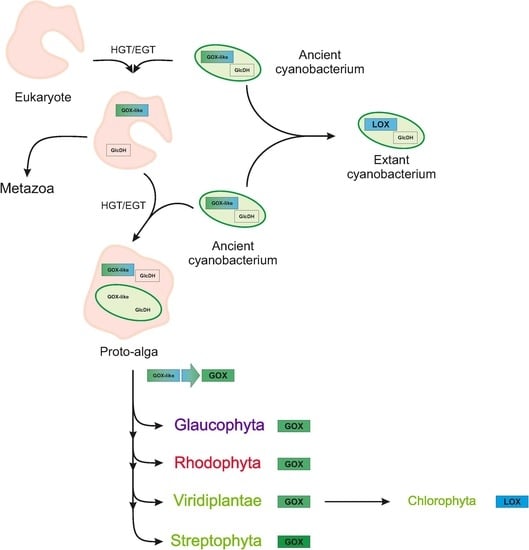
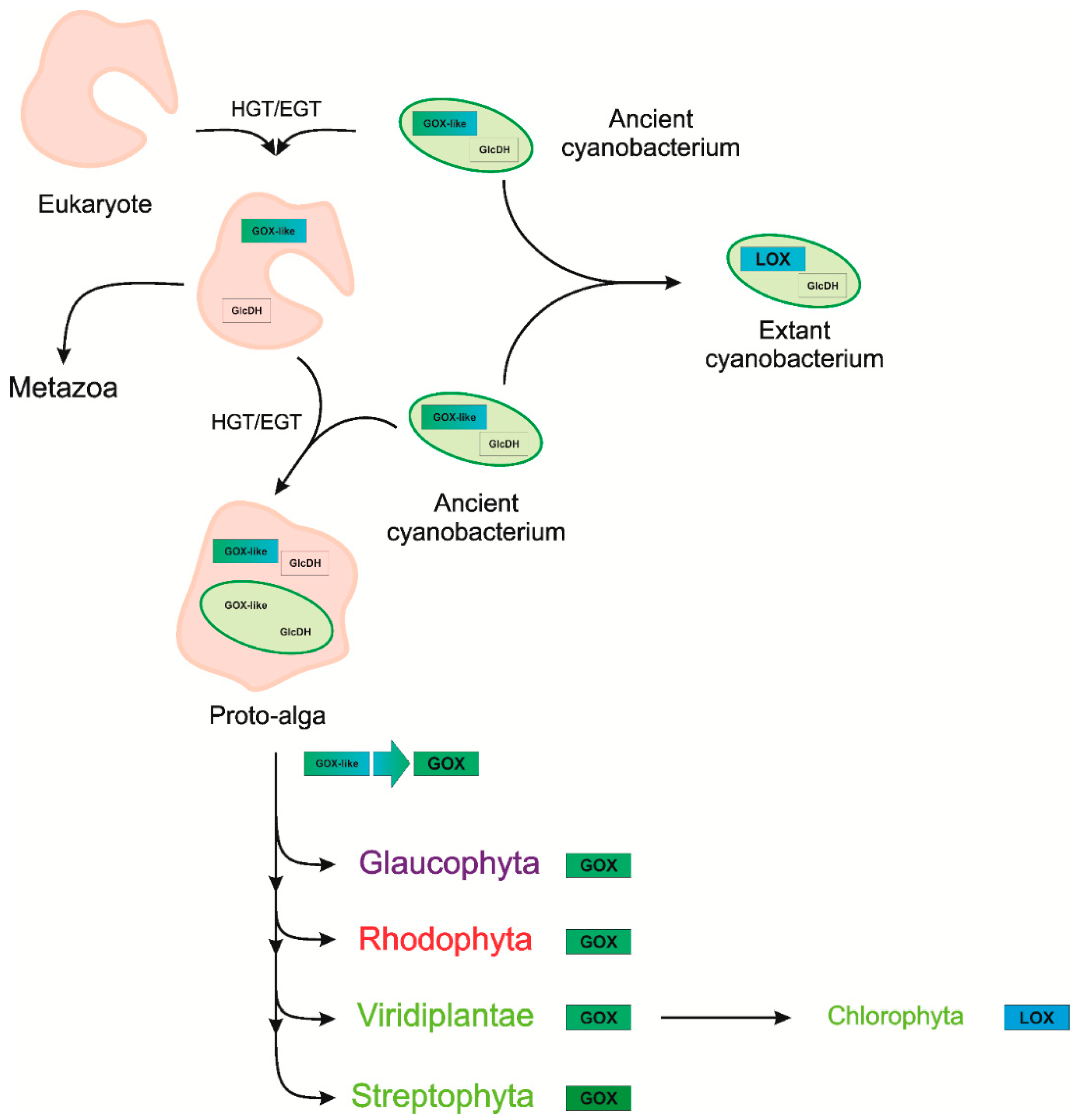
Evolution of Photorespiratory Glycolate Oxidase among Archaeplastida
Abstract
1. Introduction
2. Results
2.1. Data Mining
2.2. Monophyly of Eukaryotic and Cyanobacterial GOX-Like Proteins
2.3. Archaeplastida Except Chlorophyta Possess a GOX Protein
2.4. Ancestral GOX-Like Protein Sequence with Active Site Identical to Plant GOX
2.5. Activity of Ancestral GOX Proteins Point to Early Evolution of Preferential Glycolate Oxidation
3. Discussion
4. Materials and Methods
4.1. Phylogenetic Analysis
4.2. Ancestral Sequence Reconstruction
4.3. Sequencing of the cDNA Encoding the GOX from Spirogyra Pratensis
4.4. Estimation and Synthesis of the cDNA Encoding the GOX from Cyanophora Paradoxa
4.5. Cloning, Heterologous Expression and Purification of Recombinant GOX Proteins
4.6. Enzyme Activity Assays
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Hohmann-Marriott, M.F.; Blankenship, R.E. Evolution of photosynthesis. Annu. Rev. Plant Biol. 2011, 62, 515–548. [Google Scholar] [CrossRef]
- Soo, R.M.; Hemp, J.; Parks, D.H.; Fischer, W.W.; Hugenholtz, P. On the origins of oxygenic photosynthesis and aerobic respiration in Cyanobacteria. Science 2017, 355, 1436–1440. [Google Scholar] [CrossRef]
- Nowack, E.C.; Weber, A.P. Genomics-informed insights into endosymbiotic organelle evolution in photosynthetic Eukaryotes. Annu. Rev. Plant Biol. 2018, 69, 51–84. [Google Scholar] [CrossRef]
- Keeling, P.J. The endosymbiotic origin, diversification and fate of plastids. Philos. Trans. R. Soc. B Boil. Sci. 2010, 365, 729–748. [Google Scholar] [CrossRef] [PubMed]
- Stiller, J.W.; Reel, D.E.C.; Johnson, J.C. A single origin of plastids revisited: Convergent evolution in organellar genome content. J. Phycol. 2003, 39, 95–105. [Google Scholar] [CrossRef]
- Leliaert, F.; Smith, D.R.; Moreau, H.; Herron, M.D.; Verbruggen, H.; Delwiche, C.F.; De Clerck, O. Phylogeny and Molecular Evolution of the Green Algae. Crit. Rev. Plant Sci. 2012, 31, 1–46. [Google Scholar] [CrossRef]
- Nishiyama, T.; Sakayama, H.; de Vries, J.; Buschmann, H.; Saint-Marcoux, D.; Ullrich, K.K.; Haas, F.B.; Vanderstraeten, L.; Becker, D.; Lang, D.; et al. The Chara genome: Secondary complexity and implications for plant terrestrialization. Cell 2018, 174, 448–464. [Google Scholar] [CrossRef]
- Bowes, G.; Ogren, W.; Hageman, R. Phosphoglycolate production catalyzed by ribulose diphosphate carboxylase. Biochem. Biophys. Res. Commun. 1971, 45, 716–722. [Google Scholar] [CrossRef]
- Ogren, W.L. Affixing the O to Rubisco: Discovering the source of photorespiratory glycolate and its regulation. Photosynth. Res. 2003, 76, 53–63. [Google Scholar] [CrossRef]
- Tcherkez, G.G.B.; Farquhar, G.D.; Andrews, T.J. Despite slow catalysis and confused substrate specificity, all ribulose bisphosphate carboxylases may be nearly perfectly optimized. Proc. Natl. Acad. Sci. USA 2006, 103, 7246–7251. [Google Scholar] [CrossRef]
- Kelly, G.J.; Latzko, E. Inhibition of spinach-leaf phosphofructokinase by 2-phosphoglycollate. FEBS Lett. 1976, 68, 55–58. [Google Scholar] [CrossRef]
- Anderson, K.L.; Tayne, T.A.; Ward, D.M. Formation and fate of fermentation products in hot spring cyanobacterial mats. Appl. Environ. Microbiol. 1987, 53, 2343–2352. [Google Scholar] [CrossRef] [PubMed]
- Flügel, F.; Timm, S.; Arrivault, S.; Florian, A.; Stitt, M.; Fernie, A.R.; Bauwe, H. The Photorespiratory Metabolite 2-Phosphoglycolate Regulates Photosynthesis and Starch Accumulation in Arabidopsis. Plant Cell 2017, 29, 2537–2551. [Google Scholar] [CrossRef] [PubMed]
- Bauwe, H.; Hagemann, M.; Fernie, A.R. Photorespiration: Players, partners and origin. Trends Plant Sci. 2010, 15, 330–336. [Google Scholar] [CrossRef] [PubMed]
- Walker, B.J.; Vanloocke, A.; Bernacchi, C.J.; Ort, D.R. The Costs of Photorespiration to Food Production Now and in the Future. Annu. Rev. Plant Boil. 2016, 67, 107–129. [Google Scholar] [CrossRef]
- Ort, D.R.; Merchant, S.S.; Alric, J.; Barkan, A.; Blankenship, R.E.; Bock, R.; Croce, R.; Hanson, M.R.; Hibberd, J.M.; Long, S.P.; et al. Redesigning photosynthesis to sustainably meet global food and bioenergy demand. Proc. Natl. Acad. Sci. USA 2015, 112, 8529–8536. [Google Scholar] [CrossRef]
- Hagemann, M.; Bauwe, H. Photorespiration and the potential to improve photosynthesis. Curr. Opin. Chem. Biol. 2016, 35, 109–116. [Google Scholar] [CrossRef]
- Becker, B. Snow ball earth and the split of Streptophyta and Chlorophyta. Trends Plant Sci. 2013, 18, 180–183. [Google Scholar] [CrossRef]
- Eisenhut, M.; Ruth, W.; Haimovich, M.; Bauwe, H.; Kaplan, A.; Hagemann, M. The photorespiratory glycolate metabolism is essential for cyanobacteria and might have been conveyed endosymbiontically to plants. Proc. Natl. Acad. Sci. USA 2008, 105, 17199–17204. [Google Scholar] [CrossRef]
- Kern, R.; Bauwe, H.; Hagemann, M. Evolution of enzymes involved in the photorespiratory 2-phosphoglycolate cycle from cyanobacteria via algae toward plants. Photosynth. Res. 2011, 109, 103–114. [Google Scholar] [CrossRef]
- Kern, R.; Eisenhut, M.; Bauwe, H.; Weber, A.P.M.; Hagemann, M. Does the Cyanophora paradoxa genome revise our view on the evolution of photorespiratory enzymes? Plant Biol. 2013, 15, 759–768. [Google Scholar] [CrossRef] [PubMed]
- Hagemann, M.; Kern, R.; Maurino, V.G.; Hanson, D.T.; Weber, A.P.M.; Sage, R.F.; Bauwe, H. Evolution of photorespiration from cyanobacteria to land plants, considering protein phylogenies and acquisition of carbon concentrating mechanisms. J. Exp. Bot. 2016, 67, 2963–2976. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, Y.; Kanakagiri, S.; Van, K.; He, W.; Spalding, M.H. Disruption of the glycolate dehydrogenase gene in the high-CO2-requiring mutant HCR89 of Chlamydomonas reinhardtii. Can. J. Bot. 2005, 83, 820–833. [Google Scholar] [CrossRef]
- Kehlenbeck, P.; Goyal, A.; Tolbert, N.E. Factors affecting development of peroxisomes and glycolate metabolism among algae of different evolutionary lines of the Prasinophyceae. Plant Physiol. 1995, 109, 1363–1370. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Stabenau, H.; Winkler, U. Glycolate metabolism in green algae. Physiol. Plant. 2005, 123, 235–245. [Google Scholar] [CrossRef]
- Lindqvist, Y.; Brändén, C.I.; Mathews, F.S.; Lederer, F. Spinach glycolate oxidase and yeast flavocytochrome b2 are structurally homologous and evolutionarily related enzymes with distinctly different function and flavin mononucleotide binding. J. Biol. Chem. 1991, 266, 3198–3207. [Google Scholar]
- Maeda-Yorita, K.; Aki, K.; Sagai, H.; Misaki, H.; Massey, V. l-Lactate oxidase and l-lactate monooxygenase, Mechanistic variations on a common structural theme. Biochimie 1995, 77, 631–642. [Google Scholar] [CrossRef]
- Stenberg, K.; Lindqvist, Y. Three-dimensional structures of glycolate oxidase with bound active-site inhibitors. Protein Sci. 1997, 6, 1009–1015. [Google Scholar] [CrossRef]
- Leiros, I.; Wang, E.; Rasmussen, T.; Oksanen, E.; Repo, H.; Petersen, S.B.; Heikinheimo, P.; Hough, E. The 2.1 Å structure of Aerococcus viridans l-lactate oxidase (LOX). Acta Crystallogr. Sect. F Struct. Boil. Cryst. Commun. 2006, 62, 1185–1190. [Google Scholar] [CrossRef]
- Hackenberg, C.; Kern, R.; Huge, J.; Stal, L.J.; Tsuji, Y.; Kopka, J.; Shiraiwa, Y.; Bauwe, H.; Hagemann, M. Cyanobacterial lactate oxidases serve as essential partners in N2 fixation and evolved into photorespiratory glycolate oxidases in plants. Plant Cell 2011, 23, 2978–2990. [Google Scholar] [CrossRef]
- Ponce-Toledo, R.I.; Deschamps, P.; López-García, P.; Zivanovic, Y.; Benzerara, K.; Moreira, D. An early-branching freshwater cyanobacterium at the origin of plastids. Curr. Biol. 2017, 27, 386–391. [Google Scholar] [CrossRef] [PubMed]
- Esser, C.; Kuhn, A.; Groth, G.; Lercher, M.J.; Maurino, V.G. Plant and animal glycolate oxidases have a common eukaryotic ancestor and convergently duplicated to evolve long-chain 2-hydroxy acid oxidases. Mol. Biol. Evol. 2014, 31, 1089–1101. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Do, C.B.; Mahabhashyam, M.S.P.; Brudno, M.; Batzoglou, S. ProbCons, Probabilistic consistency-based multiple sequence alignment. Genome Res. 2005, 15, 330–340. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. MUSCLE, multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [PubMed]
- Griffiths, E.; Gupta, R.S. Signature sequences in diverse proteins provide evidence for the late divergence of the order Aquificales. Int. Microbiol. 2004, 7, 41–52. [Google Scholar]
- Placzek, S.; Schomburg, I.; Chang, A.; Jeske, L.; Ulbrich, M.; Tillack, J.; Schomburg, D. BRENDA in 2017, new perspectives and new tools in BRENDA. Nucleic Acids Res. 2017, 45, 380–388. [Google Scholar] [CrossRef]
- Rademacher, N.; Kern, R.; Fujiwara, T.; Mettler-Altmann, T.; Miyagishima, S.-y.; Hagemann, M.; Eisenhut, M.; Weber, A.P.M. Photorespiratory glycolate oxidase is essential for the survival of the red alga Cyanidioschyzon merolae under ambient CO2 conditions. J. Exp. Bot. 2016, 67, 3165–3175. [Google Scholar] [CrossRef]
- Stabenau, H. Microbodies from Spirogyra, organelles of a filamentous alga similar to leaf peroxisomes. Plant Physiol. 1976, 58, 693–695. [Google Scholar] [CrossRef]
- Betsche, T.; Schaller, D.; Melkonian, M. Identification and characterization of glycolate oxidase and related enzymes from the endocyanotic alga Cyanophora paradoxa and from Pea leaves. Plant Physiol. 1992, 98, 887–893. [Google Scholar] [CrossRef]
- Seki, M.; Iida, K.-I.; Saito, M.; Nakayama, H.; Yoshida, S.-I. Hydrogen Peroxide Production in Streptococcus pyogenes: Involvement of Lactate Oxidase and Coupling with Aerobic Utilization of Lactate. J. Bacteriol. 2004, 186, 2046–2051. [Google Scholar] [CrossRef] [PubMed]
- Zelitch, I.; Schultes, N.P.; Peterson, R.B.; Brown, P.; Brutnell, T.P. High glycolate oxidase activity is required for survival of maize in normal air. Plant Physiol. 2009, 149, 195–204. [Google Scholar] [CrossRef] [PubMed]
- Jones, J.M.; Morrell, J.C.; Gould, S.J. Identification and characterization of HAOX1; HAOX2; and HAOX3; three human peroxisomal 2-hydroxy acid oxidases. J. Biol. Chem. 2000, 275, 12590–12597. [Google Scholar] [CrossRef]
- Pellicer, M.T.; Badia, J.; Aguilar, J.; Baldoma, L. glc locus of Escherichia coli, Characterization of genes encoding the subunits of glycolate oxidase and the glc regulator protein. J. Bacteriol. 1996, 178, 2051–2059. [Google Scholar] [CrossRef]
- Gunshore, S.; Brush, E.J.; Hamilton, G.A. Equilibrium constants for the formation of glyoxylate thiohemiacetals and kinetic constants for their oxidation by O2 catalyzed by l-hydroxy acid oxidase. Bioorg. Chem. 1985, 13, 1–13. [Google Scholar] [CrossRef]
- Dupuis, L.; Caro, J.D.; Brachet, P.; Puigserver, A. Purification and some characteristics of chicken liver l-2-hydroxyacid oxidase A. FEBS Lett. 1990, 266, 183–186. [Google Scholar] [CrossRef][Green Version]
- Streitenberger, S.A.; López-Más, J.A.; Sánchez-Ferrer, Á.; García-Carmona, F. Use of Dye Affinity Chromatography for the Purification of Aerococcus viridans Lactate Oxidase. Biotechnol. Prog. 2002, 18, 657–659. [Google Scholar] [CrossRef]
- Barns, S.M.; Delwiche, C.F.; Palmer, J.D.; Dawson, S.C.; Hershberger, K.L.; Pace, N.R. Phylogenetic perspective on microbial life in hydrothermal ecosystems; past and present. Ciba Found. Symp. 1996, 202, 24–39. [Google Scholar]
- Da Cunha, V.; Gaia, M.; Gadelle, D.; Nasir, A.; Forterre, P. Lokiarchaea are close relatives of Euryarchaeota; not bridging the gap between prokaryotes and eukaryotes. PLoS Genet. 2017, 13, e1006810. [Google Scholar] [CrossRef]
- Schulz, F.; Eloe-Fadrosh, E.A.; Bowers, R.M.; Jarett, J.; Nielsen, T.; Ivanova, N.N.; Kyrpides, N.C.; Woyke, T. Towards a balanced view of the bacterial tree of life. Microbiome 2017, 5, 140. [Google Scholar] [CrossRef]
- Koonin, E.V.; Wolf, Y.I. Genomics of bacteria and archaea, the emerging dynamic view of the prokaryotic world. Nucleic Acids Res. 2008, 36, 6688–6719. [Google Scholar] [CrossRef] [PubMed]
- Gabaldón, T.; Snel, B.; van Zimmeren, F.; Hemrika, W.; Tabak, H.; Huynen, M.A. Origin and evolution of the peroxisomal proteome. Biol. Direct 2006, 1, 8. [Google Scholar] [CrossRef] [PubMed]
- Millard, A.; Scanlan, D.J.; Gallagher, C.; Marsh, A.; Taylor, P.C. Unexpected evolutionary proximity of eukaryotic and cyanobacterial enzymes responsible for biosynthesis of retinoic acid and its oxidation. Mol. BioSyst. 2014, 10, 380–383. [Google Scholar] [CrossRef]
- Boldt, R.; Edner, C.; Kolukisaoglu, U.; Hagemann, M.; Weckwerth, W.; Wienkoop, S.; Morgenthal, K.; Bauwe, H. D-Glycerate 3-kinase; the last unknown enzyme in the photorespiratory cycle in Arabidopsis; belongs to a novel kinase family. Plant Cell 2005, 17, 2413–2420. [Google Scholar] [CrossRef] [PubMed]
- Maruyama, S.; Matsuzaki, M.; Misawa, K.; Nozaki, H. Cyanobacterial contribution to the genomes of the plastid-lacking protists. BMC Evol. Biol. 2009, 9, 197. [Google Scholar] [CrossRef] [PubMed]
- Howe, C.; Barbrook, A.; Nisbet, R.; Lockhart, P.; Larkum, A. The origin of plastids. Philos. Trans. R. Soc. B Boil. Sci. 2008, 363, 2675–2685. [Google Scholar] [CrossRef]
- Atteia, A.; Adrait, A.; Brugiere, S.; Tardif, M.; van Lis, R.; Deusch, O.; Dagan, T.; Kuhn, L.; Gontero, B.; Martin, W.; et al. A proteomic survey of Chlamydomonas reinhardtii mitochondria sheds new light on the metabolic plasticity of the organelle and on the nature of the alpha-proteobacterial mitochondrial ancestor. Mol. Biol. Evol. 2009, 26, 1533–1548. [Google Scholar] [CrossRef][Green Version]
- Nelson, E.B.; Tolbert, N.E. Glycolate dehydrogenase in green algae. Arch. Biochem. Biophys. 1970, 141, 102–110. [Google Scholar] [CrossRef]
- Frederic, S.E.; Gruber, P.J.; Tolbert, N.E. Occurrence of glycolate dehydrogenase and glycolate oxidase in green plants - Evolutionary survey. Plant Physiol. 1973, 52, 318–323. [Google Scholar] [CrossRef]
- Hagemann, M.; Fernie, A.R.; Espie, G.S.; Kern, R.; Eisenhut, M.; Reumann, S.; Bauwe, H.; Weber, A.P.M. Evolution of the biochemistry of the photorespiratory C2 cycle. Plant Biol. 2013, 15, 639–647. [Google Scholar] [CrossRef]
- Shih, P.M.; Occhialini, A.; Cameron, J.C.; Andralojc, P.J.; Parry, M.A.J.; Kerfeld, C.A. Biochemical characterization of predicted Precambrian RuBisCO. Nat. Commun. 2016, 7, 10382. [Google Scholar] [CrossRef] [PubMed]
- Pervez, M.T.; Babar, M.E.; Nadeem, A.; Aslam, M.; Awan, A.R.; Aslam, N.; Hussain, T.; Naveed, N.; Qadri, S.; Waheed, U.; et al. Evaluating the accuracy and efficiency of multiple sequence alignment methods. Evol. Bioinform. 2014, 10, 205–217. [Google Scholar] [CrossRef] [PubMed]
- Anderson, C.L.; Strope, C.L.; Moriyama, E.N. SuiteMSA, visual tools for multiple sequence alignment comparison and molecular sequence simulation. BMC Bioinform. 2011, 12, 184. [Google Scholar]
- Castresana, J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 2000, 17, 540–552. [Google Scholar] [CrossRef]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5, molecular evolutionary genetics analysis using maximum likelihood; evolutionary distance; and maximum parsimony methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar] [CrossRef] [PubMed]
- Darriba, D.; Taboada, G.L.; Doallo, R.; Posada, D. ProtTest 3, fast selection of best-fit models of protein evolution. Bioinformatics 2011, 27, 1164–1165. [Google Scholar] [CrossRef] [PubMed]
- Le, S.Q.; Gascuel, O. An improved general amino acid replacement matrix. Mol. Biol. Evol. 2008, 25, 1307–1320. [Google Scholar] [CrossRef] [PubMed]
- Ronquist, F.; Teslenko, M.; van der Mark, P.; Ayres, D.L.; Darling, A.; Hohna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2, efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef]
- Stamatakis, A. RAxML version 8, a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef]
- Yang, Z. PAML 4, phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 2007, 24, 1586–1591. [Google Scholar] [CrossRef]
- Ashkenazy, H.; Penn, O.; Doron-Faigenboim, A.; Cohen, O.; Cannarozzi, G.; Zomer, O.; Pupko, T. FastML, a web server for probabilistic reconstruction of ancestral sequences. Nucleic Acids Res. 2012, 40, W580–W584. [Google Scholar] [CrossRef] [PubMed]
- Timme, R.E.; Delwiche, C.F. Uncovering the evolutionary origin of plant molecular processes, comparison of Coleochaete (Coleochaetales) and Spirogyra (Zygnematales) transcriptomes. BMC Plant Biol. 2010, 10, 96. [Google Scholar] [CrossRef] [PubMed]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Gabaldón, T. Evolution of the Peroxisomal Proteome. Alzheimer’s Dis. 2018, 89, 221–233. [Google Scholar]
- Gabaldón, T. Evolutionary considerations on the origin of peroxisomes from the endoplasmic reticulum; and their relationships with mitochondria. Cell Mol. Life Sci. 2014, 71, 2379–2382. [Google Scholar] [CrossRef]
- Sugiura, A.; Mattie, S.; Prudent, J.; McBride, H.M. Newly born peroxisomes are a hybrid of mitochondrial and ER-derived pre-peroxisomes. Nature 2017, 542, 251–254. [Google Scholar] [CrossRef]
- Gabaldón, T. Peroxisome diversity and evolution. Philos. Trans. R. Soc. B Boil. Sci. 2010, 365, 765–773. [Google Scholar] [CrossRef] [PubMed]





| Enzyme | Organism | Reference | l-Lactate | Glycolate | ||
|---|---|---|---|---|---|---|
| Vmax (µmol min−1 mg−1) | Km (mM) | Vmax (µmol min−1 mg−1) | Km (mM) | |||
| N3-GOX | Synthetic ancestral protein | This study | 0.17 ± 0.01 | 13.73 ± 1.93 | 0.87 ± 0.05 | 11.8 ± 0.61 |
| No-LOX x | Nostoc sp. PCC 7120 | Hackenberg et al.x | 12.73 ± 1.55 | 0.04 ± 0.01 | 0.05 ± 0.02 | 0.23 ± 0.05 |
| Cr-LOX x | Chlamydomonas reinhardtii | Hackenberg et al.x | 10.59 ± 0.46 | 0.08 ± 0.03 | 0.19 ± 0.06 | 1.24 ± 0.06 |
| Cp-GOXc | Cyanophora paradoxa | This study | 6.94 ± 0.76 | 9.27 ± 1.44 | 8.98 ± 1.25 | 0.38 ± 0.05 |
| Cm-GOX y | Cyanidioschyzon merolae | Rademacher et al.y | 3.27 ± 0.35 | 14.92 ± 2.99 | 1.6 ± 0.29 | 0.9 ± 0.23 |
| Sp-GOX | Spirogyra pratensis | This study | 24.21 ± 6.73 | 15.52 ± 4.55 | 28.09 ± 2.78 | 0.94 ± 0.12 |
| At-GOX2 x | Arabidopsis thaliana | Hackenberg et al.x | 0.74 ± 0.04 | 0.36 ± 0.18 | 35.64 ± 11.16 | 1.91 ± 0.64 |
| Amino Acid Position in No-LOX | |||
|---|---|---|---|
| 82 | 112 | 212 | |
| l-Lactate oxidases | |||
| No-LOX | M | L | F |
| Cr-LOX | M | V | F |
| Glycolate oxidases | |||
| N3-GOX | T | W | V |
| Cp-GOXc | T | W | V |
| Cm-GOX | T | W | V |
| Sp-GOX | T | W | V |
| At-GOX2 | T | W | V |
| N3-GOX | No-LOX | At-GOX2 | |
|---|---|---|---|
| N3-GOX | 1 | 0.56 | 0.68 |
| No-LOX | 0.56 | 1 | 0.46 |
| At-GOX2 | 0.68 | 0.46 | 1 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kern, R.; Facchinelli, F.; Delwiche, C.; Weber, A.P.M.; Bauwe, H.; Hagemann, M. Evolution of Photorespiratory Glycolate Oxidase among Archaeplastida. Plants 2020, 9, 106. https://doi.org/10.3390/plants9010106
Kern R, Facchinelli F, Delwiche C, Weber APM, Bauwe H, Hagemann M. Evolution of Photorespiratory Glycolate Oxidase among Archaeplastida. Plants. 2020; 9(1):106. https://doi.org/10.3390/plants9010106
Chicago/Turabian StyleKern, Ramona, Fabio Facchinelli, Charles Delwiche, Andreas P. M. Weber, Hermann Bauwe, and Martin Hagemann. 2020. "Evolution of Photorespiratory Glycolate Oxidase among Archaeplastida" Plants 9, no. 1: 106. https://doi.org/10.3390/plants9010106
APA StyleKern, R., Facchinelli, F., Delwiche, C., Weber, A. P. M., Bauwe, H., & Hagemann, M. (2020). Evolution of Photorespiratory Glycolate Oxidase among Archaeplastida. Plants, 9(1), 106. https://doi.org/10.3390/plants9010106