The Effect of Abiotic Stress Conditions on Expression of Calmodulin (CaM) and Calmodulin-Like (CML) Genes in Wild-Growing Grapevine Vitis amurensis
Abstract
:1. Introduction
2. Results
2.1. Isolation, Molecular Cloning, and Sequencing of VaCaM and VaCML Transcripts
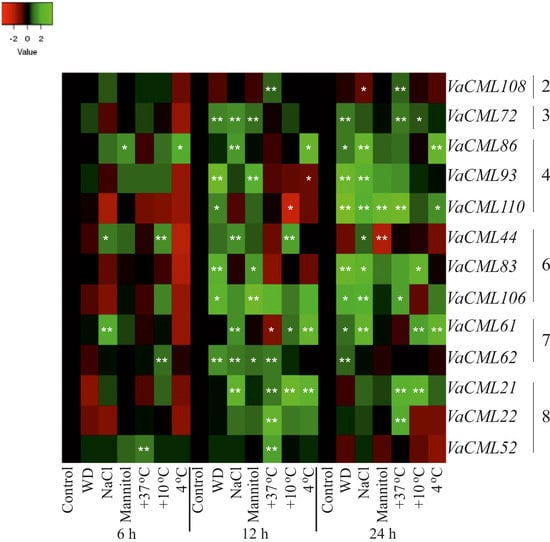
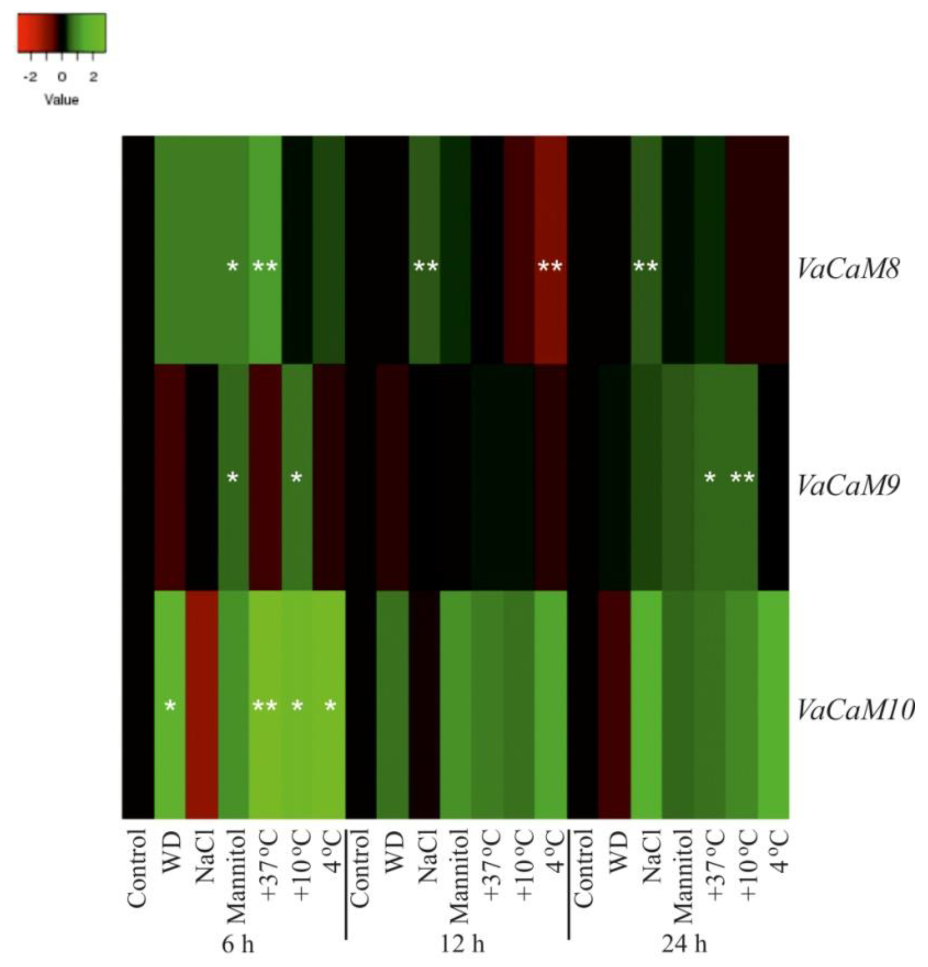
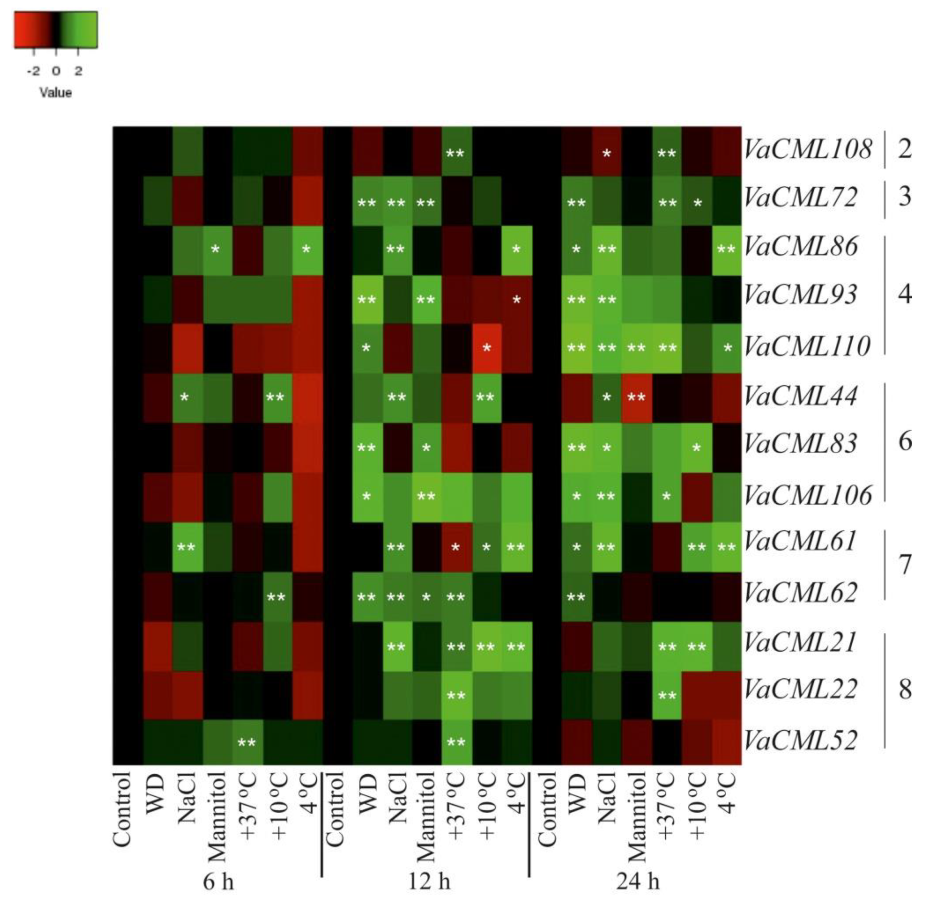
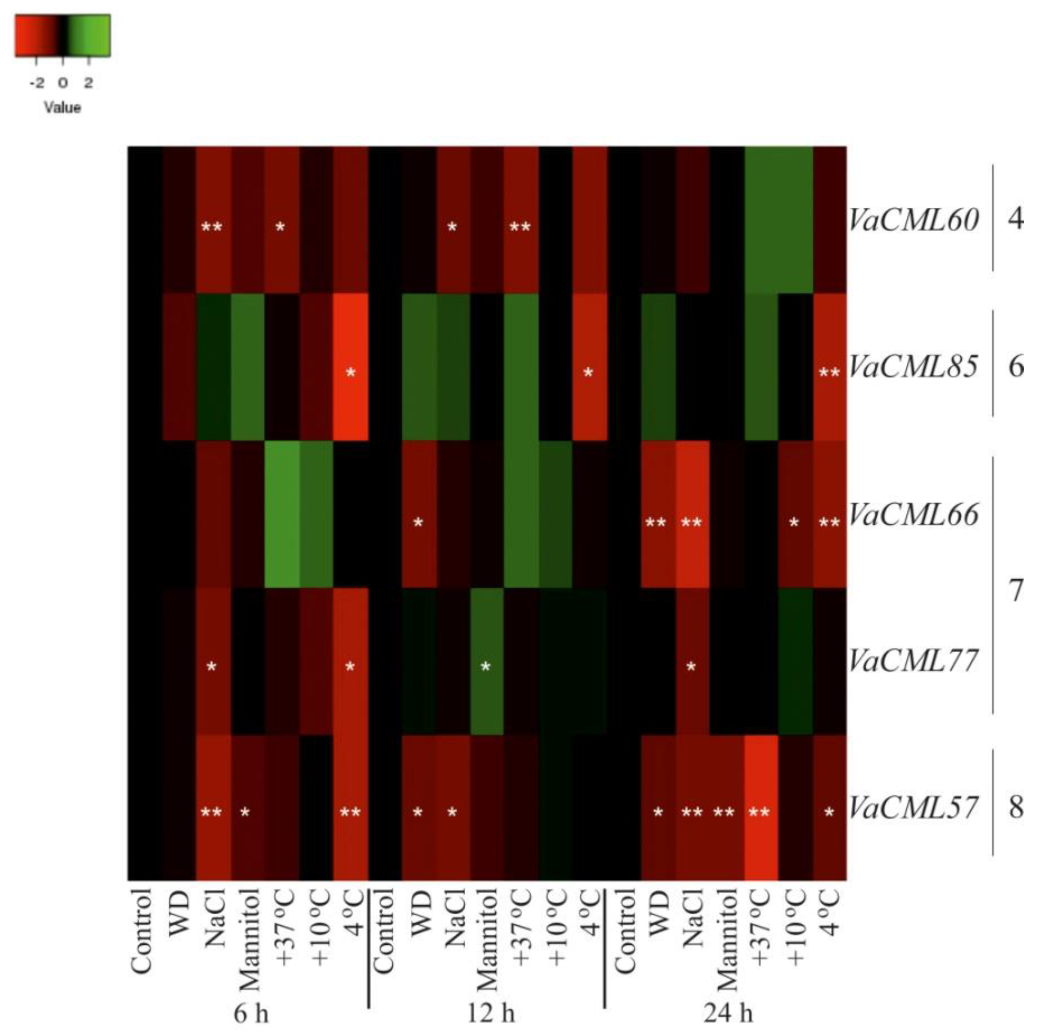
2.2. Expression of VaCaMs and VaCMLs in Response to Abiotic Stress Conditions
- (1)
- (2)
- (3)
- (4)
3. Discussion
4. Materials and Methods
4.1. Plant Material and Treatments
4.2. RNA Isolation and cDNA Preparation
4.3. Cloning and Sequencing of VaCaM and VaCML Transcripts
4.4. Expression Analysis of VaCaMs and VaCMLs
4.5. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Hashimoto, K.; Kudla, J. Calcium decoding mechanisms in plants. Biochimie 2011, 93, 2054–2059. [Google Scholar] [CrossRef] [PubMed]
- Ranty, B.; Aldon, D.; Cotelle, V.; Galaud, J.P.; Thuleau, P.; Mazars, C. Calcium sensors as key hubs in plant responses to biotic and abiotic stresses. Front. Plant Sci. 2016, 7, 327. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Halling, D.B.; Liebeskind, B.J.; Hall, A.W.; Aldrich, R.W. Conserved properties of individual Ca2+-binding sites in calmodulin. Proc. Natl. Acad. Sci. USA 2016, 113, E1216–E1225. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Choi, W.G.; Hilleary, R.; Swanson, S.J.; Kim, S.H.; Gilroy, S. Rapid, long-distance electrical and calcium signaling in plants. Annu. Rev. Plant Biol. 2016, 67, 287–307. [Google Scholar] [CrossRef]
- Kudla, J.; Becker, D.; Grill, E.; Hedrich, R.; Hippler, M.; Kummer, U.; Parniske, M.; Romeis, T.; Schumacher, K. Advances and current challenges in calcium signaling. New Phytol. 2018, 218, 414–431. [Google Scholar] [CrossRef]
- DeFalco, T.A.; Bender, K.W.; Snedden, W.A. Breaking the code: Ca2+ sensors in plant signaling. Biochem. J. 2010, 425, 27–40. [Google Scholar] [CrossRef] [Green Version]
- Mohanta, T.K.; Yadav, D.; Khan, A.L.; Hashem, A.; Abd Allah, E.F.; Al-Harrasi, A. Molecular players of EF-hand containing calcium signaling event in plants. Int. J. Mol. Sci. 2019, 20, 1476. [Google Scholar] [CrossRef] [Green Version]
- Perochon, A.; Aldon, D.; Galaud, J.P.; Ranty, B. Calmodulin and calmodulin-like proteins in plant calcium signaling. Biochimie 2011, 93, 2048–2053. [Google Scholar] [CrossRef]
- La Verde, V.; Dominici, P.; Astegno, A. Towards understanding plant calcium signaling through calmodulin-like proteins: A biochemical and structural perspective. Int. J. Mol. Sci. 2018, 19, 1331. [Google Scholar] [CrossRef] [Green Version]
- Virdi, A.S.; Singh, S.; Singh, P. Abiotic stress responses in plants: Roles of calmodulin-regulated proteins. Front. Plant Sci. 2015, 6, 809. [Google Scholar] [CrossRef] [Green Version]
- Zeng, H.; Xu, L.; Singh, A.; Wang, H.; Du, L.; Poovaiah, B.W. Involvement of calmodulin and calmodulin-like proteins in plant responses to abiotic stresses. Front. Plant Sci. 2015, 6, 600. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aldon, D.; Mbengue, M.; Mazars, C.; Galaud, J.P. Calcium signalling in plant biotic interactions. Int. J. Mol. Sci. 2018, 19, 665. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, Q.Y.; Xiong, T.T.; Li, X.P.; Chen, W.X.; Zhu, X.Y. Calcium and calcium sensors in fruit development and ripening. Sci. Hortic. 2019, 253, 412–421. [Google Scholar] [CrossRef]
- Zeng, H.; Zhang, Y.; Zhang, X.; Pi, E.; Zhu, Y. Analysis of EF-hand proteins in soybean genome suggests their potential roles in environmental and nutritional stress signaling. Front. Plant Sci. 2017, 8, 887. [Google Scholar] [CrossRef] [Green Version]
- Li, C.; Meng, D.; Zhang, J.; Cheng, L. Genome-wide identification and expression analysis of calmodulin and calmodulin-like genes in apple (Malus×domestica). Plant Physiol. Biochem. 2019, 139, 600–612. [Google Scholar] [CrossRef]
- Ma, Q.P.; Zhou, Q.Q.; Chen, C.M.; Cui, Q.Y.; Zhao, Y.X.; Wang, K.; Arkorful, E.; Chen, X.; Sun, K.; Li, X.H. Isolation and expression analysis of CsCML genes in response to abiotic stresses in the tea plant (Camellia sinensis). Sci. Rep. 2019, 9, 8211. [Google Scholar] [CrossRef]
- Rao, S.S.; El-Habbak, M.H.; Havens, W.M.; Singh, A.; Zheng, D.M.; Vaughn, L.; Haudenshield, J.S.; Hartman, G.L.; Korban, S.S.; Ghabrial, S.A. Overexpression of GmCaM4 in soybean enhances resistance to pathogens and tolerance to salt stress. Mol. Plant Pathol. 2014, 15, 145–160. [Google Scholar] [CrossRef]
- Munir, S.; Liu, H.; Xing, Y.; Hussain, S.; Ouyang, B.; Zhang, Y.; Li, H.; Ye, Z. Overexpression of calmodulin-like (ShCML44) stress-responsive gene from Solanum habrochaites enhances tolerance to multiple abiotic stresses. Sci. Rep. 2016, 6, 31772. [Google Scholar] [CrossRef]
- Yu, B.; Yan, S.; Zhou, H.; Dong, R.; Lei, J.; Chen, C.; Cao, B. Overexpression of CsCaM3 improves high temperature tolerance in cucumber. Front. Plant Sci. 2018, 9, 797. [Google Scholar] [CrossRef]
- Kalaipandian, S.; Xue, G.P.; Rae, A.L.; Glassop, D.; Bonnett, G.D.; McIntyre, L.C. Overexpression of TaCML20, a calmodulin-like gene, enhances water soluble carbohydrate accumulation and yield in wheat. Physiol. Plant. 2019, 165, 790–799. [Google Scholar] [CrossRef]
- Zhang, X.X.; Wang, T.Z.; Liu, M.; Sun, W.; Zhang, W.H. Calmodulin-like gene MtCML40 is involved in salt tolerance by regulating MtHKTs transporters in Medicago truncatula. Environ. Exp. Bot. 2019, 157, 79–90. [Google Scholar] [CrossRef]
- Yamaguchi, T.; Aharon, G.S.; Sottosanto, J.B.; Blumwald, E. Vacuolar Na+/H+ antiporter cation selectivity is regulated by calmodulin from within the vacuole in a Ca2+- and pH-dependent manner. Proc. Natl. Acad. Sci. USA 2005, 102, 16107–16112. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, S.; Jia, L.X.; Chu, H.Y.; Wu, D.; Peng, X.; Liu, X.; Zhang, J.J.; Zhao, J.F.; Chen, K.M.; Zhao, L.Q. Arabidopsis CaM1 and CaM4 promote nitric oxide production and salt resistance by Inhibiting S-Nitrosoglutathione reductase via girect binding. PLoS Genet. 2016, 12, e1006255. [Google Scholar] [CrossRef] [PubMed]
- Yoo, J.H.; Park, C.Y.; Kim, J.C.; Heo, W.D.; Cheong, M.S.; Park, H.C.; Kim, M.C.; Moon, B.C.; Choi, M.S.; Kang, Y.H.; et al. Direct interaction of a divergent CaM isoform and the transcription factor, MYB2, enhances salt tolerance in arabidopsis. J. Biol. Chem. 2005, 280, 3697–3706. [Google Scholar] [CrossRef] [Green Version]
- Vandelle, E.; Vannozzi, A.; Wong, D.; Danzi, D.; Digby, A.M.; Dal Santo, S.; Astegno, A. Identification, characterization, and expression analysis of calmodulin and calmodulin-like genes in grapevine (Vitis vinifera) reveal likely roles in stress responses. Plant Physiol. Biochem. 2018, 129, 221–237. [Google Scholar] [CrossRef] [PubMed]
- Jaillon, O.; Aury, J.M.; Noel, B.; Policriti, A.; Clepet, C.; Casagrande, A.; Choisne, N.; Aubourg, S.; Vitulo, N.; Jubin, C.; et al. The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature 2007, 449, 463–467. [Google Scholar]
- Velasco, R.; Zharkikh, A.; Troggio, M.; Cartwright, D.A.; Cestaro, A.; Pruss, D.; Pindo, M.; Fitzgerald, L.M.; Vezzulli, S.; Reid, J.; et al. A high quality draft consensus sequence of the genome of a heterozygous grapevine variety. PLoS ONE 2007, 2, e1326. [Google Scholar] [CrossRef] [Green Version]
- Grimplet, J.; Adam-Blondon, A.-F.; Bert, P.-F.; Bitz, O.; Cantu, D.; Davies, C.; Delrot, S.; Pezzotti, M.; Rombauts, S.; Cramer, G.R. The grapevine gene nomenclature system. BMC Genom. 2014, 15, 1077. [Google Scholar] [CrossRef] [Green Version]
- Database of Protein Domains, Families and Functional sites. Available online: http://prosite.expasy.org/ (accessed on 25 November 2019).
- Chung, E.; Park, J.M.; Oh, S.K.; Joung, Y.H.; Lee, S.; Choi, D. Molecular and biochemical characterization of the Capsicum annuum calcium-dependent protein kinase 3 (CaCDPK3) gene induced by abiotic and biotic stresses. Planta 2004, 220, 286–295. [Google Scholar] [CrossRef]
- Prediction of Lipid Modification Sites. Available online: http://lipid.biocuckoo.org/webserver.php (accessed on 25 November 2019).
- Compute pI/Mw Tool. Available online: http://web.expasy.org/compute_pi/ (accessed on 25 November 2019).
- Dubrovina, A.S.; Kiselev, K.V.; Khristenko, V.S. Expression of calcium-dependent protein kinase (CDPK) genes under abiotic stress conditions in wild-growing grapevine Vitis amurensis. J. Plant Physiol. 2013, 170, 1491–1500. [Google Scholar] [CrossRef]
- Liu, L.; Li, H. Review: Research progress in amur grape, Vitis amurensis Rupr. Can. J. Plant Sci. 2013, 93, 565–575. [Google Scholar] [CrossRef] [Green Version]
- Kiselev, K.V.; Dubrovina, A.S.; Shumakova, O.A.; Karetin, Y.A.; Manyakhin, A.Y. Structure and expression profiling of a novel calcium-dependent protein kinase gene, CDPK3a, in leaves, stems, grapes, and cell cultures of wild-growing grapevine Vitis amurensis Rupr. Plant Cell Rep. 2013, 32, 431–442. [Google Scholar] [CrossRef] [PubMed]
- Dubrovina, A.S.; Kiselev, K.V. The role of calcium-dependent protein kinase genes VaCPK1 and VaCPK26 in the response of Vitis amurensis (in vitro) and Arabidopsis thaliana (in vivo) to abiotic stresses. Russ. J. Genet. 2019, 55, 319–329. [Google Scholar] [CrossRef]
- Grape Genome Database. Available online: http://genomes.cribi.unipd.it/grape (accessed on 25 November 2019).
- The Arabidopsis Information Resource. Available online: https://www.arabidopsis.org (accessed on 25 November 2019).
- Falquet, L.; Pagni, M.; Bucher, P.; Hulo, N.; Sigrist, C.J.; Hofmann, K.; Bairoch, A. The PROSITE database, its status in 2002. Nucleic Acids Res. 2002, 30, 235–238. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.; Zheng, Y.; Li, H.; Luo, X.; He, Z.; Cao, S.; Shi, Y.; Zhao, Q.; Xue, Y.; Zuo, Z.; et al. GPS-Lipid: A robust tool for the prediction of multiple lipid modification sites. Sci Rep. 2016, 16, 28249. [Google Scholar] [CrossRef]
- Kiselev, K.V.; Tyunin, A.P.; Ogneva, Z.V.; Dubrovina, A.S. Age-associated alterations in somatic mutation level in Arabidopsis thaliana. Plant Growth Regul. 2015, 75, 493–501. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Sievers, F.; Wilm, A.; Dineen, D.; Gibson, T.J.; Karplus, K.; Li, W.; Lopez, R.; McWilliam, H.; Remmert, M.; Söding, J.; et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011, 11, 539. [Google Scholar] [CrossRef]
- Dubrovina, A.S.; Aleynova, O.A.; Manyakhin, A.Y.; Kiselev, K.V. The role of calcium-dependent protein kinase genes CPK16, CPK25, CPK30, and CPK32 in stilbene biosynthesis and the stress resistance of grapevine Vitis amurensis Rupr. Appl. Biochem. Microbiol. 2018, 54, 410–417. [Google Scholar] [CrossRef]
- Kiselev, K.V.; Ogneva, Z.V.; Suprun, A.R.; Grigorchuk, V.P.; Dubrovina, A.S. Action of ultraviolet-C radiation and p-coumaric acid on stilbene accumulation and expression of stilbene biosynthesis-related genes in the grapevine Vitis amurensis Rupr. Acta Physiol Plant. 2019, 41, 28. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C (T)) method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Babicki, S.; Arndt, D.; Marcu, A.; Liang, Y.; Grant, J.R.; Maciejewski, A.; Wishart, D.S. Heatmapper: Web-enabled heat mapping for all. Nucleic Acids Res. 2016, 44, W147–W153. [Google Scholar] [CrossRef] [PubMed]
- Heatmapper. Available online: http://heatmapper.ca (accessed on 25 November 2019).
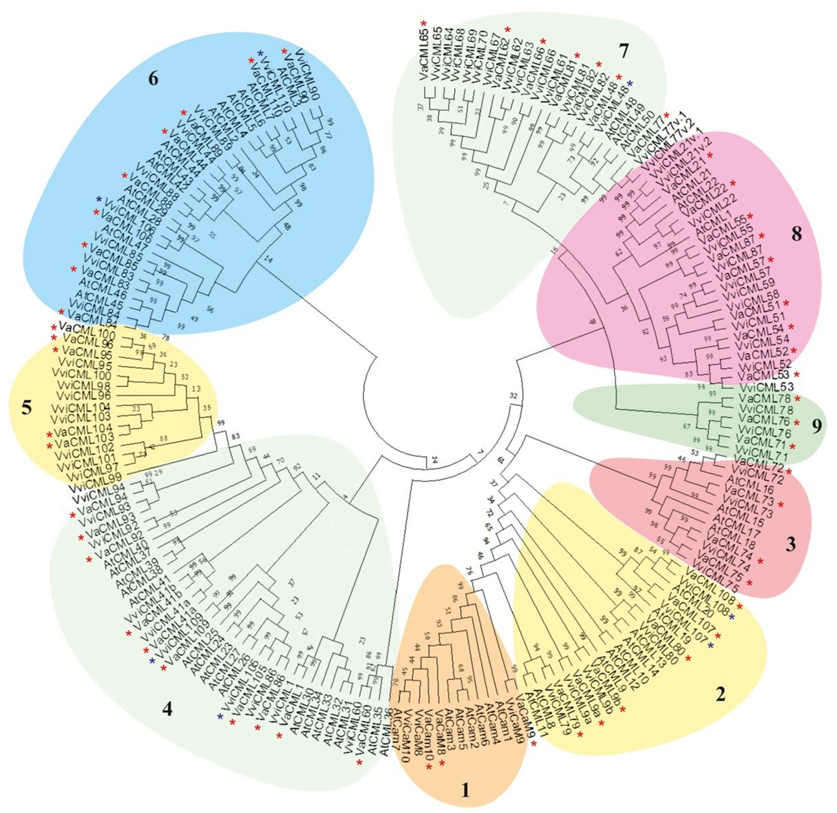

| Vitis amurensis cDNA Clone | Vitis vinifera Gene Prediction | Protein I/S (%) | No. of EF Hands | N-Myrist | N-Palmit | MW (kDa) | Group | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Transcript Name | ID GeneBank | CDS (bp) | Amino Acids | Gene Name | Gene ID | CDS (bp) | Amino Acids | ||||||
| VaCaM8 | MN515154 | 450 | 149 | VviCaM8 | VIT_208s0040g00470.6 | 450 | 149 | 100/100 | 4 | - | - | 16.8 | 1 |
| VaCaM9 | MN478368 | 450 | 149 | VviCaM9 | VIT_217s0000g00580.1 | 450 | 149 | 100/100 | 4 | - | - | 16.9 | 1 |
| VaCaM10 | MN515156 | 450 | 149 | VviCaM10 | VIT_206s0009g01910.1 | 462 | 153 | 100/100 | 4 | - | - | 16.8 | 1 |
| VaCML9a | MN515159 | 462 | 153 | VviCML9a | VIT_214s0030g02150.1 | 462 | 153 | 97/98 | 4 | - | - | 17.5 | 2 |
| VaCML9b | MN515160 | 450 | 149 | VviCML9b | VIT_200s0179g00280.1 | 450 | 149 | 100/100 | 4 | - | Yes | 17.0 | 2 |
| VaCML79 | MN515161 | 450 | 149 | VviCML79 | VIT_205s0020g04420.1 | 450 | 149 | 100/100 | 4 | - | - | 17.0 | 2 |
| VaCML80 | MN515162 | 444 | 147 | VviCML80 | VIT_202s0241g00140.1 | 444 | 147 | 100/100 | 3 | Yes | - | 16.6 | 2 |
| VaCML107 | MN562253 | 438 | 145 | VviCML107 | VIT_207s0031g00700.3 | 438 | 145 | 99/98 | 3 | - | Yes | 16.8 | 2 |
| VaCML108 | MN562252 | 522 | 173 | VviCML108 | VIT_205s0094g01240.3 | 522 | 173 | 99/99 | 4 | - | - | 19.9 | 2 |
| VaCML72 | MN515163 | 483 | 160 | VviCML72 | VIT_217s0000g04460.1 | 483 | 160 | 99/99 | 4 | Yes | - | 17.6 | 3 |
| VaCML73 | MN515164 | 489 | 162 | VviCML73 | VIT_201s0011g02470.1 | 489 | 162 | 100/100 | 4 | - | - | 17.8 | 3 |
| VaCML74 | MN537892 | 492 | 163 | VviCML74 | VIT_216s0039g01880.1 | 492 | 163 | 100/100 | 4 | - | - | 18.2 | 3 |
| VaCML75 | MN537893 | 492 | 163 | VviCML75 | VIT_202s0012g02060.1 | 492 | 163 | 100/100 | 4 | - | - | 18.1 | 3 |
| VaCML1 | MN537894 | 552 | 183 | VviCML1 | VIT_203s0063g00530.1 | 549 | 182 | 98/98 | 4 | - | - | 21.0 | 4 |
| VaCML41a | MN537895 | 561 | 186 | VviCML41a | VIT_204s0023g01100.1 | 558 | 185 | 97/98 | 3 | Yes | - | 21.0 | 4 |
| VaCML41b | MN537896 | 576 | 191 | VviCML41b | VIT_218s0001g11830.1 | 576 | 191 | 98/98 | 3 | - | - | 21.2 | 4 |
| VaCML44 | MN537897 | 489 | 162 | VviCML44 | VIT_218s0001g01630.1 | 489 | 162 | 99/99 | 4 | - | - | 18.3 | 4 |
| VaCML60 | MN537898 | 669 | 222 | VviCML60 | VIT_208s0007g05790.1 | 669 | 222 | 98/99 | 4 | - | - | 24.3 | 4 |
| VaCML86 | MN540577 | 492 | 163 | VviCML86 | VIT_217s0000g02480.1 | 282 | 93 | 98/98 a | 4 | - | - | 18.1 | 4 |
| VaCML88 | MN540578 | 579 | 192 | VviCML88 | VIT_208s0056g00290.1 | 579 | 192 | 100/100 | 3 | Yes | - | 21.4 | 4 |
| VaCML89 | MN540579 | 663 | 220 | VviCML89 | VIT_211s0016g05740.1 | 771 | 256 | 99/100 | 4 | - | Yes | 24.5 | 4 |
| VaCML90 | MN540580 | 465 | 154 | VviCML90 | VIT_207s0289g00040.1 | 465 | 154 | 100/100 | 4 | - | Yes | 17.5 | 4 |
| VaCML91 | VviCML91 | VIT_207s0141g00300.1 | |||||||||||
| VaCML92 | MN540581 | 507 | 168 | VviCML92 | VIT_218s0122g00180.1 | 507 | 168 | 100/100 | 4 | - | Yes | 18.4 | 4 |
| VaCML93 | MN540582 | 429 | 142 | VviCML93 | VIT_214s0171g00150.1 | 429 | 142 | 99/99 | 4 | - | - | 15.9 | 4 |
| VaCML94 | MN540583 | 432 | 143 | VviCML94 | VIT_217s0000g01630.1 | 432 | 143 | 99/100 | 4 | - | - | 16.1 | 4 |
| VaCML105 | MN562248 | 453 | 150 | VviCML105 | VIT_214s0006g01400.1 | 453 | 150 | 100/100 | 4 | - | Yes | 16.5 | 4 |
| VaCML106 | MN562254 | 255 | 84 | VviCML106 | VIT_208s0007g08830.1 | 255 | 84 | 100/100 | 2 | - | - | 9.5 | 4 |
| VaCML109 | MN562249 | 615 | 204 | VviCML109 | VIT_215s0048g00790.1 | 615 | 204 | 99/99 | 4 | - | Yes | 22.9 | 4 |
| VaCML110 | MN562246 | 648 | 215 | VviCML110 | VIT_205s0102g00450.1 | 645 | 214 | 99/99 | 4 | - | Yes | 24.6 | 4 |
| VaCML95 | MN540584 | 423 | 140 | VviCML95 | VIT_201s0010g03000.1 | 423 | 140 | 98/99 | 4 | - | Yes | 16.1 | 5 |
| VaCML96 | MN540585 | 423 | 140 | VviCML96 | VIT_201s0010g02990.1 | 423 | 140 | 96/97 | 4 | - | Yes | 16.1 | 5 |
| nd | VviCML97 | VIT_201s0010g02940.1 | 423 | - | - | 16.1 | 5 | ||||||
| nd | VviCML98 | VIT_201s0010g02970.1 | 423 | - | - | 16.1 | 5 | ||||||
| nd | VviCML99 | VIT_201s0010g03010.1 | 423 | - | - | 16.1 | 5 | ||||||
| VaCML100 | MN540586 | 423 | 140 | VviCML100 | VIT_201s0010g02980.1 | 423 | 140 | 96/97 | 4 | - | Yes | 16.1 | 5 |
| nd | VviCML101 | VIT_201s0010g02930.1 | 423 | - | - | 16.1 | 5 | ||||||
| nd | VviCML102 | VIT_201s0010g02950.1 | 423 | - | - | 16.1 | 5 | ||||||
| VaCML103 | MN540587 | 423 | 140 | VviCML103 | VIT_201s0010g03040.1 | 423 | 140 | 95/96 | 4 | - | Yes | 16.1 | 5 |
| VaCML104 | MN540588 | 423 | 140 | VviCML104 | VIT_201s0010g03020.1 | 423 | 140 | 96/97 | 4 | - | Yes | 16.1 | 5 |
| VaCML55 | MN540589 | 294 | 97 | VviCML55 | VIT_218s0001g10670.1 | 294 | 97 | 100/100 | 2 | - | Yes | 11.4 | 6 a |
| VaCML83 | MN540590 | 543 | 180 | VviCML83 | VIT_201s0026g02590.1 | 543 | 180 | 98/98 | 2 | - | Yes | 20.5 | 6 a |
| VaCML84 | MN540591 | 471 | 156 | VviCML84 | VIT_214s0108g01000.1 | 471 | 156 | 99/99 | 2 | - | Yes | 18.3 | 6 a |
| VaCML85 | MN540592 | 591 | 196 | VviCML85 | VIT_217s0000g06325.1 | 591 | 196 | 99/99 | 2 | - | Yes | 22.0 | 6 a |
| VaCML87 | MN540593 | 294 | 97 | VviCML87 | VIT_207s0031g00760.1 | 294 | 97 | 99/98 | 2 | - | Yes | 11.5 | 6 a |
| VaCML51 | MN540594 | 288 | 95 | VviCML51 | VIT_218s0001g10605.1 | 288 | 95 | 96/98 | 2 | - | - | 10.6 | 6 b |
| VaCML52 | MN540595 | 279 | 92 | VviCML52 | VIT_218s0001g10600.1 | 279 | 92 | 100/100 | 2 | - | - | 10.6 | 6 b |
| VaCML53 | MN540596 | 270 | 89 | VviCML53 | VIT_218s0001g10595.1 | 270 | 89 | 98/100 | 2 | - | - | 10.6 | 6 b |
| VaCML54 | MN540597 | 270 | 89 | VviCML54 | VIT_218s0001g10645.1 | 270 | 89 | 98/98 | 2 | - | Yes | 10.3 | 6 b |
| nd | VviCML56 | VIT_218s0001g10630.1 | 6 b | ||||||||||
| VaCML57 | MN540598 | 294 | 97 | VviCML57 | VIT_218s0001g10620.1 | 285 | 94 | 97/96 | 2 | - | - | 11.2 | 6 b |
| nd | VviCML58 | VIT_218s0001g10640.1 | 435 | 6 b | |||||||||
| nd | VviCML59 | VIT_218s0001g10610.1 | 300 | 6 b | |||||||||
| nd | VviCML21v.1 | VIT_219s0015g01200.1 | 708 | ||||||||||
| VaCML21v.2 | MN540599 | 699 | 232 | VviCML21v.2 | VIT_219s0015g01200.2 | 699 | 232 | 99/99 | 4 | Yes | Yes | 26.4 | 7 a |
| VaCML22 | MN540602 | 729 | 242 | VviCML22 | VIT_205s0029g00070.1 | 747 | 248 | 97/97 | 4 | Yes | Yes | 27.3 | 7 a |
| VaCML62 | MN540605 | 414 | VviCML62 | VIT_212s0059g00360.1 | 414 | 97/98 | 2 | Yes | - | 15.5 | 7 a | ||
| nd | VviCML63 | VIT_212s0059g00320.1 | 414 | 7 a | |||||||||
| nd | VviCML64 | VIT_212s0059g00370.1 | 414 | 7 a | |||||||||
| VaCML65 | MN540606 | 414 | 137 | VviCML65 | VIT_212s0059g00430.1 | 414 | 137 | 99/99 | 2 | Yes | - | 15.6 | 7 a |
| VaCML66 | MN540607 | 420 | 139 | VviCML66 | VIT_213s0156g00120.1 | 420 | 139 | 100/100 | 2 | Yes | - | 15.5 | 7 a |
| nd | VviCML67 | VIT_212s0059g00340.1 | 414 | 7 a | |||||||||
| nd | VviCML68 | VIT_212s0059g00420.1 | 414 | 7 a | |||||||||
| nd | VviCML69 | VIT_212s0059g00400.1 | 414 | 7 a | |||||||||
| nd | VviCML70 | VIT_212s0059g00350.1 | 414 | 7 a | |||||||||
| VaCML48 | MN562247 | 678 | 225 | VviCML48 | VIT_206s0080g00450.1 | 678 | 225 | 100/100 | 2 | - | - | 25.3 | 7 b |
| VaCML61 | p.s.b | VviCML61 | VIT_205s0077g00300.1 | 909 | 302 | 100/100 | 2 | Yes | - | 33.5 | 7 b | ||
| nd | VviCML77v.1 | VIT_200s0252g00130.1 | 711 | 7 b | |||||||||
| VaCML77v.2 | MN540608 | 831 | 276 | VviCML77v.2 | VIT_200s0252g00130.2 | 831 | 276 | 98/98 | 2 | Yes | - | 29.7 | |
| VaCML78 | MN540610 | 1119 | 372 | VviCML78 | VIT_210s0071g00670.1 | 1119 | 372 | 98/99 | 5 | Yes | - | 43.4 | 7 b |
| VaCML81 | MN540611 | 1425 | 474 | VviCML81 | VIT_204s0008g06280.1 | 1425 | 474 | 99/99 | 4 | - | Yes | 54.2 | 7 b |
| VaCML82 | MN540612 | 1065 | 354 | VviCML82 | VIT_211s0118g00540.1 | 1065 | 354 | 99/100 | 5 | - | - | 40.6 | 7 b |
| VaCML71 | MN548771 | 594 | 72 | VviCML71 | VIT_219s0014g04650.1 | 453 | 150 | 100/100 a | 0 | - | - | 8.0 | 8 |
| VaCML76 | MN540595 | 363 | 120 | VviCML76 | VIT_202s0012g00660.1 | 363 | 120 | 99/99 | 1 | Yes | - | 13.5 | 8 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dubrovina, A.S.; Aleynova, O.A.; Ogneva, Z.V.; Suprun, A.R.; Ananev, A.A.; Kiselev, K.V. The Effect of Abiotic Stress Conditions on Expression of Calmodulin (CaM) and Calmodulin-Like (CML) Genes in Wild-Growing Grapevine Vitis amurensis. Plants 2019, 8, 602. https://doi.org/10.3390/plants8120602
Dubrovina AS, Aleynova OA, Ogneva ZV, Suprun AR, Ananev AA, Kiselev KV. The Effect of Abiotic Stress Conditions on Expression of Calmodulin (CaM) and Calmodulin-Like (CML) Genes in Wild-Growing Grapevine Vitis amurensis. Plants. 2019; 8(12):602. https://doi.org/10.3390/plants8120602
Chicago/Turabian StyleDubrovina, Alexandra S., Olga A. Aleynova, Zlata V. Ogneva, Andrey R. Suprun, Alexey A. Ananev, and Konstantin V. Kiselev. 2019. "The Effect of Abiotic Stress Conditions on Expression of Calmodulin (CaM) and Calmodulin-Like (CML) Genes in Wild-Growing Grapevine Vitis amurensis" Plants 8, no. 12: 602. https://doi.org/10.3390/plants8120602
APA StyleDubrovina, A. S., Aleynova, O. A., Ogneva, Z. V., Suprun, A. R., Ananev, A. A., & Kiselev, K. V. (2019). The Effect of Abiotic Stress Conditions on Expression of Calmodulin (CaM) and Calmodulin-Like (CML) Genes in Wild-Growing Grapevine Vitis amurensis. Plants, 8(12), 602. https://doi.org/10.3390/plants8120602