A Cytosolic Protein Kinase STY46 in Arabidopsis thaliana Is Involved in Plant Growth and Abiotic Stress Response
Abstract
1. Introduction
2. Results
2.1. In Silico Sequence and Expression Analysis of STY46
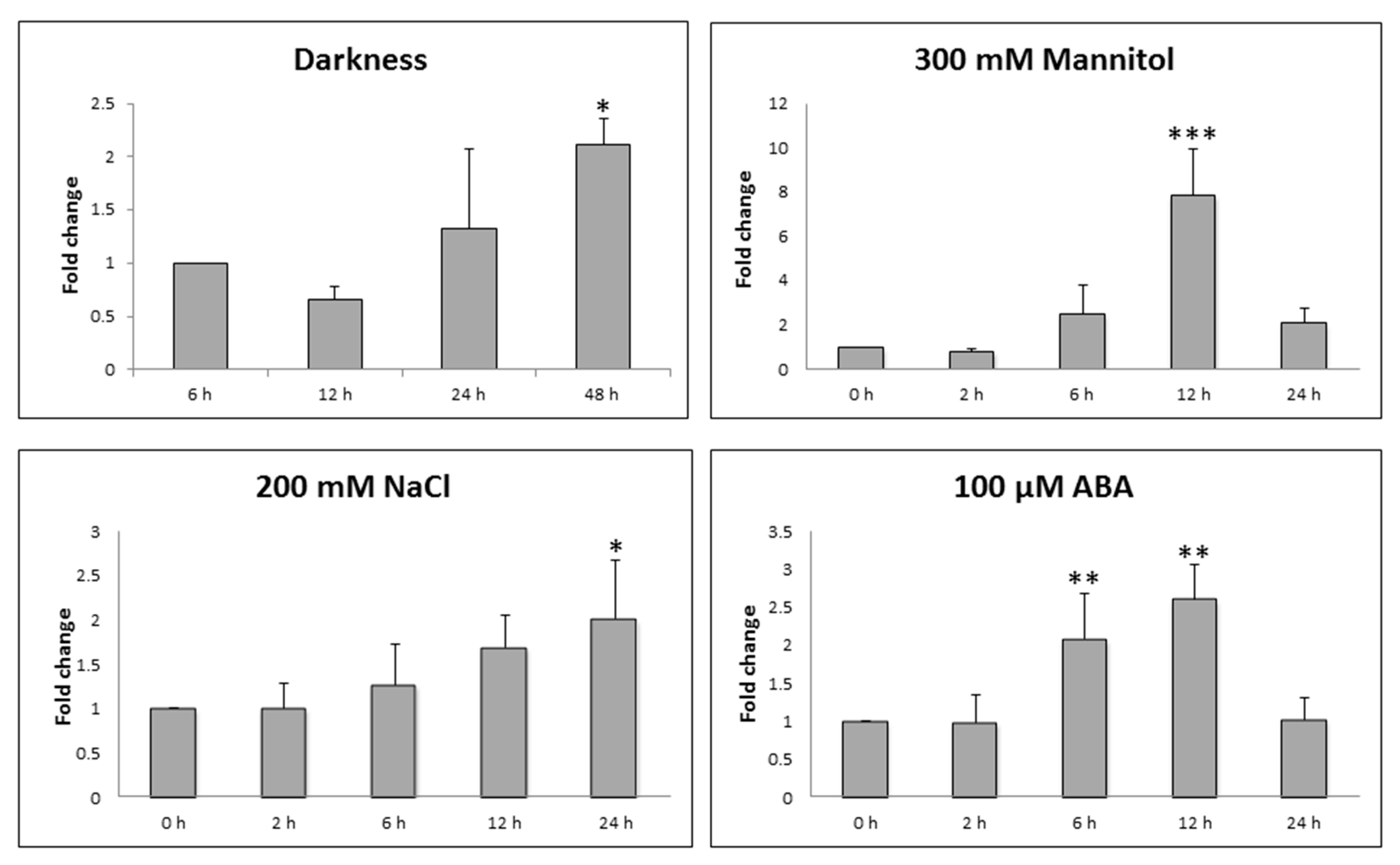
2.2. Expression Analysis of STY46 in Arabidopsis thaliana Rosettes
2.3. Generation of STY46 Transgenic Plants
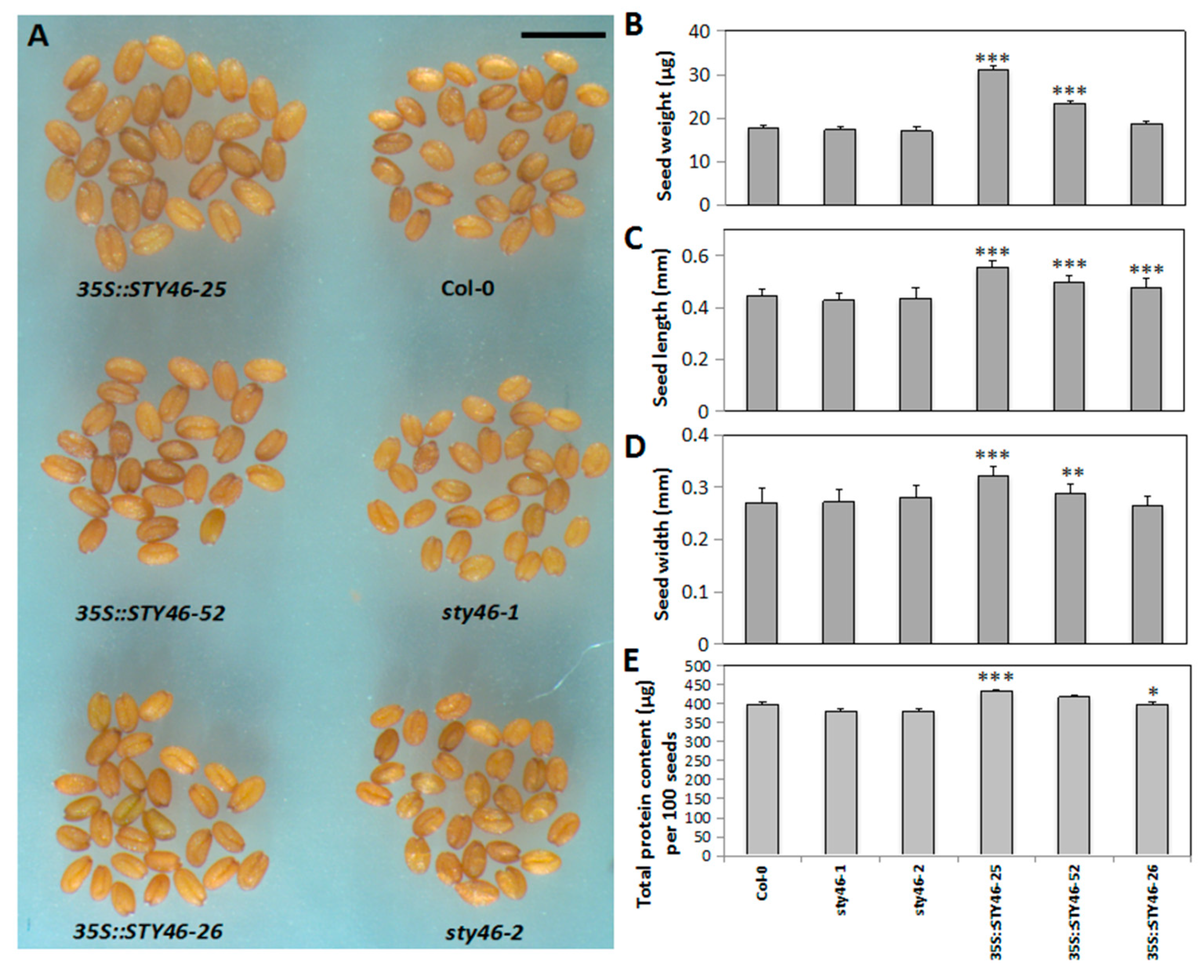
2.4. Characterization of STY46 Transgenic Plants under Control Conditions
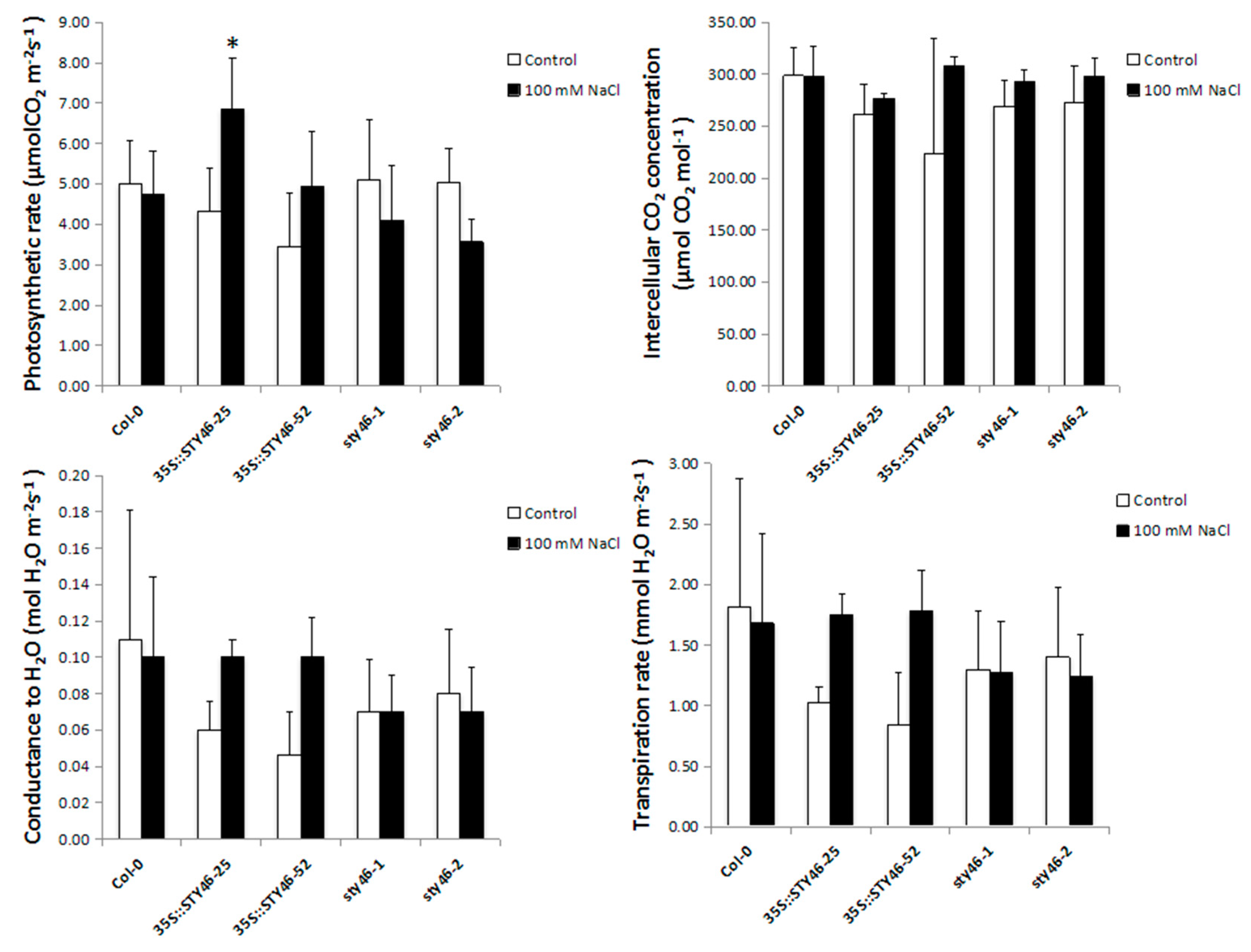
2.5. Characterization of STY46 Transgenic Plants under Stress
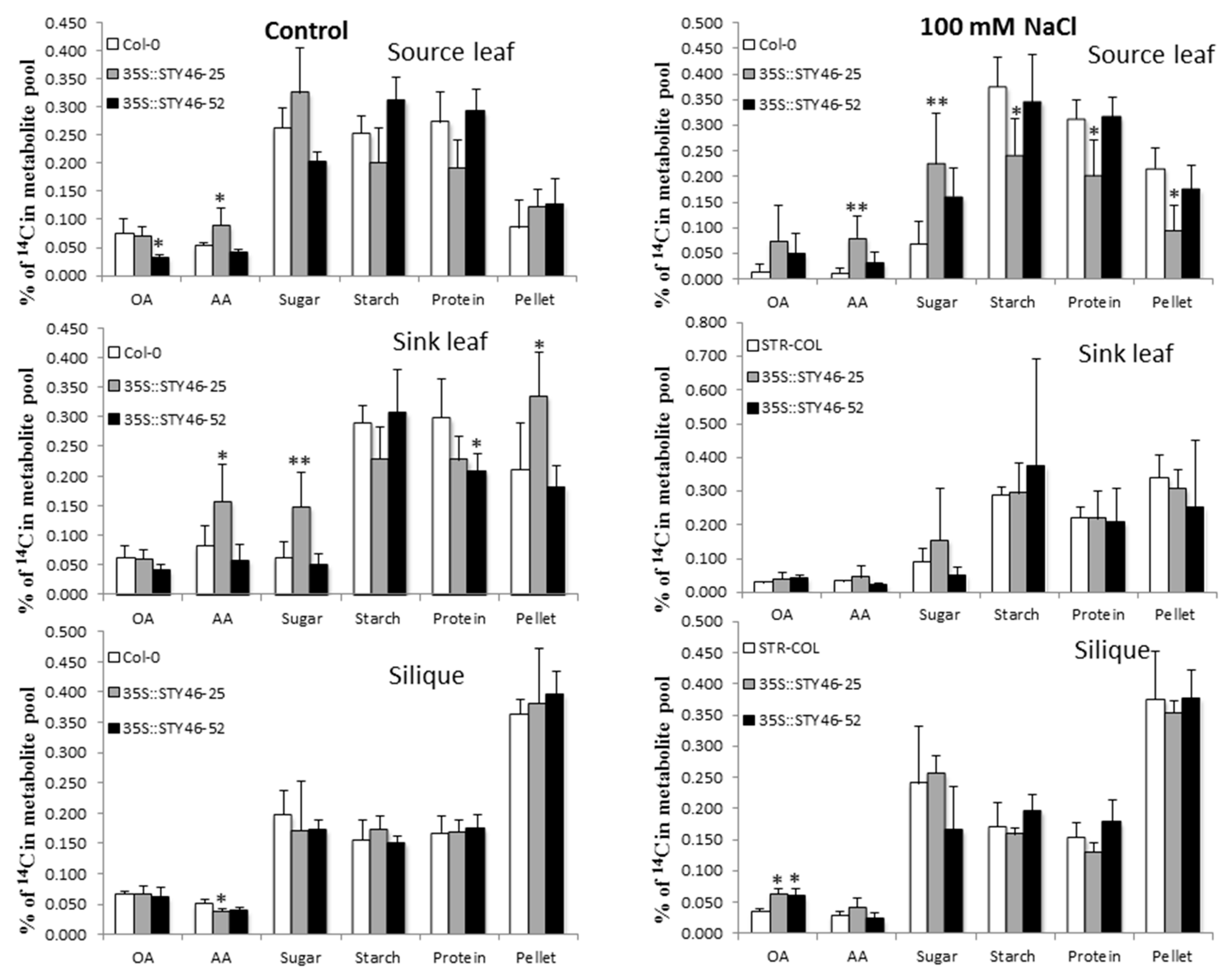
2.6. 14CO2 Partitioning and Allocation in Stress-Treated Plant Tissues
3. Discussion
3.1. Expression Analysis of STY46 under Stressed Conditions
3.2. STY46 Has a Role in Regulating Growth of Arabidopsis Source and Sink Tissues
3.3. The Role of STY46 in Abiotic Stress Response
4. Materials and Methods
4.1. Analysis of STY46 T-DNA Mutant Lines
4.2. Generation of STY46 Overexpressing Transgenic Arabidopsis Lines
4.3. Stress Treatments
4.4. Genomic DNA Extraction
4.5. Quantitative Real-Time Reverse Transcript-PCR
4.6. Carbohydrate Analysis
4.7. 14CO2 Pulse-Chase Labeling and Fractionation of 14CO2-Labelled Plant Tissue
4.8. Photosynthesis Measurements
4.9. Assay of Rosette Size and Biomass
4.10. Assay of Seed Size and Seed Weight
4.11. Total Protein Extraction and Quantification
4.12. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Krasavina, M.S.; Burmistrova, N.A.; Raldugina, G.N. The role of carbohydrates in plant resistance to abiotic stresses. In Emerging Technologies and Management of Crop Stress Tolerance; Elsevier: Amsterdam, The Netherlands, 2014; pp. 229–270. [Google Scholar]
- Hare, P.D.; Cress, W.A.; Van Staden, J. Dissecting the roles of osmolyte accumulation during stress. Plant Cell Environ. 1998, 21, 535–553. [Google Scholar] [CrossRef]
- Thomashow, M.F. Plant cold acclimation: Freezing tolerance genes and regulatory mechanisms. Annu. Rev. Plant Physiol. 1999, 50, 571–599. [Google Scholar] [CrossRef] [PubMed]
- Wanner, L.A.; Junttila, O. Cold-induced freezing tolerance in Arabidopsis. Plant. Physiol. 1999, 120, 391–399. [Google Scholar] [CrossRef] [PubMed]
- Graf, A.; Smith, A.M. Starch and the clock: The dark side of plant productivity. Trends Plant Sci. 2011, 16, 169–175. [Google Scholar] [CrossRef]
- Dong, S.; Beckles, D.M. Dynamic changes in the starch-sugar interconversion within plant source and sink tissues promote a better abiotic stress response. J. Plant Physiol. 2019, 234–235, 80–93. [Google Scholar] [CrossRef]
- Thalmann, M.; Santelia, D. Starch as a determinant of plant fitness under abiotic stress. New Phytol. 2017, 214, 943–951. [Google Scholar] [CrossRef]
- Smith, A.M.; Stitt, M. Coordination of carbon supply and plant growth. Plant Cell Environ. 2007, 30, 1126–1149. [Google Scholar] [CrossRef]
- Sulpice, R.; Pyl, E.T.; Ishihara, H.; Trenkamp, S.; Steinfath, M.; Witucka-Wall, H.; Gibon, Y.; Usadel, B.; Poree, F.; Piques, M.C.; et al. Starch as a major integrator in the regulation of plant growth. Proc. Natl. Acad. Sci. USA 2009, 106, 10348–10353. [Google Scholar] [CrossRef]
- Stitt, M.; Lunn, J.; Usadel, B. Arabidopsis and primary photosynthetic metabolism-more than the icing on the cake. Plant J. 2010, 61, 1067–1091. [Google Scholar] [CrossRef]
- Gupta, A.K.; Kaur, N. Sugar signalling and gene expression in relation to carbohydrate metabolism under abiotic stresses in plants. J. Biosci. 2005, 30, 761–776. [Google Scholar] [CrossRef]
- Brouquisse, R.; James, F.; Raymond, P.; Pradet, A. Study of glucose starvation in excised maize root tips. Plant Physiol. 1991, 96, 619–626. [Google Scholar] [CrossRef]
- Araujo, W.L.; Tohge, T.; Ishizaki, K.; Leaver, C.J.; Fernie, A.R. Protein degradation—An alternative respiratory substrate for stressed plants. Trends Plant Sci. 2011, 16, 489–498. [Google Scholar] [CrossRef] [PubMed]
- Izumi, M.; Hidema, J.; Makino, A.; Ishida, H. Autophagy Contributes to Nighttime Energy Availability for Growth in Arabidopsis. Plant Physiol. 2013, 161, 1682–1693. [Google Scholar] [CrossRef] [PubMed]
- Ishihara, H.; Obata, T.; Sulpice, R.; Fernie, A.R.; Stitt, M. Quantifying Protein Synthesis and Degradation in Arabidopsis by Dynamic (CO2)-C-13 Labeling and Analysis of Enrichment in Individual Amino Acids in Their Free Pools and in Protein. Plant Physiol. 2015, 168, 74–93. [Google Scholar] [CrossRef] [PubMed]
- Pilkington, S.M.; Encke, B.; Krohn, N.; Hohne, M.; Stitt, M.; Pyl, E.T. Relationship between starch degradation and carbon demand for maintenance and growth in Arabidopsis thaliana in different irradiance and temperature regimes. Plant Cell Environ. 2015, 38, 157–171. [Google Scholar] [CrossRef] [PubMed]
- Lee, E.J.; Matsumura, Y.; Soga, K.; Hoson, T.; Koizumi, N. Glycosyl hydrolases of cell wall are induced by sugar starvation in Arabidopsis. Plant Cell Physiol. 2007, 48, 405–413. [Google Scholar] [CrossRef]
- Patro, L.; Mohapatra, P.K.; Biswal, U.C.; Biswal, B. Dehydration induced loss of photosynthesis in Arabidopsis leaves during senescence is accompanied by the reversible enhancement in the activity of cell wall β-glucosidase. J. Photochem. Photobiol. B Biol. 2014, 137, 49–54. [Google Scholar] [CrossRef]
- Geigenberger, P. Regulation of Starch Biosynthesis in Response to a Fluctuating Environment. Plant Physiol. 2011, 155, 1566–1577. [Google Scholar] [CrossRef]
- Morkunas, I.; Borek, S.; Formela, M.; Ratajczak, L. Plant responses to sugar starvation. In Carbohydrates-Comprehensive Studies on Glycobiology and Glycotechnology; IntechOpen: Rijeka, Croatia, 2012; pp. 409–438. [Google Scholar]
- Arias, M.C.; Pelletier, S.; Hilliou, F.; Wattebled, F.; Renou, J.P.; D’Hulst, C. From dusk till dawn: The Arabidopsis thaliana sugar starving responsive network. Front. Plant Sci. 2014, 5, 482. [Google Scholar] [CrossRef]
- Weston, D.J.; Gunter, L.E.; Rogers, A.; Wullschleger, S.D. Connecting genes, coexpression modules, and molecular signatures to environmental stress phenotypes in plants. BMC Syst. Biol. 2008, 2, 16. [Google Scholar] [CrossRef]
- Yu, D.H.; Lim, J.H.; Wang, X.L.; Liang, F.M.; Xiao, G.H. Enhanced construction of gene regulatory networks using hub gene information. BMC Bioinform. 2017, 18, 186. [Google Scholar] [CrossRef]
- Rudrabhatla, P.; Reddy, M.M.; Rajasekharan, R. Genome-wide analysis and experimentation of plant serine/threonine/tyrosine-specific protein kinases. Plant Mol. Biol. 2006, 60, 293–319. [Google Scholar] [CrossRef] [PubMed]
- Lamberti, G.; Gugel, I.L.; Meurer, J.; Soll, J.; Schwenkert, S. The Cytosolic Kinases STY8, STY17, and STY46 Are Involved in Chloroplast Differentiation in Arabidopsis. Plant Physiol. 2011, 157, 70–85. [Google Scholar] [CrossRef]
- Beier, M.P.; Obara, M.; Taniai, A.; Sawa, Y.; Ishizawa, J.; Yoshida, H.; Tomita, N.; Yamanaka, T.; Ishizuka, Y.; Kudo, S.; et al. Lack of ACTPK1, an STY kinase, enhances ammonium uptake and use, and promotes growth of rice seedlings under sufficient external ammonium. Plant J. 2018, 93, 992–1006. [Google Scholar] [CrossRef] [PubMed]
- Ramachandiran, I.; Vijayakumar, A.; Ramya, V.; Rajasekharan, R. Arabidopsis serine/threonine/tyrosine protein kinase phosphorylates oil body proteins that regulate oil content in the seeds. Sci. Rep. 2018, 8, 1154. [Google Scholar] [CrossRef] [PubMed]
- Rudrabhatla, P.; Rajasekharan, R. Developmentally regulated dual-specificity kinase from peanut that is induced by abiotic stresses. Plant Physiol. 2002, 130, 380–390. [Google Scholar] [CrossRef][Green Version]
- Rudrabhatla, P.; Rajasekharan, R. Mutational analysis of stress-responsive peanut dual specificity protein kinase-Identification of tyrosine residues involved in regulation of protein kinase activity. J. Biol. Chem. 2003, 278, 17328–17335. [Google Scholar] [CrossRef]
- Parthibane, V.; Iyappan, R.; Vijayakumar, A.; Venkateshwari, V.; Rajasekharan, R. Serine/threonine/tyrosine protein kinase phosphorylates oleosin, a regulator of lipid metabolic functions. Plant Physiol. 2012, 159, 95–104. [Google Scholar] [CrossRef]
- Law, Y.-S.; Ngan, L.; Yan, J.; Kwok, L.Y.; Sun, Y.; Cheng, S.; Schwenkert, S.; Lim, B.L. Multiple Kinases Can Phosphorylate the N-Terminal Sequences of Mitochondrial Proteins in Arabidopsis thaliana. Front. Plant Sci. 2018, 9, 982. [Google Scholar] [CrossRef]
- Zimmermann, P.; Hirsch-Hoffmann, M.; Hennig, L.; Gruissem, W. GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiol. 2004, 136, 2621–2632. [Google Scholar] [CrossRef]
- Thitisaksakul, M.; Jimenez, R.C.; Arias, M.C.; Beckles, D.M. Effects of environmental factors on cereal starch biosynthesis and composition. J. Cereal Sci. 2012, 56, 67–80. [Google Scholar] [CrossRef]
- Dong, S.; Zhang, J.; Beckles, D.M. A pivotal role for starch in the reconfiguration of (14)C-partitioning and allocation in Arabidopsis thaliana under short-term abiotic stress. Sci. Rep. 2018, 8, 9314. [Google Scholar] [CrossRef]
- De Zelicourt, A.; Colcombet, J.; Hirt, H. The Role of MAPK Modules and ABA during Abiotic Stress Signaling. Trends Plant Sci. 2016, 21, 677–685. [Google Scholar] [CrossRef] [PubMed]
- Ishitani, M.; Xiong, L.M.; Stevenson, B.; Zhu, J.K. Genetic analysis of osmotic and cold stress signal transduction in Arabidopsis: Interactions and convergence of abscisic acid-dependent and abscisic acid-independent pathways. Plant Cell 1997, 9, 1935–1949. [Google Scholar] [CrossRef] [PubMed]
- Kaeppler, S.M.; Kaeppler, H.F.; Rhee, Y. Epigenetic aspects of somaclonal variation in plants. Plant Mol. Biol. 2000, 43, 179–188. [Google Scholar] [CrossRef]
- Hills, M.J. Control of storage-product synthesis in seeds. Curr. Opin. Plant Biol. 2004, 7, 302–308. [Google Scholar] [CrossRef]
- Baud, S.; Boutin, J.P.; Miquel, M.; Lepiniec, L.; Rochat, C. An integrated overview of seed development in Arabidopsis thaliana ecotype WS. Plant Physiol. Biochem. 2002, 40, 151–160. [Google Scholar] [CrossRef]
- Fait, A.; Angelovici, R.; Less, H.; Ohad, I.; Urbanczyk-Wochniak, E.; Fernie, A.R.; Galili, G. Arabidopsis seed development and germination is associated with temporally distinct metabolic switches. Plant Physiol. 2006, 142, 839–854. [Google Scholar] [CrossRef]
- Berger, F. Endosperm: The crossroad of seed development. Curr. Opin. Plant Biol. 2003, 6, 42–50. [Google Scholar] [CrossRef]
- Brown, R.C.; Lemmon, B.E.; Nguyen, H. Events during the first four rounds of mitosis establish three developmental domains in the syncytial endosperm of Arabidopsis thaliana. Protoplasma 2003, 222, 167–174. [Google Scholar] [CrossRef]
- Morley-Smith, E.R.; Pike, M.J.; Findlay, K.; Köckenberger, W.; Hill, L.M.; Smith, A.M.; Rawsthorne, S. The transport of sugars to developing embryos is not via the bulk endosperm in oilseed rape seeds. Plant Physiol. 2008, 147, 2121–2130. [Google Scholar] [CrossRef] [PubMed]
- Fujii, H.; Verslues, P.E.; Zhu, J.K. Identification of two protein kinases required for abscisic acid regulation of seed germination, root growth, and gene expression in Arabidopsis. Plant Cell 2007, 19, 485–494. [Google Scholar] [CrossRef] [PubMed]
- Krasensky, J.; Jonak, C. Drought, salt, and temperature stress-induced metabolic rearrangements and regulatory networks. J. Exp. Bot. 2012, 63, 1593–1608. [Google Scholar] [CrossRef] [PubMed]
- Tarkowski, L.P.; Van den Ende, W. Cold tolerance triggered by soluble sugars: A multifaceted countermeasure. Front. Plant Sci. 2015, 6, 203. [Google Scholar] [CrossRef] [PubMed]
- Keunen, E.; Peshev, D.; Vangronsveld, J.; Van den Ende, W.; Cuypers, A. Plant sugars are crucial players in the oxidative challenge during abiotic stress: Extending the traditional concept. Plant Cell Environ. 2013, 36, 1242–1255. [Google Scholar] [CrossRef]
- Rolland, F.; Baena-Gonzalez, E.; Sheen, J. Sugar sensing and signaling in plants: Conserved and novel mechanisms. Annu. Rev. Plant Biol. 2006, 57, 675–709. [Google Scholar] [CrossRef]
- Yu, S.M.; Lo, S.F.; Ho, T.H.D. Source-Sink Communication: Regulated by Hormone, Nutrient, and Stress Cross-Signaling. Trends Plant Sci. 2015, 20, 844–857. [Google Scholar] [CrossRef]
- MacNeill, G.J.; Mehrpouyan, S.; Minow, M.A.A.; Patterson, J.A.; Tetlow, I.J.; Emes, M.J. Starch as a source, starch as a sink: The bifunctional role of starch in carbon allocation. J. Exp. Bot. 2017, 68, 4433–4453. [Google Scholar] [CrossRef]
- Kempa, S.; Krasensky, J.; Dal Santo, S.; Kopka, J.; Jonak, C. A Central Role of Abscisic Acid in Stress-Regulated Carbohydrate Metabolism. PLoS ONE 2008, 3, e3935. [Google Scholar] [CrossRef]
- Hoagland, D.R.; Arnon, D.I. The Water-Culture Method for Growing Plants Without Soil. Circ. Calif. Agric. Exp. Stn. 1950, 347, 32. [Google Scholar]
- Richards, E.; Reichardt, M.; Rogers, S. Preparation of Genomic DNA from Plant Tissue. Curr. Protoc. Mol. Boil. 1994, 27. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(T)(-Delta Delta C) method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Sade, N.; Rubio-Wilhelmi, M.D.; Umnajkitikorn, K.; Blumwald, E. Stress-induced senescence and plant tolerance to abiotic stress. J. Exp. Bot. 2018, 69, 845–853. [Google Scholar] [CrossRef] [PubMed]
- Abràmoff, M.D.; Magalhães, P.J.; Ram, S.J. Image processing with ImageJ. Biophotonics Int. 2004, 11, 36–42. [Google Scholar]
- Focks, N.; Benning, C. wrinkled1: A novel, low-seed-oil mutant of Arabidopsis with a deficiency in the seed-specific regulation of carbohydrate metabolism. Plant Physiol. 1998, 118, 91–101. [Google Scholar] [CrossRef]
- Eisa, A.; Bölter, B.; Schwenkert, S. The ACT domain in chloroplast precursor–phosphorylating STY kinases binds metabolites and allosterically regulates kinase activity. J. Biol. Chem. 2019, 294, 17278–17288. [Google Scholar] [CrossRef]




© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dong, S.; Zhang, F.; Beckles, D.M. A Cytosolic Protein Kinase STY46 in Arabidopsis thaliana Is Involved in Plant Growth and Abiotic Stress Response. Plants 2020, 9, 57. https://doi.org/10.3390/plants9010057
Dong S, Zhang F, Beckles DM. A Cytosolic Protein Kinase STY46 in Arabidopsis thaliana Is Involved in Plant Growth and Abiotic Stress Response. Plants. 2020; 9(1):57. https://doi.org/10.3390/plants9010057
Chicago/Turabian StyleDong, Shaoyun, Fenglan Zhang, and Diane M. Beckles. 2020. "A Cytosolic Protein Kinase STY46 in Arabidopsis thaliana Is Involved in Plant Growth and Abiotic Stress Response" Plants 9, no. 1: 57. https://doi.org/10.3390/plants9010057
APA StyleDong, S., Zhang, F., & Beckles, D. M. (2020). A Cytosolic Protein Kinase STY46 in Arabidopsis thaliana Is Involved in Plant Growth and Abiotic Stress Response. Plants, 9(1), 57. https://doi.org/10.3390/plants9010057





