Constitutive Expression of Arabidopsis Senescence Associated Gene 101 in Brachypodium distachyon Enhances Resistance to Puccinia brachypodii and Magnaporthe oryzae
Abstract
:1. Introduction
2. Results
2.1. Overexpression of AtSAG101 in B. distachyon
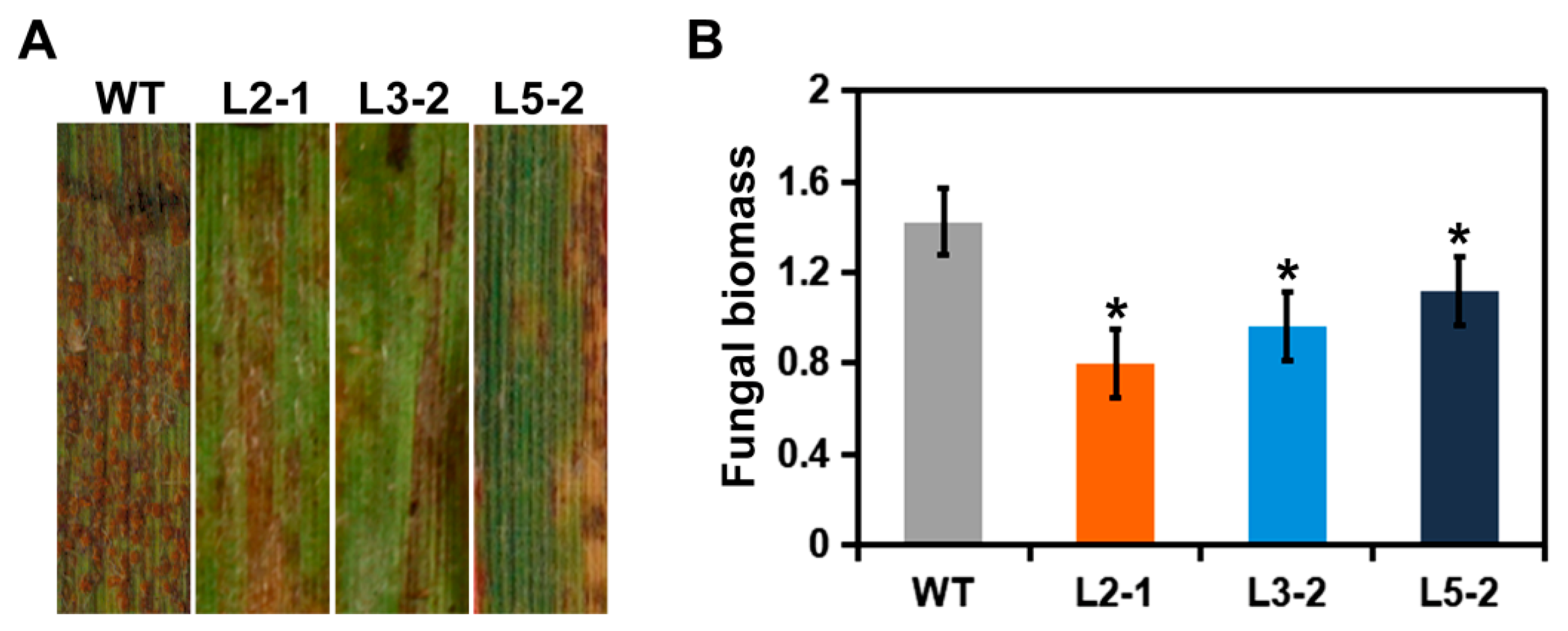
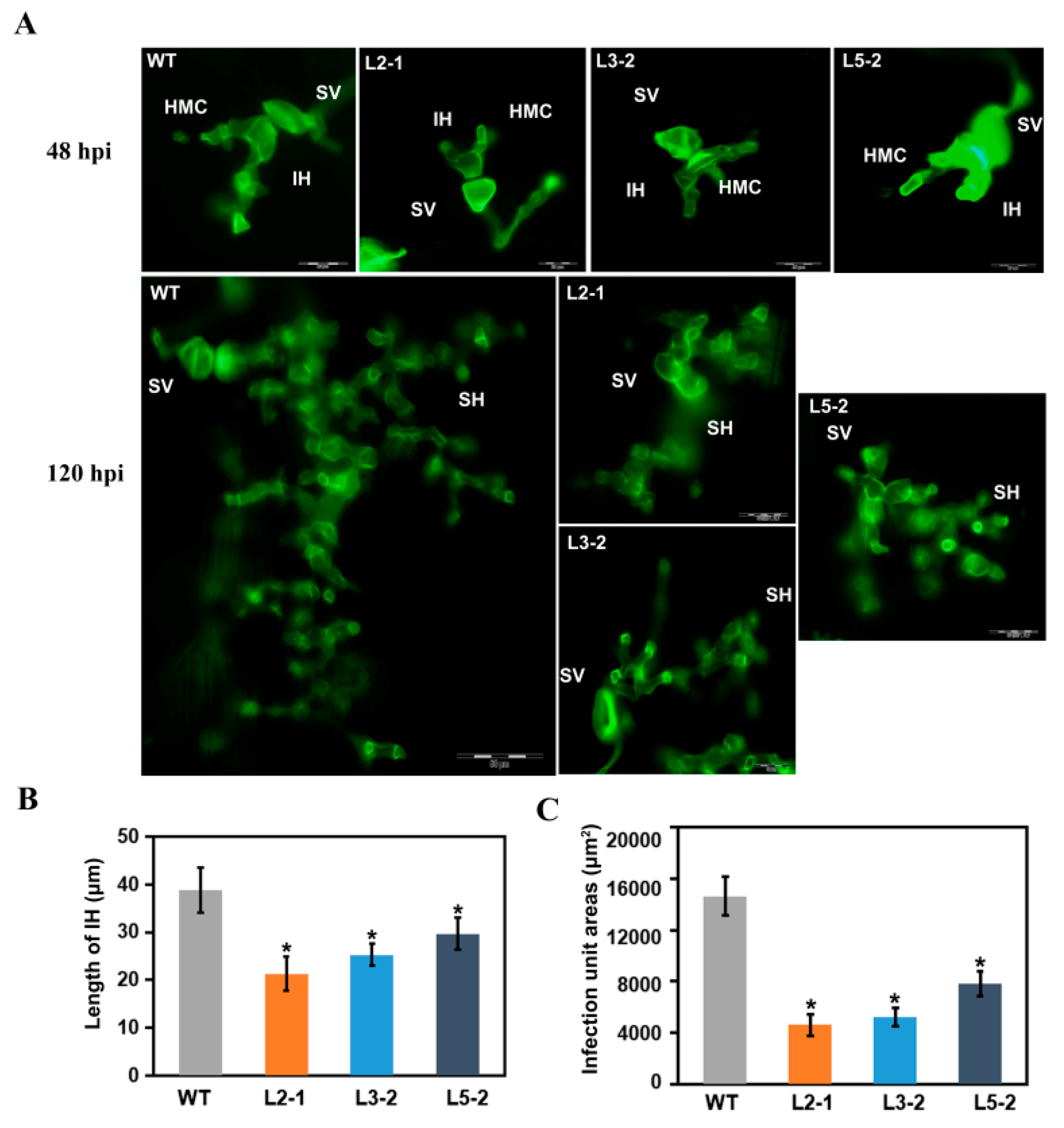
2.2. AtSAG101 Transgenic Plants Induce Resistance to P. brachypodii
2.3. SA Levels Are Increased in AtSAG101-Overexpressing B. distachyon Leaves
2.4. AtSAG101 Transgenic Plants Produce Resistance to Magnaporthe oryzae
3. Discussion
4. Materials and Methods
4.1. Plant and Fungal Materials
4.2. Gene Cloning and Agrobacterium Transformation
4.3. Histochemical Staining
4.4. Endogenous SA Level Analysis
4.5. Histological Observation of Fungal Growth
4.6. PCR Analysis
4.7. Biomass Analysis
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Wang, Y.; Tyler, B.M.; Wang, Y. Defense and counter defense during plant-pathogenic Oomycete infection. Annu. Rev. Microbiol. 2019, 73, 1–30. [Google Scholar] [CrossRef] [PubMed]
- Durner, J.; Shah, J.; Klessig, D.F. Salicylic acid and disease resistance in plants. Trends Plant Sci. 1997, 2, 266–274. [Google Scholar] [CrossRef]
- Vlot, A.C.; Dempsey, D.M.A.; Klessig, D.F. Salicylic acid, a multifaceted hormone to combat disease. Annu. Rev. Phytopathol. 2009, 47, 177–206. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Feys, B.J.; Moisan, L.J.; Newman, M.A.; Parker, J.E. Direct interaction between the Arabidopsis disease resistance signaling proteins, EDS1 and PAD4. EMBO J. 2001, 20, 5400–5411. [Google Scholar] [CrossRef]
- Feys, B.J.; Wiermer, M.; Bhat, R.A.; Moisan, L.J.; Medina-Escobar, N.; Neu, C.; Cabral, A.; Parker, J.E. Arabidopsis SENESCENCE-ASSOCIATED GENE101 stabilizes and signals within an ENHANCED DISEASE SUSCEPTIBILITY1 complex in plant innate immunity. Plant Cell 2005, 17, 2601–2613. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, S.; Jeong, R.D.; Venugopal, S.C.; Lapchyk, L.; Navarre, D.; Kachroo, A.; Kachroo, P. SAG101 forms a ternary complex with EDS1 and PAD4 and is required for resistance signaling against turnip crinkle virus. PLoS Pathog. 2011, 7, e1002318. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gantner, J.; Ordon, J.; Kretschmer, C.; Guerois, R.; Stuttmann, J. An EDS1-SAG101 Complex is Essential for TNL-mediated Immunity in Nicotiana benthamiana. Plant Cell 2019, 31, 2456–2474. [Google Scholar] [CrossRef] [Green Version]
- Yu, S.; Tian, L. Breeding Major Cereal Grains through the Lens of Nutrition Sensitivity. Mol. Plant 2018, 11, 23–30. [Google Scholar] [CrossRef] [Green Version]
- Garvin, D.F.; Gu, Y.Q.; Hasterok, R.; Hazen, S.P.; Jenkins, G.; Mockler, T.C.; Mur, L.A.; Vogel, J.P. Development of genetic and genomic research resources for Brachypodium distachyon, a new model system for grass crop research. Crop Sci. 2008, 48, S-69. [Google Scholar] [CrossRef]
- Draper, J.; Mur, L.A.; Jenkins, G.; Ghosh-Biswas, G.C.; Bablak, P.; Hasterok, R.; Routledge, A.P. Brachypodium distachyon. A new model system for functional genomics in grasses. Plant Physiol. 2001, 127, 1539–1555. [Google Scholar] [CrossRef]
- Dean, R.; Van Kan, J.A.; Pretorius, Z.A.; Hammond-Kosack, K.E.; Di Pietro, A.; Spanu, P.D.; Rudd, J.J.; Dickman, M.; Kahmann, R.; Ellis, J.; et al. The Top 10 fungal pathogens in molecular plant pathology. Mol. Plant Pathol. 2012, 13, 414–430. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fitzgerald, T.L.; Powell, J.J.; Schneebeli, K.; Hsia, M.M.; Gardiner, D.M.; Bragg, J.N.; McIntyre, C.L.; Manners, J.M.; Ayliffe, M.; Watt, M.; et al. Brachypodium as an emerging model for cereal–pathogen interactions. Ann. Bot. 2015, 115, 717–731. [Google Scholar] [CrossRef] [PubMed]
- Parker, D.; Beckmann, M.; Enot, D.P.; Overy, D.P.; Rios, Z.C.; Gilbert, M.; Talbot, N.; Draper, J. Rice blast infection of Brachypodium distachyon as a model system to study dynamic host/pathogen interactions. Nat. Protoc. 2008, 3, 435. [Google Scholar] [CrossRef]
- Zambino, P.J.; Szabo, L.J. Phylogenetic relationships of selected cereal and grass rusts based on rDNA sequence analysis. Mycologia 1993, 85, 401–414. [Google Scholar] [CrossRef]
- Joglekar, S.; Suliman, M.; Bartsch, M.; Halder, V.; Maintz, J.; Bautor, J.; Zeier, J.; Parker, J.E.; Kombrink, E. Chemical activation of EDS1/PAD4 signaling leading to pathogen resistance in Arabidopsis. Plant Cell Physiol. 2018, 59, 1592–1607. [Google Scholar] [CrossRef]
- Zhou, N.; Tootle, T.L.; Tsui, F.; Klessig, D.F.; Glazebrook, J. PAD4 functions upstream from salicylic acid to control defense responses in Arabidopsis. Plant Cell 1998, 10, 1021–1030. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Collier, S.M.; Hamel, L.P.; Moffett, P. Cell death mediated by the N-terminal domains of a unique and highly conserved class of NB-LRR protein. Mol. Plant-Microbe Interact. 2011, 24, 918–931. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cui, H.; Gobbato, E.; Kracher, B.; Qiu, J.; Bautor, J.; Parker, J.E. A core function of EDS1 with PAD4 is to protect the salicylic acid defense sector in Arabidopsis immunity. New Phytol. 2017, 213, 1802–1817. [Google Scholar] [CrossRef] [Green Version]
- Lapin, D.; Kovacova, V.; Sun, X.; Dongus, J.A.; Bhandari, D.; von Born, P.; Bautor, J.; Guarneri, N.; Rzemieniewski, J.; Stuttmann, J.; et al. A coevolved EDS1-SAG101-NRG1 module mediates cell death signaling by TIR-domain immune receptors. Plant Cell 2019, 31, 2430–2455. [Google Scholar] [CrossRef] [Green Version]
- Jirage, D.; Tootle, T.L.; Reuber, T.L.; Frost, L.N.; Feys, B.J.; Parker, J.E.; Ausubel, F.M.; Glazebrook, J. Arabidopsis thaliana PAD4 encodes a lipase-like gene that is important for salicylic acid signaling. Proc. Natl. Acad. Sci. USA 1999, 96, 13583–13588. [Google Scholar] [CrossRef] [Green Version]
- Lipka, V.; Dittgen, J.; Bednarek, P.; Bhat, R.; Wiermer, M.; Stein, M.; Landtag, J.; Brandt, W.; Rosahl, S.; Scheel, D.; et al. Pre-and postinvasion defenses both contribute to nonhost resistance in Arabidopsis. Science 2005, 310, 1180–1183. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, B.; Wang, N.; Song, N.; Wang, W.; Wang, J.; Wang, X.; Kang, Z. Overexpression of AtPAD4 in transgenic Brachypodium distachyon enhances resistance to Puccinia brachypodii. Plant Biol. 2017, 19, 868–874. [Google Scholar] [CrossRef] [PubMed]
- Vogel, J.P.; Garvin, D.F.; Leong, O.M.; Hayden, D.M. Agrobacterium-mediated transformation and inbred line development in the model grass Brachypodium distachyon. Plant Cell Tissue Organ Cult. 2006, 84, 199–211. [Google Scholar] [CrossRef]
- Sundaresan, V.; Springer, P.; Volpe, T.; Haward, S.; Jones, J.D.; Dean, C.; Ma, H.; Martienssen, R. Patterns of gene action in plant development revealed by enhancer trap and gene trap transposable elements. Genes Dev. 1995, 9, 1797–1810. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Segarra, G.; Jáuregui, O.; Casanova, E.; Trillas, I. Simultaneous quantitative LC–ESI-MS/MS analyses of salicylic acid and jasmonic acid in crude extracts of Cucumis sativus under biotic stress. Phytochemistry 2006, 67, 395–401. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]



© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, N.; Song, N.; Tang, Z.; Wang, X.; Kang, Z.; Dai, L.; Wang, B. Constitutive Expression of Arabidopsis Senescence Associated Gene 101 in Brachypodium distachyon Enhances Resistance to Puccinia brachypodii and Magnaporthe oryzae. Plants 2020, 9, 1316. https://doi.org/10.3390/plants9101316
Wang N, Song N, Tang Z, Wang X, Kang Z, Dai L, Wang B. Constitutive Expression of Arabidopsis Senescence Associated Gene 101 in Brachypodium distachyon Enhances Resistance to Puccinia brachypodii and Magnaporthe oryzae. Plants. 2020; 9(10):1316. https://doi.org/10.3390/plants9101316
Chicago/Turabian StyleWang, Ning, Na Song, Zejun Tang, Xiaojie Wang, Zhensheng Kang, Liangying Dai, and Bing Wang. 2020. "Constitutive Expression of Arabidopsis Senescence Associated Gene 101 in Brachypodium distachyon Enhances Resistance to Puccinia brachypodii and Magnaporthe oryzae" Plants 9, no. 10: 1316. https://doi.org/10.3390/plants9101316
APA StyleWang, N., Song, N., Tang, Z., Wang, X., Kang, Z., Dai, L., & Wang, B. (2020). Constitutive Expression of Arabidopsis Senescence Associated Gene 101 in Brachypodium distachyon Enhances Resistance to Puccinia brachypodii and Magnaporthe oryzae. Plants, 9(10), 1316. https://doi.org/10.3390/plants9101316




