H. Genetic diversity of root system architecture in response to drought stress in grain legumes. J. Exp. Bot. 2018, 69, 3267–3277. [Google Scholar] [CrossRef] [PubMed]
- Fahad, S.; Bajwa, A.A.; Nazir, U.; Anjum, S.A.; Farooq, A.; Zohaib, A.; Sadia, S.; Nasim, W.; Adkins, S.; Saud, S.; et al. Crop production under drought and heat stress: Plant responses and management option. Front Plant Sci. 2017, 8, 1147. [Google Scholar] [CrossRef] [PubMed]
- Reynolds, M.P. Physiological approaches to wheat breeding. In Bread Wheat: Improvement and Production; Curtis, B.C., Rajaram, S., Gomez Macpherson, H., Eds.; Food Agr Org: Rome, Italy, 2002; pp. 118–140. [Google Scholar]
- Talebi, R.; Fayaz, F.; Naji, N. Effective selection criteria for assessing drought stress tolerance in durum wheat (Triticum durum Desf.). Gen. App. Plant Physiol. 2009, 35, 64–74. [Google Scholar]
- Zaheri, A.; Bahraminejad, S. Assessment of drought tolerance in oat (Avena sativa L.) genotypes. Ann. Biol. Res. 2012, 3, 2194–2201. [Google Scholar]
- Wituszynska, W.; Slesak, I.; Vanderauwera, S.; Szechynska-Hebda, M.; Kornas, A.; Van Der Kelen, K.; Mühlenbock, P.; Karpinska, B.; Mackowski, S.; Van Breusegem, F.; et al. Lesion Simulating Disease1, Enhanced Disease Susceptibility1, and Phytoalexin Deficient4 conditionally regulate cellular signaling homeostasis, photosynthesis, water use efficiency, and seed yield in Arabidopsis. Plant Physiol. 2013, 161, 1795–1805. [Google Scholar] [CrossRef]
- Dyda, M.; Wąsek, I.; Tyrka, M.; Wędzony, M.; Szechyńska-Hebda, M. Local and systemic regulation of PSII efficiency in triticale infected by the hemibiotrophic pathogen Microdochium nivale. Physiol. Plant 2019, 165, 711–727. [Google Scholar] [CrossRef]
- Stanley, C.D. Proper use and data interpretation for plant- and soil water status measuring instrumentation: Introductory remarks. Hort Sci. 1990, 25, 1534. [Google Scholar] [CrossRef]
- Richards, R.A. Defining selection criteria to improve yield under drought. Plant Grow. Regul. 1996, 20, 157–166. [Google Scholar] [CrossRef]
- Colombi, T.; Herrmann, A.M.; Vallenback, P.; Keller, T. Cortical cell diameter is key to energy costs of root growth in wheat. Plant Physiol. 2019, 180, 2049–2060. [Google Scholar] [CrossRef]
- Colombi, T.; Torres, L.C.; Walter, A.; Keller, T. Feedbacks between soil penetration resistance, root architecture and water uptake limit water accessibility and crop growth—A vicious circle. Sci. Total Environ. 2018, 626, 1026–1035. [Google Scholar] [CrossRef] [PubMed]
- Rapacz, M.; Kościelniak, J.; Jurczyk, B.; Adamska, A.; Wójcik, M. Different patterns of physiological and molecular response to drought in seedlings of malt- and feed-type barleys (Hordeum vulgare). J. Agron. Crop Sci. 2010, 196, 9–19. [Google Scholar] [CrossRef]
- Hochholdinger, F. Untapping root system architecture for crop improvement. J. Exp. Bot. 2016, 67, 4431–4433. [Google Scholar] [CrossRef] [PubMed]
- Fry, E.L.; Evans, A.L.; Sturrock, C.J.; Bullock, J.M.; Bardgett, R.D. Root architecture governs plasticity in response to drought. Plant Soil 2018, 433, 189–200. [Google Scholar] [CrossRef]
- Wasaya, A.; Zhang, X.; Fang, Q.; Zongzheng, Y. Root phenotyping for drought tolerance: A review. Agronomy 2018, 8, 241. [Google Scholar] [CrossRef]
- Chaichi, M.; Sanjarian, F.; Razavi, K.; Gonzalez-Hernandez, J.L. Analysis of transcriptional responses in root tissue of bread wheat landrace (Triticum aestivum L.) reveals drought avoidance mechanisms under water scarcity. PLoS ONE 2019, 14, e0212671. [Google Scholar] [CrossRef]
- Li, Y.C.; Meng, F.R.; Zhang, C.Y.; Zhang, N.; Sun, M.S.; Ren, J.P.; Niu, H.B.; Wang, X.; Yin, J. Comparative analysis of water stress-responsive transcriptomes in drought-susceptible and -tolerant wheat (Triticum aestivum L.). J Plant Biol. 2012, 55, 349–360. [Google Scholar] [CrossRef]
- Ma, J.; Li, R.; Wang, H.; Li, D.; Wang, X.; Zhang, Y.; Zhen, W.; Duan, H.; Yan, G.; Li, Y. Transcriptomics analyses reveal wheat responses to drought stress during reproductive stages under field conditions. Fron. Plant Sci. 2017, 8, 592. [Google Scholar] [CrossRef]
- Kulkarni, M.; Soolanayakanahally, R.; Ogawa, S.; Uga, Y.; Selvaraj, M.G.; Kagale, S. Drought response in wheat: Key genes and regulatory mechanisms controlling root system architecture and transpiration efficiency. Front. Chem. 2017, 5, 106. [Google Scholar] [CrossRef]
- Benny, J.; Pisciotta, A.; Caruso, T.; Martinelli, F. Identification of key genes and its chromosome regions linked to drought responses in leaves across different crops through meta-analysis of RNA-Seq data. BMC Plant Biol. 2019, 19, 194. [Google Scholar] [CrossRef]
- Janiak, A.; Kwaśniewski, M.; Szarejko, I. Gene expression regulation in roots under drought. J. Exp. Bot. 2017, 67, 1003–1014. [Google Scholar] [CrossRef] [PubMed]
- Dudziak, K.; Zapalska, M.; Börner, A.; Szczerba, H.; Kowalczyk, K.; Nowak, M. Analysis of wheat gene expression related to the oxidative stress response and signal transduction under short-term osmotic stress. Sci. Rep. 2019, 9, 2743. [Google Scholar] [CrossRef] [PubMed]
- Iquebal, M.A.; Sharma, P.; Jasrotia, R.S.; Jaiswal, S.; Kaur, A.; Saroha, M.; Angadi, U.B.; Sheoran, S.; Singh, R.; Singh, G.P.; et al. RNAseq analysis reveals drought-responsive molecular pathways with candidate genes and putative molecular markers in root tissue of wheat. Sci. Rep. 2019, 9, 13917. [Google Scholar] [CrossRef] [PubMed]
- Hillel, D.; van Bavel, C.H.M. Simulation of profile water storage as related to soil hydraulic properties. Soil Sci. Soc. Am. J. 1976, 40, 807–815. [Google Scholar] [CrossRef]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).