The NAC Protein from Tamarix hispida, ThNAC7, Confers Salt and Osmotic Stress Tolerance by Increasing Reactive Oxygen Species Scavenging Capability
Abstract
:1. Introduction
2. Results
2.1. Bioinformatics Analysis of ThNAC7
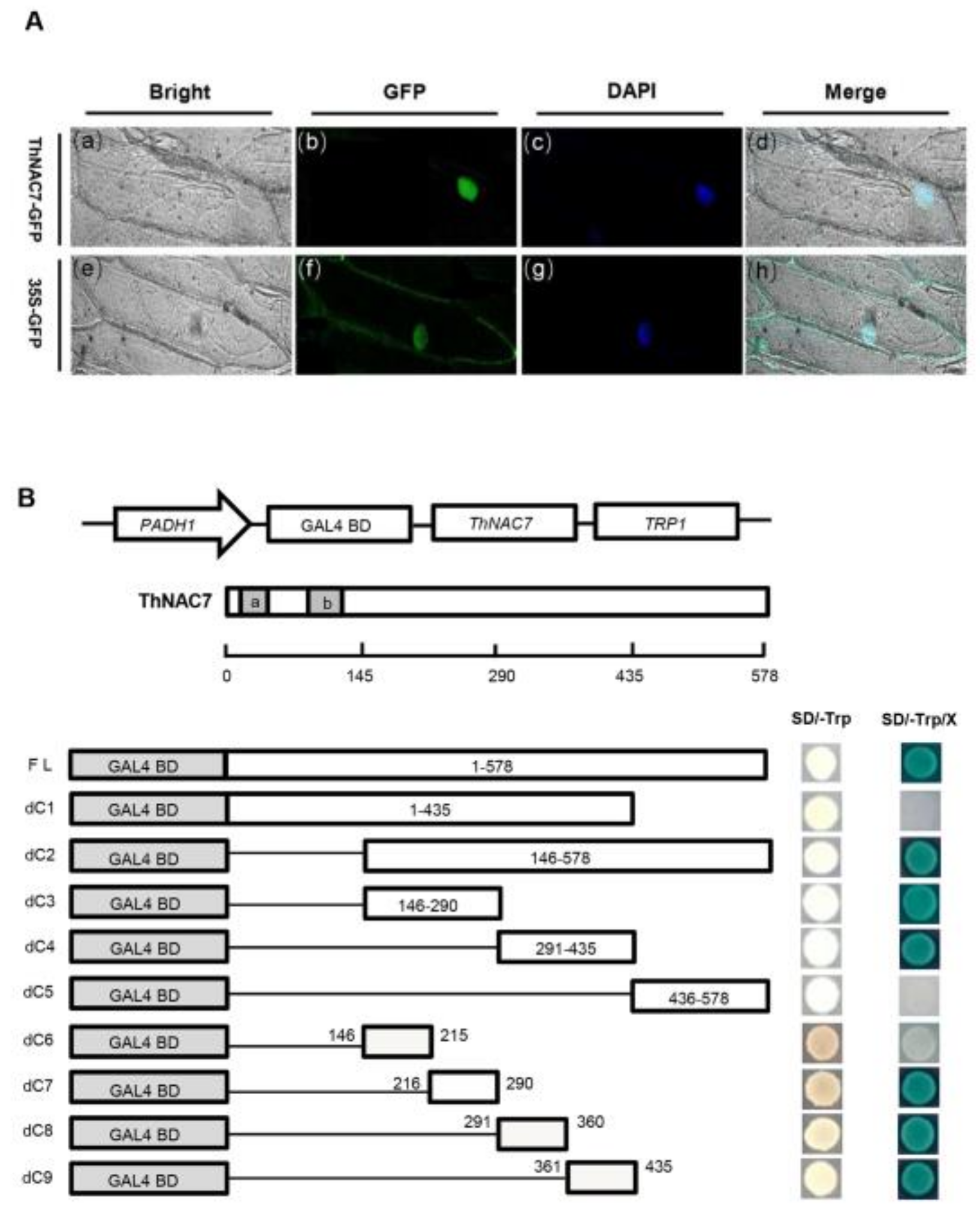
2.2. ThNAC7 Is a Nuclear Localization Protein with Transactivation Activity
2.3. The Expression of ThNAC7 Is Induced by Osmotic and Salt Stress
2.4. Overexpression of ThNAC7 in Arabidopsis Confers Osmotic and Salt Stress Tolerance
2.5. Transient Overexpression or Knockdown of ThNAC7 in T. hispida Plants
2.6. The ROS Scavenging Capability Was Improved by Overexpressing ThNAC7
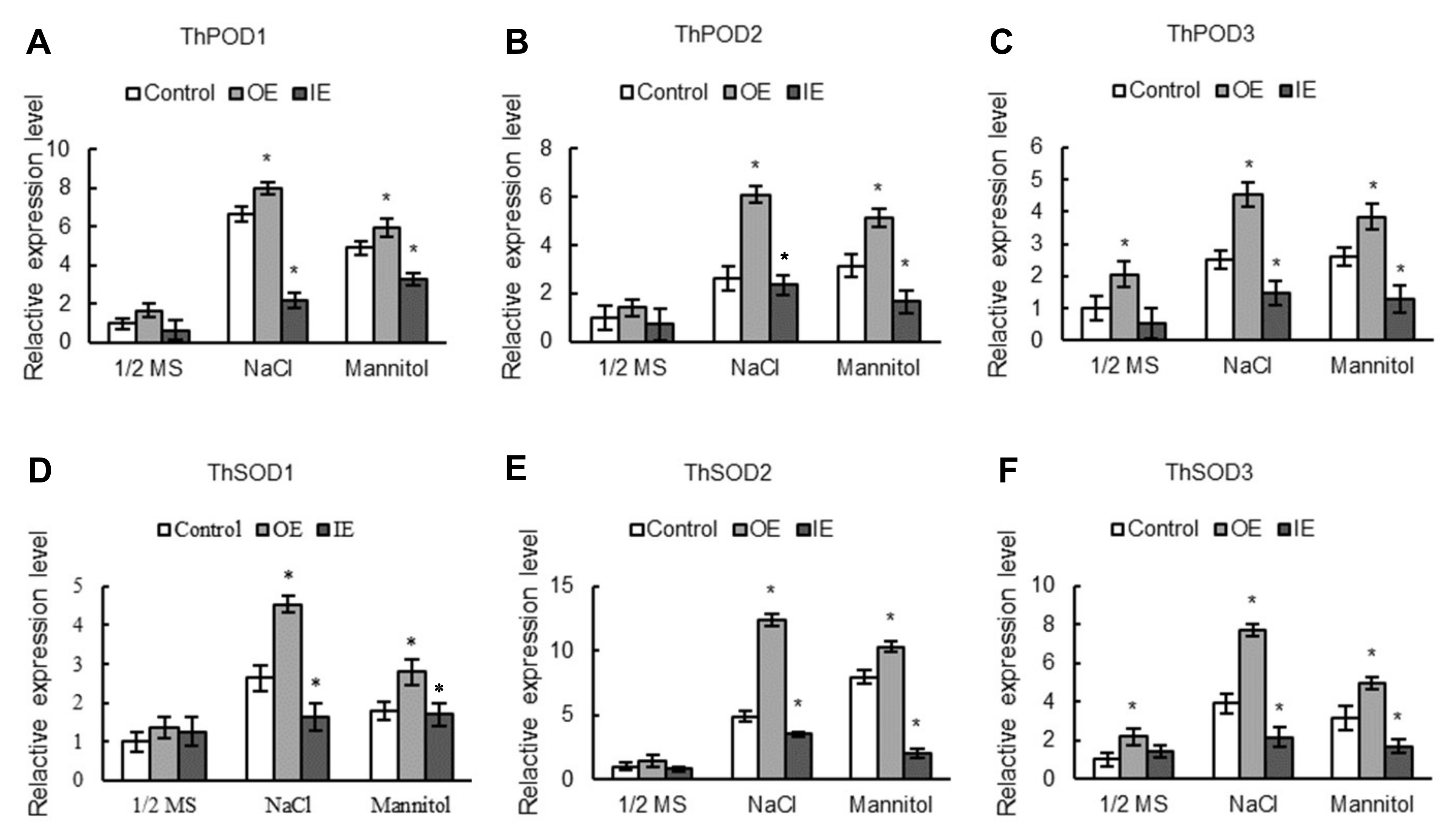
2.7. The Expression Levels of SODs and PODs Are Induced by ThNAC7
2.8. Overexpression of ThNAC7 Reduces Water Loss Rate, Cell Death and Malondialdehyde Contents
2.9. ThNAC7 Positively Affects Proline Biosynthesis
2.10. Identification of the Genes Regulated by ThNAC7 Using RNA-Seq
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. Plasmid Construction and Plant Transformation
4.3. Subcellular Location of ThNAC7 Proteins
4.4. Transactivational Activity of ThNAC7
4.5. Stress Tolerance Assays
4.6. Physiological Measurements
4.7. qRT-PCR Assay
4.8. RNA-Sequencing Analysis
4.9. Statistical Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Yamaguchi-Shinozaki, K.; Shinozaki, K. Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annu. Rev. Plant Biol. 2006, 57, 781–803. [Google Scholar] [CrossRef] [PubMed]
- Xiong, L.; Schumaker, K.S.; Zhu, J.K. Cell signaling during cold, drought, and salt stress. Plant Cell 2002, 14 (Suppl. 1), S165–S183. [Google Scholar] [CrossRef] [PubMed]
- Tak, H.; Negi, S.; Ganapathi, T.R. Banana NAC transcription factor MusaNAC042 is positively associated with drought and salinity tolerance. Protoplasma 2017, 254, 803–816. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Ji, X.; Nie, X.; Qu, M.; Zheng, L.; Tan, Z.; Zhao, H.; Huo, L.; Liu, S.; Zhang, B.; et al. Arabidopsis AtbHLH112 regulates the expression of genes involved in abiotic stress tolerance by binding to their E-box and GCG-box motifs. New Phytol. 2015, 207, 692–709. [Google Scholar] [CrossRef] [PubMed]
- Guo, H.; Wang, Y.; Wang, L.; Hu, P.; Wang, Y.; Jia, Y.; Zhang, C.; Zhang, Y.; Zhang, Y.; Wang, C.; et al. Expression of the MYB transcription factor gene BplMYB46 affects abiotic stress tolerance and secondary cell wall deposition in Betula platyphylla. Plant Biotechnol. J. 2017, 15, 107–121. [Google Scholar] [CrossRef] [PubMed]
- Zhu, D.; Hou, L.; Xiao, P.; Guo, Y.; Deyholos, M.K.; Liu, X. VvWRKY30, a grape WRKY transcription factor, plays a positive regulatory role under salinity stress. Plant Sci. 2019, 280, 132–142. [Google Scholar] [CrossRef]
- Kumar, M.; Lee, S.C.; Kim, J.Y.; Kim, S.J.; Aye, S.S.; Kim, S.R. Over-expression of dehydrin gene, Osdhn1, improves drought and salt stress tolerance through scavenging of reactive oxygen species in rice (Oryza sativa L.). J. Plant Biol. 2014, 57, 383–393. [Google Scholar] [CrossRef]
- Ijaz, R.; Ejaz, J.; Gao, S.; Liu, T.; Imtiaz, M.; Ye, Z.; Wang, T. Overexpression of annexin gene AnnSp2, enhances drought and salt tolerance through modulation of ABA synthesis and scavenging ROS in tomato. Sci. Rep. 2017, 7, 12087. [Google Scholar] [CrossRef]
- Puranik, S.; Sahu, P.P.; Srivastava, P.S.; Prasad, M. NAC proteins: Regulation and role in stress tolerance. Trends Plant Sci. 2012, 17, 369–381. [Google Scholar] [CrossRef]
- Riechmann, J.L.; Heard, J.; Martin, G.; Reuber, L.; Jiang, C.; Keddie, J.; Adam, L.; Pineda, O.; Ratcliffe, O.J.; Samaha, R.R.; et al. Arabidopsis transcription factors: Genome-wide comparative analysis among eukaryotes. Science 2000, 290, 2105–2110. [Google Scholar] [CrossRef]
- Tran, L.S.; Nishiyama, R.; Yamaguchi-Shinozaki, K.; Shinozaki, K. Potential utilization of NAC transcription factors to enhance abiotic stress tolerance in plants by biotechnological approach. G.M. Crops 2010, 1, 32–39. [Google Scholar] [CrossRef] [PubMed]
- Nuruzzaman, M.; Sharoni, A.M.; Kikuchi, S. Roles of NAC transcription factors in the regulation of biotic and abiotic stress responses in plants. Front. Microbiol. 2013, 4, 248. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nakashima, K.; Takasaki, H.; Mizoi, J.; Shinozaki, K.; Yamaguchi-Shinozaki, K. NAC transcription factors in plant abiotic stress responses. Biochim. Biophys. Acta. 2012, 1819, 97–103. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Deyholos, M.K. Comprehensive transcriptional profiling of NaCl stressed Arabidopsis roots reveals novel classes of responsive genes. BMC Plant Biol. 2006, 6, 25. [Google Scholar] [CrossRef] [PubMed]
- Nuruzzaman, M.; Sharoni, A.M.; Satoh, K.; Karim, M.R.; Harikrishna, J.A.; Shimizu, T.; Sasaya, T.; Omura, T.; Haque, M.A.; Hasan, S.M.; et al. NAC transcription factor family genes are differentially expressed in rice during infections with Rice dwarf virus, Rice black-streaked dwarf virus, Rice grassy stunt virus, Rice ragged stunt virus, and Rice transitory yellowing virus. Front. Plant Sci. 2015, 6, 676. [Google Scholar] [CrossRef] [PubMed]
- Fujita, M.; Fujita, Y.; Maruyama, K.; Seki, M.; Hiratsu, K.; Ohme-Takagi, M.; Tran, L.S.; Yamaguchi-Shinozaki, K.; Shinozaki, K. Adehydration-induced NAC protein, RD26, is involved in a novel ABA-dependent stress-signaling pathway. Plant J. 2004, 39, 863–876. [Google Scholar] [CrossRef] [PubMed]
- Tran, L.S.; Nakashima, K.; Sakuma, Y.; Simpson, S.D.; Fujita, Y.; Maruyama, K.; Fujita, M.; Seki, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Isolation and functional analysis of Arabidopsis stress-inducible NAC transcription factors that bind to a drought-responsive cis-element in the early responsive to dehydration stress 1 promoter. Plant Cell 2004, 16, 2481–2498. [Google Scholar] [CrossRef] [PubMed]
- Hu, H.; You, J.; Fang, Y.; Zhu, X.; Qi, Z.; Xiong, L. Characterization of transcription factor gene SNAC2 conferring cold and salt tolerance in rice. Plant Mol. Biol. 2008, 67, 169–181. [Google Scholar] [CrossRef]
- Fang, Y.; Liao, K.; Du, H.; Xu, Y.; Song, H.; Li, X.; Xiong, L. A stress responsive NAC transcription factor SNAC3 confers heat and drought tolerance through modulation of reactive oxygen species in rice. J. Exp. Bot. 2015, 66, 6803–6817. [Google Scholar] [CrossRef]
- Lee, D.K.; Chung, P.J.; Jeong, J.S.; Jang, G.; Bang, S.W.; Jung, H.; Kim, Y.S.; Ha, S.H.; Choi, Y.D.; Kim, J.K. The rice OsNAC6 transcription factor orchestrates multiple molecular mechanisms involving root structural adaptions and nicotianamine biosynthesis for drought tolerance. Plant Biotechnol. J. 2017, 15, 754–764. [Google Scholar] [CrossRef]
- Shim, J.S.; Oh, N.; Chung, P.J.; Kim, Y.S.; Choi, Y.D.; Kim, J.K. Overexpression of OsNAC14 Improves Drought Tolerance in Rice. Front. Plant Sci. 2018, 9, 310. [Google Scholar] [CrossRef] [PubMed]
- Nakashima, K.; Tran, L.S.; Van Nguyen, D.; Fujita, M.; Maruyama, K.; Todaka, D.; Ito, Y.; Hayashi, N.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Functional analysis of a NAC-type transcription factor OsNAC6 involved in abiotic and biotic stress-responsive gene expression in rice. Plant J. 2007, 51, 617–630. [Google Scholar] [CrossRef] [PubMed]
- Xue, G.P.; Way, H.M.; Richardson, T.; Drenth, J.; Joyce, P.A.; McIntyre, C.L. Overexpression of TaNAC69 leads to enhanced transcript levels of stress up-regulated genes and dehydration tolerance in bread wheat. Mol. Plant 2011, 4, 697–712. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Wang, B.; Li, Z.; Peng, Z.; Zhang, J. TsNAC1 Is a Key Transcription Factor in Abiotic Stress Resistance and Growth. Plant Physiol. 2018, 176, 742–756. [Google Scholar] [CrossRef] [PubMed]
- Negi, S.; Tak, H.; Ganapathi, T.R. Expression analysis of MusaNAC68 transcription factor and its functional analysis by overexpression in transgenic banana plants. Plant Cell Tiss. Organ Cult. 2016, 125, 59–70. [Google Scholar] [CrossRef]
- Jia, D.; Jiang, Q.; van Nocker, S.; Gong, X.; Ma, F. An apple (Malus domestica) NAC transcription factor enhances drought tolerance in transgenic apple plants. Plant Physiol Biochem. 2019, 139, 504–512. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Gao, C.; Wang, L.; Zheng, L.; Yang, C.; Wang, Y. Comprehensive transcriptional profiling of NaHCO3-stressed Tamarix hispida roots reveals networks of responsive genes. Plant Mol. Biol. 2014, 84, 145–157. [Google Scholar] [CrossRef]
- Demidchik, V.; Straltsova, D.; Medvedev, S.S.; Pozhvanov, G.A.; Sokolik, A.; Yurin, V. Stress-induced electrolyte leakage: The role of K+-permeable channels and involvement in programmed cell death and metabolic adjustment. J. Exp. Bot. 2014, 65, 1259–1270. [Google Scholar] [CrossRef]
- Hao, Y.J.; Song, Q.X.; Chen, H.W.; Zou, H.F.; Wei, W.; Kang, X.S.; Ma, B.; Zhang, W.K.; Zhang, J.S.; Chen, S.Y. Plant NAC-type transcription factor proteins contain a NARD domain for repression of transcriptional activation. Planta 2010, 232, 1033–1043. [Google Scholar] [CrossRef]
- Petrov, V.; Hille, J.; Mueller-Roeber, B.; Gechev, T.S. ROS-mediated abiotic stress-induced programmed cell death in plants. Front. Plant Sci. 2015, 6, 69. [Google Scholar] [CrossRef]
- Zang, D.; Wang, L.; Zhang, Y.; Zhao, H.; Wang, Y. ThDof1.4 and ThZFP1 constitute a transcriptional regulatory cascade involved in salt or osmotic stress in Tamarix hispida. Plant Mol. Biol. 2017, 94, 495–507. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Chu, Z.; Luo, J.; Zhou, Y.; Cai, Y.; Lu, Y.; Xia, J.; Kuang, H.; Ye, Z.; Ouyang, B. The C2H2 zinc-finger protein SlZF3 regulates AsA synthesis and salt tolerance by interacting with CSN5B. Plant Biotechnol. J. 2018, 16, 1201–1213. [Google Scholar] [CrossRef] [PubMed]
- Mao, C.; Ding, J.; Zhang, B.; Xi, D.; Ming, F. OsNAC2 positively affects salt-induced cell death and binds to the OsAP37 and OsCOX11 promoters. Plant J. 2018, 94, 454–468. [Google Scholar] [CrossRef]
- Wang, L.; Li, Z.; Lu, M.; Wang, Y. ThNAC13, a NAC Transcription Factor from Tamarix hispida, Confers Salt and Osmotic Stress Tolerance to Transgenic Tamarix and Arabidopsis. Front. Plant Sci. 2017, 8, 635. [Google Scholar] [CrossRef] [PubMed]
- Niu, F.; Wang, C.; Yan, J.; Guo, X.; Wu, F.; Yang, B.; Deyholos, M.K.; Jiang, Y.Q. Functional characterization of NAC55 transcription factor from oilseed rape (Brassica napus L.) as a novel transcriptional activator modulating reactive oxygen species accumulation and cell death. Plant Mol. Biol. 2016, 92, 89–104. [Google Scholar] [CrossRef] [PubMed]
- Jin, C.; Li, K.Q.; Xu, X.Y.; Zhang, H.P.; Chen, H.X.; Chen, Y.H.; Hao, J.; Wang, Y.; Huang, X.S.; Zhang, S.L. A Novel NAC Transcription Factor, PbeNAC1, of Pyrus betulifolia Confers Cold and Drought Tolerance via Interacting with PbeDREBs and Activating the Expression of Stress-Responsive Genes. Front. Plant Sci. 2017, 8, 1049. [Google Scholar] [CrossRef] [PubMed]
- Yan, J.; Tong, T.; Li, X.; Chen, Q.; Dai, M.; Niu, F.; Yang, M.; Deyholos, M.K.; Yang, B.; Jiang, Y.Q. A Novel NAC-Type Transcription Factor, NAC87, from Oilseed Rape Modulates Reactive Oxygen Species Accumulation and Cell Death. Plant Cell Physiol. 2018, 59, 290–303. [Google Scholar] [CrossRef] [PubMed]
- Silva-Ortega, C.O.; Ochoa-Alfaro, A.E.; Reyes-Agüero, J.A.; Aguado-Santacruz, G.A.; Jiménez-Bremont, J.F. Salt stress increases the expression of Pp5Ccs gene and induces proline accumulation in cactus pear. Plant Physiol. Biochem. 2008, 46, 82–92. [Google Scholar] [CrossRef] [PubMed]
- Kavi Kishor, P.B.; Sreenivasulu, N. Is proline accumulation per se correlated with stress tolerance or is proline homeostasis a more critical issue? Plant Cell Environ. 2014, 37, 300–311. [Google Scholar] [CrossRef]
- Forrest, K.L.; Bhave, M. Major intrinsic proteins (MIPs) in plants: A complex gene family with major impacts on plant phenotype. Funct. Integr. Genomics 2007, 7, 263–289. [Google Scholar] [CrossRef]
- Guo, L.; Wang, Z.Y.; Lin, H.; Cui, W.E.; Chen, J.; Liu, M.; Chen, Z.L.; Qu, L.J.; Gu, H. Expression and functional analysis of the rice plasma-membrane intrinsic protein gene family. Cell Res. 2006, 16, 277–286. [Google Scholar] [CrossRef] [PubMed]
- Park, C.J.; Shin, R.; Park, J.M.; Lee, G.J.; You, J.S.; Paek, K.H. Induction of pepper cDNA encoding a lipid transfer protein during the resistance response to tobacco mosaic virus. Plant Mol. Biol. 2002, 48, 243–254. [Google Scholar] [CrossRef] [PubMed]
- Cameron, K.D.; Teece, M.A.; Smart, L.B. Increased accumulation of cuticular wax and expression of lipid transfer protein in response to periodic drying events in leaves of tree tobacco. Plant Physiol. 2006, 140, 176–183. [Google Scholar] [CrossRef] [PubMed]
- Guo, L.; Yang, H.; Zhang, X.; Yang, S. Lipid transfer protein 3 as a target of MYB96 mediates freezing and drought stress in Arabidopsis. J. Exp. Bot. 2013, 64, 1755–1767. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Q.; Zhang, J.; Gao, X.; Tong, J.; Xiao, L.; Li, W.; Zhang, H. The Arabidopsis AP2/ERF transcription factor RAP2.6 participates in ABA, salt and osmotic stress responses. Gene 2010, 457, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Krishnaswamy, S.; Verma, S.; Rahman, M.H.; Kav, N.N. Functional characterization of four APETALA2-family genes (RAP2.6, RAP2.6L, DREB19 and DREB26) in Arabidopsis. Plant Mol. Biol. 2011, 75, 107–127. [Google Scholar] [CrossRef] [PubMed]
- Pandey, G.K.; Grant, J.J.; Cheong, Y.H.; Kim, B.G.; Li, L.; Luan, S. ABR1, an APETALA2-domain transcription factor that functions as a repressor of ABA response in Arabidopsis. Plant Physiol. 2005, 139, 1185–1193. [Google Scholar] [CrossRef]
- Song, C.; Je, J.; Hong, J.K.; Lim, C.O. Ectopic expression of an Arabidopsis dehydration-responsive element-binding factor DREB2C improves salt stress tolerance in crucifers. Plant Cell Rep. 2014, 33, 1239–1254. [Google Scholar] [CrossRef]
- Babitha, K.C.; Ramu, S.V.; Pruthvi, V.; Mahesh, P.; Nataraja, K.N.; Udayakumar, M. Co-expression of AtbHLH17 and AtWRKY28 confers resistance to abiotic stress in Arabidopsis. Transgenic Res. 2013, 22, 327–341. [Google Scholar] [CrossRef]
- Balazadeh, S.; Siddiqui, H.; Allu, A.D.; Matallana-Ramirez, L.P.; Caldana, C.; Mehrnia, M.; Zanor, M.I.; Köhler, B.; Mueller-Roeber, B. A gene regulatory network controlled by the NAC transcription factor ANAC092/AtNAC2/ORE1 during salt-promoted senescence. Plant J. 2010, 62, 250–264. [Google Scholar] [CrossRef]
- Ji, X.Y.; Zheng, L.; Liu, Y.J.; Nie, X.G.; Liu, S.N.; Wang, Y.C. A transient transformation system for the functional characterization of genes involved in stress response. Plant Mol. Biol. Rep. 2014, 32, 732–739. [Google Scholar] [CrossRef]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef] [PubMed]
- Lichtenthaler, H.K. Chlorophylls and carotenoids: Pigments of photosynthetic biomembranes. Methods Enzymol. 1987, 148, 350–382. [Google Scholar] [CrossRef]
- Fryer, M.J.; Oxborough, K.; Mullineaux, P.M.; Baker, N.R. Imaging of photo-oxidative stress responses in leaves. J. Exp. Bot. 2002, 53, 1249–1254. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Wang, L.; Meng, H.; Wen, H.; Fan, Y.; Zhao, J. Maize ABP9 enhances tolerance to multiple stresses in transgenic Arabidopsis by modulating ABA signaling and cellular levels of reactive oxygen species. Plant Mol. Biol. 2011, 75, 365–378. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Feng, Z.; Wang, X.; Wang, X.; Zhang, X. DEGseq: A R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics 2010, 26, 136–138. [Google Scholar] [CrossRef]
- Dal Santo, S.; Stampfl, H.; Krasensky, J.; Kempa, S.; Gibon, Y.; Petutschnig, E.; Rozhon, W.; Heuck, A.; Clausen, T.; Jonak, C. Stress-induced GSK3 regulates the redox stress response by phosphorylating glucose-6-phosphate dehydrogenase in Arabidopsis. Plant Cell 2012, 24, 3380–3392. [Google Scholar] [CrossRef]
- Bates, L.S.; Waldren, R.P.; Teare, I.D. Rapid determination of free proline for water-stress studies. Plant Soil 1973, 39, 205–207. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Wang, Y.; Gao, C.; Liang, Y.; Wang, C.; Yang, C.; Liu, G. A novel bZIP gene from Tamarix hispida mediates physiological responses to salt stress in tobacco plants. J. Plant Physiol. 2010, 167, 222–230. [Google Scholar] [CrossRef]







| Locus ID | Gene Name | FDR | Fold Change | Annotations-Description |
|---|---|---|---|---|
| AT5G42180 | PER64 | 2.62 × 10−8 | 6.65 | peroxidase |
| AT1G43160 | RAP2.6 | 6.88 × 10−16 | 4.12 | ethylene-responsive transcription factor RAP2-6 |
| AT1G09540 | MYB61 | 1.02 × 10−6 | 3.44 | myb domain protein 61 |
| AT5G64750 | ABR1 | 8.68 × 10−20 | 3.04 | ethylene-responsive transcription factor ABR1 |
| AT2G45180 | LTP | 1.23 × 10−21 | 2.78 | lipid transfer protein (LTP) family protein |
| AT2G32510 | MAPKKK17 | 5.09 × 10−9 | 2.38 | mitogen-activated protein kinase kinasekinase 17 |
| AT1G52690 | LEA | 0.004977 | 2.31 | Late embryogenesis abundant protein (LEA) family protein |
| AT1G21910 | DREB26 | 2.03 × 10−6 | 2.24 | ethylene-responsive transcription factor ERF012 |
| AT4G29930 | bHLH27 | 1.36 × 10−7 | 2.2 | transcription factor bHLH27 |
| AT1G05100 | MAPKKK18 | 0.008566 | 2.18 | mitogen-activated protein kinase kinasekinase 18 |
| AT4G22470 | LTP | 6.33 × 10−10 | 2.12 | lipid transfer protein (LTP) family protein |
| AT5G59310 | LTP4 | 1.96 × 10−7 | 2.11 | non-specific lipid-transfer protein 4 |
| AT2G37180 | RD28 | 6.49 × 10−44 | 2.1 | aquaporin PIP2-3 |
| AT5G12020 | HSP17.6II | 0.008487 | 2.1 | class II heat shock protein 17.6 |
| AT2G20880 | ERF053 | 0.00016 | 2.09 | ethylene-responsive transcription factor ERF053 |
| AT2G40340 | DREB2C | 0.000646 | 2.04 | dehydration-responsive element-binding protein 2C |
| AT5G61890 | ERF114 | 0.001014 | 2.03 | ethylene-responsive transcription factor ERF114 |
| AT4G05100 | AtMYB74 | 2.65 × 10−7 | 2.01 | myb domain protein 74 |
| AT3G18550 | BRC1 | 0.000907 | 1.93 | transcription factor TCP18 |
| AT4G18170 | WRKY28 | 2.97 × 10−7 | 1.75 | DNA binding-like protein |
| AT5G13330 | RAP2.6L | 3.84 × 10−12 | 1.71 | ethylene-responsive transcription factor ERF113 |
| AT1G67260 | TCP1 | 2.94 × 10−7 | 1.71 | transcription factor TCP1 |
| AT1G71030 | ATMYBL2 | 2.28 × 10−8 | 1.64 | putative myb family transcription factor |
| AT5G39610 | ATNAC2 | 9.09 × 10−6 | 1.63 | NAC-domain transcription factor |
| AT2G22200 | ERF056 | 4.68 × 10−5 | 1.63 | ethylene-responsive transcription factor ERF056 |
| AT1G75490 | DREB2D | 0.001527 | 1.6 | dehydration-responsive element-binding protein 2D |
| AT4G23400 | PIP1D | 3.65 × 10−11 | 1.57 | putative aquaporin PIP1-5 |
| AT5G40630 | - | 0.001874 | 1.56 | ubiquitin family protein |
| AT2G22770 | bHLH020 | 6.89 × 10−6 | 1.44 | putative bHLH transcription factor |
| AT2G26150 | ATHSFA2 | 2.19 × 10−6 | 1.43 | heat stress transcription factor A-2 |
| AT3G53420 | PIP2A | 1.95 × 10−13 | 1.4 | aquaporin PIP2-1 |
| AT5G56550 | OXS3 | 1.04 × 10−10 | 1.37 | protein OXIDATIVE STRESS 3 |
| AT2G21650 | MEE3 | 0.006908 | 1.36 | MYB transcription factor RSM1 |
| AT4G27410 | RD26 | 6.73 × 10−5 | 1.33 | NAC transcription factor RD26 |
| AT5G60890 | ATMYB34 | 1.14 × 10−7 | 1.31 | myb domain protein 34 |
| AT1G56650 | PAP1 | 2.41 × 10−5 | 1.28 | transcription factor MYB75 |
| AT1G54100 | ALDH7B4 | 1.61 × 10−8 | 1.28 | aldehyde dehydrogenase 7B4 |
| AT5G28770 | ATbZIP63 | 1.08 × 10−13 | 1.27 | basic leucine zipper 63 |
| AT5G50915 | bHLH137 | 3.16 × 10−5 | 1.21 | transcription factor bHLH137 |
| AT2G36830 | TIP | 2.04 × 10-10 | 1.21 | aquaporin TIP1-1 |
| Locus ID | FDR | Fold Change | Annotations-Description |
|---|---|---|---|
| AT5G42180 | 2.62 × 10−8 | 6.65 | peroxidase |
| AT5G24240 | 0.01 | 4.44 | phosphatidylinositol 3- and 4-kinase/ubiquitin family protein |
| AT5G57550 | 3.65 × 10−20 | 4.35 | probable xyloglucan endotransglucosylase/hydrolase protein 25 |
| AT1G75750 | 3.37 × 10−38 | 4.26 | GA-responsive GAST1 protein-like protein |
| AT2G38240 | 2.86 × 10−13 | 4.21 | 2-oxoglutarate (2OG) and Fe (II)-dependent oxygenase-like protein |
| AT1G43160 | 6.88 × 10−16 | 4.12 | ethylene-responsive transcription factor RAP2-6 |
| AT4G22485 | 5.48 × 10−4 | −4.81 | protease inhibitor/seed storage/LTP family protein |
| AT2G34390 | 6.8 × 10−14 | −4.7 | aquaporin NIP2-1 |
| AT1G50750 | 5.5 × 10−9 | −3.71 | plant mobile domain family protein |
| AT2G26010 | 1.24 × 10−5 | −3.07 | plant defensin 1.3 |
| AT4G29610 | 2.42 × 10−14 | −2.73 | cytidine/deoxycytidylate deaminase family protein |
| AT5G52750 | 2.14 × 10−8 | −1.36 | alpha/beta-hydrolases superfamily protein |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
He, Z.; Li, Z.; Lu, H.; Huo, L.; Wang, Z.; Wang, Y.; Ji, X. The NAC Protein from Tamarix hispida, ThNAC7, Confers Salt and Osmotic Stress Tolerance by Increasing Reactive Oxygen Species Scavenging Capability. Plants 2019, 8, 221. https://doi.org/10.3390/plants8070221
He Z, Li Z, Lu H, Huo L, Wang Z, Wang Y, Ji X. The NAC Protein from Tamarix hispida, ThNAC7, Confers Salt and Osmotic Stress Tolerance by Increasing Reactive Oxygen Species Scavenging Capability. Plants. 2019; 8(7):221. https://doi.org/10.3390/plants8070221
Chicago/Turabian StyleHe, Zihang, Ziyi Li, Huijun Lu, Lin Huo, Zhibo Wang, Yucheng Wang, and Xiaoyu Ji. 2019. "The NAC Protein from Tamarix hispida, ThNAC7, Confers Salt and Osmotic Stress Tolerance by Increasing Reactive Oxygen Species Scavenging Capability" Plants 8, no. 7: 221. https://doi.org/10.3390/plants8070221
APA StyleHe, Z., Li, Z., Lu, H., Huo, L., Wang, Z., Wang, Y., & Ji, X. (2019). The NAC Protein from Tamarix hispida, ThNAC7, Confers Salt and Osmotic Stress Tolerance by Increasing Reactive Oxygen Species Scavenging Capability. Plants, 8(7), 221. https://doi.org/10.3390/plants8070221





