Genome-Wide Identification of Barley ABC Genes and Their Expression in Response to Abiotic Stress Treatment
Abstract
1. Introduction
2. Materials and Methods
2.1. Identification of ABC Gene Family Members in Barley
2.2. Multiple Sequence Alignment and Phylogenetic Analysis of the ABC Gene Family in Barley
2.3. Analysis of ABC Gene Structure and Chromosome Location in Barley
2.4. Construction of ABC Gene Expression Profiles in Barley
2.5. Stress Treatment, Total RNA Extraction, and qRT-PCR Analysis
3. Results
3.1. Identification and Physicochemical Properties of the ABC Gene Family in Barley
3.2. Phylogenetic Analysis of the ABC Gene Family in Barley
3.3. Chromosome Mapping of the ABC Gene Family in Barley
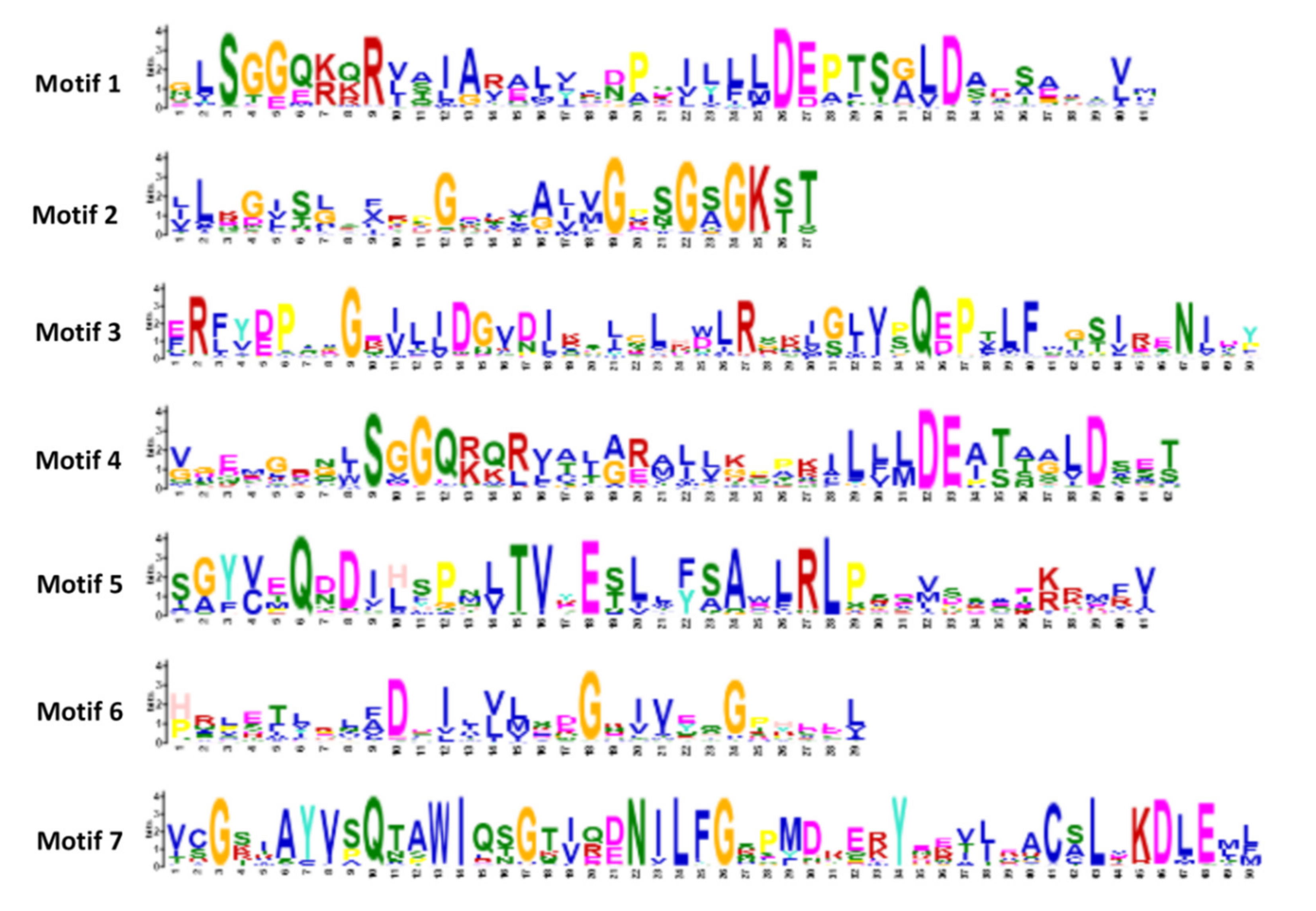
3.4. Analysis of Exon-Intron Structure and Conserved Domain of the ABC Gene Family in Barley
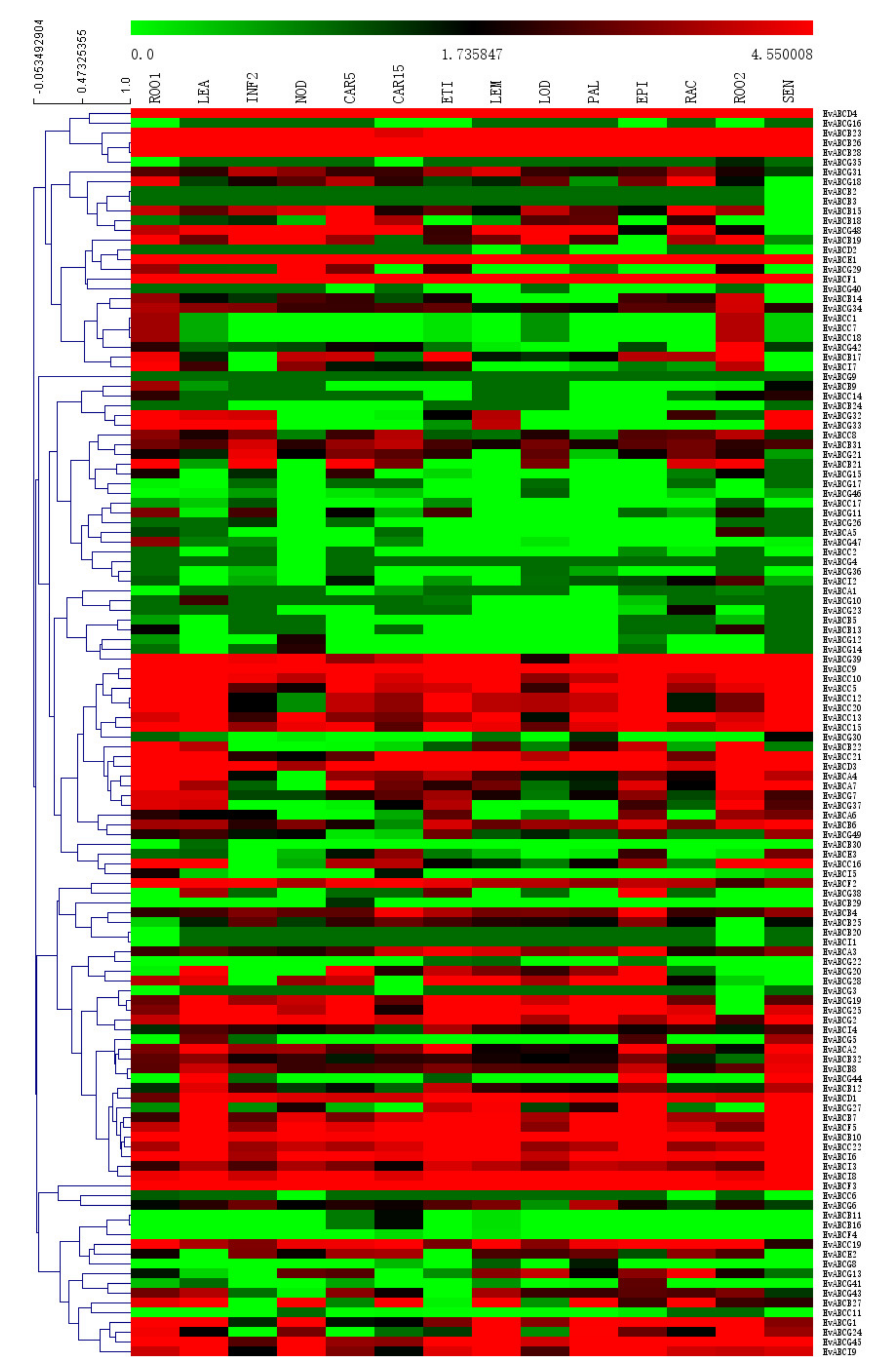
3.5. Tissue-Specific Expression of ABC Genes in Barley
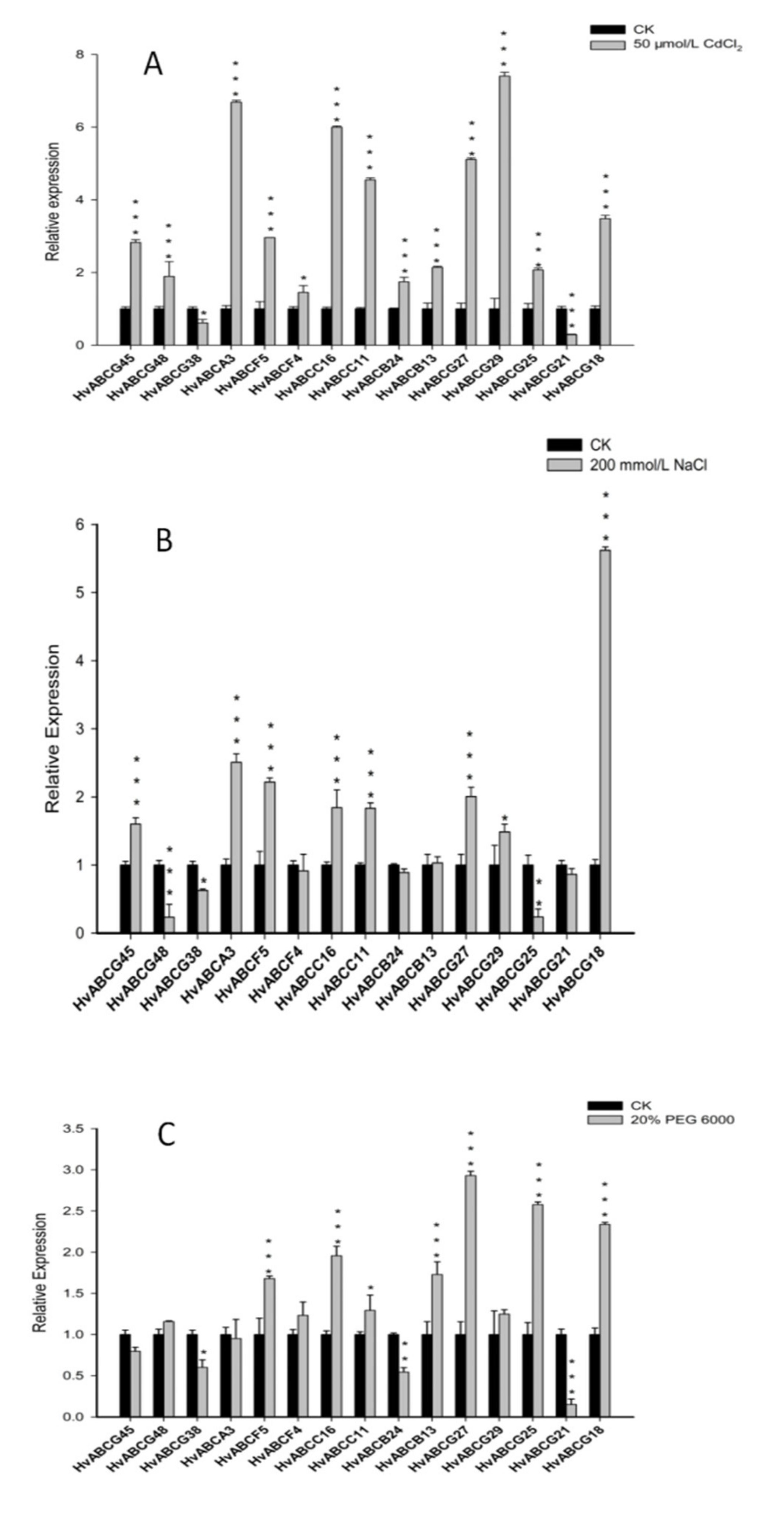
3.6. Expression Analysis of ABC Genes in Barley in Response to Abiotic Stress
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Higgins, C.F.; Linton, K.J. The ATP switch model for ABC transporters. Nat. Struct. Mol. Biol. 2004, 11, 918–926. [Google Scholar] [CrossRef] [PubMed]
- Davidson, A.L.; Dassa, E.; Orelle, C.; Chen, J. Structure, function, and evolution of bacterial ATP-binding cassette systems. Microbiol. Mol. Biol. Rev. 2008, 72, 317–364. [Google Scholar] [CrossRef] [PubMed]
- Lane, T.S.; Rempe, C.S.; Davitt, J.; Staton, M.E.; Peng, Y.; Soltis, D.E.; Melkonian, M.; Deyholos, M.; Leebens-Mack, J.H.; Chase, M. Diversity of ABC transporter genes across the plant kingdom and their potential utility in biotechnology. BMC Biotechnol. 2017, 16, 47. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.Q.; Zhao, Y.F. Structure and mechanism of ABC transporter. Chin. Bull. Life Sci. 2017, 29, 223–229. [Google Scholar]
- Jasinski, M.; Ducos, E.; Martinoia, E.; Boutry, M. The ATP-binding cassette transporters: Structure, function and gene family comparison between rice and Arabidopsis. Plant Physiol. 2003, 131, 1169. [Google Scholar] [CrossRef]
- Rea, P.A. Plant ATP-binding cassette transporters. Annu. Rev. Plant Biol. 2007, 58, 347–375. [Google Scholar] [CrossRef]
- Schneider, E.; Hunke, S. ATP-binding-cassette (ABC) transport systems: Functional and structural aspects of the ATP-hydrolyzing subunits/domains. FEMS Microbiol. Rev. 1998, 22, 1–20. [Google Scholar] [CrossRef]
- Mcfarlane, H.E.; Shin, J.J.H.; Bird, D.A.; Samuels, A.L. Arabidopsis ABCG transporters, which are required for export of diverse cuticular lipids, dimerize in different combinations. Plant Cell 2010, 22, 3066–3075. [Google Scholar] [CrossRef]
- Higgins, C.F. ABC transporters: From microorganisms to man. Annu. Rev. Cell Biol. 1992, 8, 67–113. [Google Scholar] [CrossRef]
- Dong, J.; Lai, R.; Jennings, J.L.; Link, A.J.; Hinnebusch, A.G. The novel ATP-binding cassette protein ARB1 is a shuttling factor that stimulates 40s and 60s ribosome biogenesis. Mol. Cell. Biol. 2005, 25, 9859–9873. [Google Scholar] [CrossRef]
- Dong, J.; Lai, R.; Nielsen, K.; Fekete, C.A.; Qiu, H.; Hinnebusch, A.G. The essential ATP-binding cassette protein RLI1 functions in translation by promoting preinitiation complex assembly. J. Biol. Chem. 2004, 279, 42157. [Google Scholar] [CrossRef] [PubMed]
- Garcia, O.; Bouige, P.; Forestier, C.; Dassa, E. Inventory and comparative analysis of rice and Arabidopsis ATP-binding cassette (ABC) systems. Mol. Biol. 2004, 343, 249–265. [Google Scholar] [CrossRef] [PubMed]
- Kärblane, K.; Gerassimenko, J.; Nigul, L.; Piirsoo, A.; Smialowska, A.; Vinkel, K.; Kylsten, P.; Ekwall, K.; Swoboda, P.; Truve, E.; et al. ABCE1 is a highly conserved RNA silencing suppressor. PLoS ONE 2015, 10, e0116702. [Google Scholar] [CrossRef] [PubMed]
- Kovalchuk, A.; Driessen, A.J. Phylogenetic analysis of fungal ABC transporters. BMC Genomics 2010, 11, 177. [Google Scholar] [CrossRef] [PubMed]
- Sanchez-Fernandez, R.; Davies, T.G.; Coleman, J.O.; Rea, P.A. The Arabidopsis thaliana ABC protein superfamily, a complete inventory. J. Biol. Chem. 2001, 276, 30231–30244. [Google Scholar] [CrossRef]
- Zhang, X.D.; Zhao, K.X.; Yang, Z.M. Identification of genomic ATP binding cassette (ABC) transporter genes and Cd-responsive ABCs in Brassica napus. Gene 2018, 664, 139–151. [Google Scholar] [CrossRef]
- Pang, K.; Li, Y.; Liu, M.; Meng, Z.; Yu, Y. Inventory and general analysis of the ATP-binding cassette (ABC) gene superfamily in maize (Zea mays L.). Gene 2013, 526, 411–428. [Google Scholar] [CrossRef]
- Verrier, P.J.; Bird, D.; Burla, B.; Dassa, E.; Forestier, C.; Geisler, M.; Klein, M.; Kolukisaoglu, U.; Lee, Y.; Martinoia, E.; et al. Plant ABC proteins--a unified nomenclature and updated inventory. Trends Plant Sci. 2008, 13, 151–159. [Google Scholar] [CrossRef]
- Annilo, T.; Chen, Z.Q.; Shulenin, S.; Costantino, J.; Thomas, L.; Lou, H.; Stefanov, S.; Dean, M. Evolution of the vertebrate ABC gene family: Analysis of gene birth and death. Genomics 2006, 88, 1–11. [Google Scholar] [CrossRef]
- Dermauw, W.; Van Leeuwen, T. The ABC gene family in arthropods: Comparative genomics and role in insecticide transport and resistance. Insect. Biochem. Mol. Biol. 2014, 45, 89–110. [Google Scholar] [CrossRef]
- Kang, J.; Park, J.; Choi, H.; Burla, B.; Kretzschmar, T.; Lee, Y.; Martinoia, E. Plant ABC Transporters. Arabidopsis Book 2011, 9, e0153. [Google Scholar] [CrossRef] [PubMed]
- Roy, S.J.; Tucker, E.J.; Tester, M.A. Genetic analysis of abiotic stress tolerance in crops. Curr. Opin. Plant Biol. 2011, 14, 232–239. [Google Scholar] [CrossRef] [PubMed]
- Orsi, C.H.; Tanksley, S.D. Natural variation in an ABC transporter gene associated with seed size evolution in tomato species. PLoS Genet. 2009, 5, e1000347. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.H.; Zhao, H.J.; Liu, Q.L.; Frank, T.; Engel, K.H.; An, G.; Shu, Q.Y. Mutations of the multi-drug resistance-associated protein ABC transporter gene 5 result in reduction of phytic acid in rice seeds. Theor. Appl. Genet. 2009, 119, 75–83. [Google Scholar] [CrossRef]
- Crouzet, J.; Trombik, T.; Fraysse, A.S.; Boutry, M. Organization and function of the plant pleiotropic drug resistance ABC transporter family. FEBS Lett. 2006, 580, 1123–1130. [Google Scholar] [CrossRef]
- Hwang, J.U.; Song, W.Y.; Hong, D.; Ko, D.; Yamaoka, Y.; Jang, S.; Yim, S. Plant ABC transporters enable many unique aspects of a terrestrial plant’s lifestyle. Mol. Plant 2016, 9, 338–355. [Google Scholar] [CrossRef]
- Tong, T.; Fang, Y.X.; Zhang, Z.L.; Zheng, J.J.; Xue, D.W. Genome-wide identification, phylogenetic and expression analysis of SBP-box gene family in barley (Hordeum vulgare L.). Plant Growth Regul. 2020, 90, 1–13. [Google Scholar] [CrossRef]
- Finn, R.D.; Tate, J.; Mistry, J.; Coggill, P.C.; Sammut, S.J.; Hotz, H.R.; Ceric, G.; Forslund, K.; Eddy, S.R.; Sonnhammer, E.L.; et al. The Pfam protein families database. Nucleic Acids Res. 2008, 36, D281–D288. [Google Scholar] [CrossRef]
- Marchler-Bauer, A.; Derbyshire, M.K.; Gonzales, N.R.; Lu, S.; Chitsaz, F.; Geer, L.Y.; Geer, R.C.; He, J.; Gwadz, M.; Hurwitz, D.I.; et al. CDD: NCBI’s conserved domain database. Nucleic Acids Res. 2015, 43, D222–D226. [Google Scholar] [CrossRef]
- Letunic, I.; Doerks, T.; Bork, P. SMART 7: Recent updates to the protein domain annotation resource. Nucleic Acids Res. 2012, 40, D302–D305. [Google Scholar] [CrossRef]
- Artimo, P.; Jonnalagedda, M.; Arnold, K.; Baratin, D.; Csardi, G.; de Castro, E.; Duvaud, S.; Flegel, V.; Fortier, A.; Gasteiger, E.; et al. ExPASy: SIB bioinformatics resource portal. Nucleic Acids Res. 2012, 40, W597–W603. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.Y.; Sun, W.J.; Ma, Z.T. Genome-wide investigation of the AP2/ERF gene family in tartary buckwheat (Fagopyum Tataricum). BMC Plant Biol. 2019, 19, 84. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 5, 1792–1797. [Google Scholar] [CrossRef]
- Mcwilliam, H.; Li, W.; Uludag, M. Analysis Tool Web Services from the EMBL-EBI. Nucleic Acids Res. 2013, 41, W597–W600. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. MUSCLE: A multiple sequence alignment method with reduced time and space complexity. BMC Bioinform. 2004, 5, 113. [Google Scholar] [CrossRef] [PubMed]
- Hu, B.; Jin, J.; Guo, A.Y.; Zhang, H.; Gao, G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 2014, 31, 1296–1297. [Google Scholar] [CrossRef]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef]
- Horton, P.; Park, K.J.; Obayashi, T.; Nakai, K. Protein subcellular localization prediction with WOLF PSORT. In Proceedings of the 4th Asia-Pacific Bioinformatics Conference, Taipei, Taiwan, 13–16 February 2006. [Google Scholar]
- Zhang, X.; Tong, T.; Tian, B.; Fang, Y.; Xue, D. Physiological, biochemical and molecular responses of barley seedlings to Aluminum stress. Phyton 2019, 88, 253–260. [Google Scholar] [CrossRef]
- Tong, C.; Wang, X.; Yu, J.; Wu, J.; Liu, S. Comprehensive analysis of RNA-seq data reveals the complexity of the transcriptome in Brassica rapa. BMC Genomics 2013, 14, 689. [Google Scholar] [CrossRef]
- Theodoulou, F.L. Plant ABC Transporters. Biochim. Biophys. Acta 2000, 1465, 79–103. [Google Scholar] [CrossRef]
- Song, W.Y.; Mendoza-Cózatl, D.G.; Lee, Y.; Schroeder, J.I.; Ahn, S.N.; Lee, H.S.; Wicker, T.; Martinoia, E. Phytochelatin-metal(loid) transport into vacuoles shows different substrate preferences in barley and Arabidopsis. Plant Cell Environ. 2013, 37, 1192–1201. [Google Scholar] [CrossRef] [PubMed]
- Sousa, C.A.; Hanselaer, S.; Soares, E.V. ABCC subfamily vacuolar transporters are involved in Pb (Lead) detoxification in Saccharomyces cerevisiae. Appl. Biochem. Biotechnol. 2014, 175, 65–74. [Google Scholar] [CrossRef] [PubMed]
- Theodoulou, F.L.; Kerr, I.D. ABC transporter research: Going strong 40 years on. Biochem. Soc. Trans. 2015, 43, 1033–1040. [Google Scholar] [CrossRef] [PubMed]
- Martinoia, E.; Klein, M.; Geisler, M.; Bovet, L.; Forestier, C.; Kolukisaoglu, U.; Müller-Röber, B.; Schulz, B. Multifunctionality of plant ABC transporters—More than just detoxifiers. Planta 2002, 214, 345–355. [Google Scholar] [CrossRef]
- Martin, C.; Berridge, G.; Mistry, P.; Higgins, C.; Charlton, P.; Callaghan, R. Drug binding sites on P-glycoprotein are altered by ATP binding prior to nucleotide hydrolysis. Biochemistry 2000, 39, 11901–11906. [Google Scholar] [CrossRef]
- Broehan, G.; Kroeger, T.; Lorenzen, M.; Merzendorfer, H. Functional analysis of the ATP-binding cassette (ABC) transporter gene family of Tribolium castaneum. BMC Genomics 2013, 14, 1–18. [Google Scholar] [CrossRef]
- Xu, X.; Qiu, J.; Xu, Y.; Xu, C.W. Molecular evolution and expression analysis of subfamily ABCB transporter genes in rice. Chin. J. Rice Sci. 2012, 26, 127–136. [Google Scholar]
- Akifumi, S.; Nobukazu, S.; Shusei, S.; Yasukazu, N.; Satoshi, T.; Kazufumi, Y. Genome-wide analysis of ATP-binding cassette (ABC) proteins in a model legume plant, Lotus japonicus: Comparison with Arabidopsis ABC protein family. DNA Res. 2006, 13, 205–228. [Google Scholar]
- Çakır, B.; Kılıçkaya, O. Whole-genome survey of the putative ATP-binding cassette transporter family genes in Vitis vinifera. PLoS ONE 2013, 8, e78860. [Google Scholar] [CrossRef]
- Amoako, O.P.; Ayaka, M.; Mami, S.; Enrico, M.; Stefan, R.; Koh, A.; Daisuke, S.; Shungo, O.; Shogo, M.; Katsuhiro, S. Genome-wide analysis of ATP binding cassette (ABC) transporters in tomato. PLoS ONE 2018, 13, e0200854. [Google Scholar]
- Okamoto, K.; Ueda, H.; Shimada, T.; Tamura, K.; Hara-Nishimura, I. An ABC transporter B family protein, ABCB19, is required for cytoplasmic streaming and gravitropism of the inflorescence stems. Plant Signal Behav. 2016, 11, e1010947. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Liu, L.; Mo, H.; Qian, L.; Cao, Y.; Cui, S.; Li, X.; Ma, L.; Merks, R.M.H.J.P.O. The ATP-Binding cassette transporter ABCB19 regulates postembryonic organ separation in Arabidopsis. PLoS ONE 2013, 8, e60809. [Google Scholar] [CrossRef] [PubMed]
- Cecchetti, V.; Brunetti, P.; Napoli, N.; Fattorini, L.; Altamura, M.M.; Costantino, P.; Cardarelli, M. ABCB1 and ABCB19 auxin transporters have synergistic effects on early and late Arabidopsis anther development. J. Integr. Plant Biol. 2015, 57, 1089–1098. [Google Scholar] [CrossRef] [PubMed]
- Chai, C.; Subudhi, P.K. Comprehensive analysis and expression profiling of the OsLAX and OsABCB auxin transporter gene families in Rice (Oryza sativa) under phytohormone stimuli and abiotic stresses. Front. Plant Sci. 2016, 3, 593. [Google Scholar] [CrossRef]
- Shen, C.J.; Bai, Y.H.; Wang, B.J.; Zhang, S.N.; Wu, Y.R.; Chen, M.; Jiang, D.A.; Qi, Y.H. Expression profile of PIN, AUX/LAX and PGP auxin transporter gene families in Sorghum bicolor under phytohormone and abiotic stress. FEBS J. 2010, 277, 2954–2969. [Google Scholar] [CrossRef]
- Kim, D.Y.; Bovet, L.; Kushnir, S.; Noh, E.W.; Martinoia, E.; Lee, Y. AtATM3 is involved in heavy metal resistance in Arabidopsis. Physiol. Plant. 2006, 140, 922–932. [Google Scholar] [CrossRef]
- Jayita, S.; Atreyee, S.; Kamala, G.; Bhaskar, G. Molecular phylogenetic study and expression analysis of ATP-binding cassette transporter gene family in Oryza sativa in response to salt stress. Comput. Biol. Chem. 2015, 54, 18–32. [Google Scholar]
- Huang, C.F.; Yamaji, N.; Mitani, N.; Yano, M.; Nagamura, Y.; Ma, J.F. A bacterial-type ABC transporter is involved in aluminum tolerance in rice. Plant Cell 2009, 21, 655–667. [Google Scholar] [CrossRef]
- Bird, D.A. The role of ABC transporters in cuticular lipid secretion. Plant Sci. 2008, 174, 563–569. [Google Scholar] [CrossRef]
- Pighin, J.A.; Zheng, H.; Balakshin, L.J.; Goodman, I.P.; Western, T.L.; Jetter, R.; Kunst, L.; Samuels, A.L. Plant cuticular lipid export requires an ABC transporter. Science 2004, 306, 702–704. [Google Scholar] [CrossRef]
- Von, R.L.E.; Degenkolb, T.; Zerjeski, M. Profiling of Arabidopsis secondary metabolites by capillary liquid chromatography coupled to electrospray ionization quadrupole time-of-flight mass spectrometry. Plant Physiol. 2004, 134, 548–559. [Google Scholar]
- Kim, D.Y.; Jin, J.Y.; Alejandro, S.; Martinoia, E.; Lee, Y. Overexpression of AtABCG36 improves drought and salt stress resistance in Arabidopsis. Physiol Plant. 2010, 139, 170–180. [Google Scholar] [CrossRef] [PubMed]
- Moons, A. Ospdr9, which encodes a PDR-type ABC transporter, is induced by heavy metals, hypoxic stress and redox perturbations in rice roots. FEBS Lett. 2003, 553, 370–376. [Google Scholar] [CrossRef]


| Gene Name | Primer | Forward Primer Sequence (5′-3′) | Reverse Primer Sequence (5′-3′) |
|---|---|---|---|
| HvABCG45 | ABC1 | GGCGGAACTGCTGTCATCT | AGTCGGCAACACCCTTTCT |
| HvABCG48 | ABC2 | CAGCCTGGGTTCGTTTGAG | TCGGAGTGATCGCCGTTGT |
| HvABCG38 | ABC3 | GGTTTGGATGCTCGTGCTG | TGATTTGACCGCCTCTTTT |
| HvABCA3 | ABC4 | TGGGCTCATTCCACCTACA | CCGTCAATGTTTCCCAGAG |
| HvABCF5 | ABC5 | TGGCTGGAAGAAACACTGAA | TCGGGTCTGCACATACTGGTC |
| HvABCF4 | ABC6 | CACATGCAGAACAAGACCCTC | GCTTCGCAGATCCATGACC |
| HvABCC16 | ABC7 | GCCATTCGGCGACCATACA | CACGAGCAAGCTGAACACG |
| HvABCC11 | ABC8 | GTCCTTGACGCTGATACTGG | TAGCACTGGTGCCTCCTCC |
| HvABCB24 | ABC9 | TGATACTGGGATTTGGTTAGG | CGAATGGCACTGAGAATGAG |
| HvABCB13 | ABC10 | CGTTCAACTCGGAGGACAAGA | CCATGCAGCGTACCACAGG |
| HvABCG27 | ABC11 | GAGGGAGGCAGCGTCAAGCA | GCAGGATGGCGAACTGGTTG |
| HvABCG29 | ABC12 | AGGGCTTCCCGTTGTAGGTG | TCGCATCCGTCATCACCATG |
| HvABCG25 | ABC13 | GTTCTGGATCGAGATGGGTGT | GAAGATGGTCGCCAGGATGA |
| HvABCG21 | ABC14 | ATACCGGCATACTGGCTGTGG | CCAGCACTCGCTCCTTCACC |
| HvABCG18 | ABC15 | TGCTCACCGCCAACTCATTC | TCCTTGCTCGCCACGAAGT |
| HvActin | Actin | TGGATCGGAGGGTCCATCCT | GCACTTCCTGTGGACGATCGCTG |
| Motif | Protein Sequence |
|---|---|
| Motif1 | GLSGGQKQRVAIARALLABPSILLLDEPTSGLDAESAAIVM |
| Motif2 | LLKGISLSFRPGELVALVGPSGSGKST |
| Motif3 | ERFYDPTAGEILJDGVDIKSJGLHWLRSKJGJVPQEPTLFMGSIRENIDY |
| Motif4 | VGEMGRNLSGGQRQRVALARAJLKPPKILLLDEATAALDSET |
| Motif5 | SGYVEQBDIHSPNLTVYESLLFSAWLRLPSDVSSAEKRMFV |
| Motif6 | HRLETLRLFDDILVLSDGKIVEQGPHEEL |
| Motif7 | VCGSIAYVSQTAWIQSGTIQDNILFGSPMDRERYEEVJEACSLVKDLEML |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Z.; Tong, T.; Fang, Y.; Zheng, J.; Zhang, X.; Niu, C.; Li, J.; Zhang, X.; Xue, D. Genome-Wide Identification of Barley ABC Genes and Their Expression in Response to Abiotic Stress Treatment. Plants 2020, 9, 1281. https://doi.org/10.3390/plants9101281
Zhang Z, Tong T, Fang Y, Zheng J, Zhang X, Niu C, Li J, Zhang X, Xue D. Genome-Wide Identification of Barley ABC Genes and Their Expression in Response to Abiotic Stress Treatment. Plants. 2020; 9(10):1281. https://doi.org/10.3390/plants9101281
Chicago/Turabian StyleZhang, Ziling, Tao Tong, Yunxia Fang, Junjun Zheng, Xian Zhang, Chunyu Niu, Jia Li, Xiaoqin Zhang, and Dawei Xue. 2020. "Genome-Wide Identification of Barley ABC Genes and Their Expression in Response to Abiotic Stress Treatment" Plants 9, no. 10: 1281. https://doi.org/10.3390/plants9101281
APA StyleZhang, Z., Tong, T., Fang, Y., Zheng, J., Zhang, X., Niu, C., Li, J., Zhang, X., & Xue, D. (2020). Genome-Wide Identification of Barley ABC Genes and Their Expression in Response to Abiotic Stress Treatment. Plants, 9(10), 1281. https://doi.org/10.3390/plants9101281






