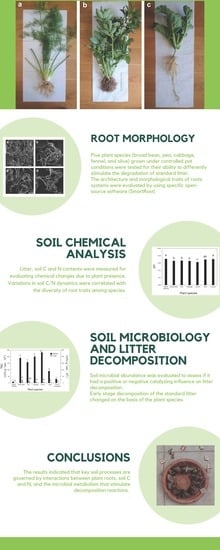
Structural and Functional Organization of the Root System: A Comparative Study on Five Plant Species
Abstract
1. Introduction
2. Results
2.1. Plant Traits and Root Image Processing
2.2. Standard Litter Decomposition
2.3. Soil Chemical Analysis
2.4. Microbial Counts
3. Discussion
4. Materials and Methods
4.1. Experimental Site, Orchard Management, and Soil Sampling
4.2. Plant Traits and Root Architecture
4.3. Standard Litter Decomposition
4.3.1. Tea Bags Installation and Final Sampling
4.3.2. Calculation of Decomposition Index
4.4. Litter and Soil Chemical Analysis
4.5. Total Bacterial and Fungal Enumeration
4.6. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Berendse, F. Litter Decomposability - A Neglected Component of Plant Fitness. J. Ecol. 1994, 82, 187. [Google Scholar] [CrossRef]
- Vivanco, L.; Austin, A.T. Intrinsic effects of species on leaf litter and root decomposition: A comparison of temperate grasses from North and South America. Oecologia 2006, 150, 97–107. [Google Scholar] [CrossRef] [PubMed]
- Bray, S.R.; Kitajima, K.; Mack, M.C. Temporal dynamics of microbial communities on decomposing leaf litter of 10 plant species in relation to decomposition rate. Soil Biol. Biochem. 2012, 49, 30–37. [Google Scholar] [CrossRef]
- Cornwell, W.K.; Cornelissen, J.H.C.; Amatangelo, K.; Dorrepaal, E.; Eviner, V.T.; Godoy, O.; Hobbie, S.E.; Hoorens, B.; Kurokawa, H.; Pérez-Harguindeguy, N.; et al. Plant species traits are the predominant control on litter decomposition rates within biomes worldwide. Ecol. Lett. 2008, 11, 1065–1071. [Google Scholar] [CrossRef]
- Hobbie, S.E. Plant species effects on nutrient cycling: Revisiting Litter Feedbacks. Trends Ecol. Evol. 2015, 30, 357–363. [Google Scholar] [CrossRef]
- Beatty, S.W.; Sholes, O.D.V. Leaf litter effect on plant species composition of deciduous forest treefall pits. Can. J. For. Res. 1988, 18, 553–559. [Google Scholar] [CrossRef]
- Hobbie, S.E. Temperature and plant species control over litter decomposition in Alaskan tundra. Ecol. Monogr. 1996, 66, 503–522. [Google Scholar] [CrossRef]
- Jensen, K.; Gutekunst, K. Effects of litter on establishment of grassland plant species: The role of seed size and successional status. Basic Appl. Ecol. 2003, 4, 579–587. [Google Scholar] [CrossRef]
- Ayres, E.; Dromph, K.M.; Bardgett, R.D. Do plant species encourage soil biota that specialise in the rapid decomposition of their litter? Soil Biol. Biochem. 2006, 38, 183–186. [Google Scholar] [CrossRef]
- Wardle, D.A.; Yeates, G.W.; Barker, G.M.; Bonner, K.I. The influence of plant litter diversity on decomposer abundance and diversity. Soil Biol. Biochem. 2006, 38, 1052–1062. [Google Scholar] [CrossRef]
- Ayres, E.; Steltzer, H.; Berg, S.; Wall, D.H. Soil biota accelerate decomposition in high-elevation forests by specializing in the breakdown of litter produced by the plant species above them. J. Ecol. 2009, 97, 901–912. [Google Scholar] [CrossRef]
- Meisner, A.; de Boer, W.; Cornelissen, J.H.C.; van der Putten, W.H. Reciprocal effects of litter from exotic and congeneric native plant species via soil nutrients. PLoS ONE 2012, 7. [Google Scholar] [CrossRef]
- Wedin, D.A.; Tilman, D. Species effects on nitrogen cycling: A test with perennial grasses. Oecologia 1990, 84, 433–441. [Google Scholar] [CrossRef] [PubMed]
- Murphy, K.L.; Klopatek, J.M.; Klopatek, C.C. The effects of litter quality and climate on decomposition along an elevational gradient. Ecol. Appl. 1998, 8, 1061–1071. [Google Scholar] [CrossRef]
- Osanai, Y.; Flittner, A.; Janes, J.K.; Theobald, P.; Pendall, E.; Newton, P.C.D.; Hovenden, M.J. Decomposition and nitrogen transformation rates in a temperate grassland vary among co-occurring plant species. Plant Soil 2012, 350, 365–378. [Google Scholar] [CrossRef]
- Meier, C.L.; Bowman, W.D. Links between plant litter chemistry, species diversity, and below-ground ecosystem function. Proc. Natl. Acad. Sci. USA 2008, 105, 19780–19785. [Google Scholar] [CrossRef]
- Horodecki, P.; Jagodziński, A.M. Tree species effects on litter decomposition in pure stands on afforested post-mining sites. For. Ecol. Manage. 2017, 406, 1–11. [Google Scholar] [CrossRef]
- Cornelissen, J.H.C. An Experimental Comparison of Leaf Decomposition Rates in a Wide Range of Temperate Plant Species and Types. J. Ecol. 1996, 84, 573. [Google Scholar] [CrossRef]
- Osono, T.; Azuma, J.; Hirose, D. Plant species effect on the decomposition and chemical changes of leaf litter in grassland and pine and oak forest soils. Plant Soil 2014, 376, 411–421. [Google Scholar] [CrossRef]
- Keuskamp, J.A.; Dingemans, B.J.J.; Lehtinen, T.; Sarneel, J.M.; Hefting, M.M. Tea Bag Index: A novel approach to collect uniform decomposition data across ecosystems. Methods Ecol. Evol. 2013, 4, 1070–1075. [Google Scholar] [CrossRef]
- Didion, M.; Repo, A.; Liski, J.; Forsius, M.; Bierbaumer, M.; Djukic, I. Towards harmonizing leaf litter decomposition studies using standard tea bags-a field study and model application. Forests 2016, 7, 1–12. [Google Scholar] [CrossRef]
- Djukic, I.; Kepfer-Rojas, S.; Schmidt, I.K.; Larsen, K.S.; Beier, C.; Berg, B.; Verheyen, K.; Caliman, A.; Paquette, A.; Gutiérrez-Girón, A.; et al. Early stage litter decomposition across biomes. Sci. Total Environ. 2018. [Google Scholar] [CrossRef] [PubMed]
- Sofo, A.; Nicoletta Mininni, A.; Ricciuti, P. Comparing the effects of soil fauna on litter decomposition and organic matter turnover in sustainably and conventionally managed olive orchards. Geoderma 2020, 372. [Google Scholar] [CrossRef]
- Sofo, A.; Manfreda, S.; Fiorentino, M.; Dichio, B.; Xiloyannis, C. The olive tree: A paradigm for drought tolerance in Mediterranean climates. Hydrol. Earth Syst. Sci. 2007, 12, 2811–2835. [Google Scholar] [CrossRef]
- Sofo, A.; Vitti, A.; Nuzzaci, M.; Tataranni, G.; Scopa, A.; Vangronsveld, J.; Remans, T.; Falasca, G.; Altamura, M.M.; Degola, F.; et al. Correlation between hormonal homeostasis and morphogenic responses in Arabidopsis thaliana seedlings growing in a Cd/Cu/Zn multi-pollution context. Physiol. Plant. 2013. [Google Scholar] [CrossRef]
- Sarneel, J.M.J.; Veen, G.F.C. Legacy effects of altered flooding regimes on decomposition in a boreal floodplain. Plant Soil 2017, 421, 57–66. [Google Scholar] [CrossRef]
- Tarshis, L.G.; Tarshis, G.I. Higher plants: Structural diversity of roots. In Measuring Roots: An Updated Approach; Springer: Berlin/Heidelberg, Germany, 2012; ISBN 9783642220678. [Google Scholar]
- White, P.J. Root traits benefitting crop production in environments with limited water and nutrient availability. Ann. Bot. 2019, 124, 883–890. [Google Scholar] [CrossRef]
- Gregory, P.J. Plant Roots: Growth, Activity and Interaction with Soils; Wiley: Hoboken, NJ, USA, 2007; ISBN 9781405119061. [Google Scholar]
- Ogden, L.E. Brewing Big Data: The Tea-Bag Index. Bioscience 2017, 67, 680. [Google Scholar] [CrossRef][Green Version]
- Sofo, A.; Ricciuti, P.; Fausto, C.; Mininni, A.N.; Crecchio, C.; Scagliola, M.; Malerba, A.D.; Xiloyannis, C.; Dichio, B. The metabolic and genetic diversity of soil bacterial communities depends on the soil management system and C/N dynamics: The case of sustainable and conventional olive groves. Appl. Soil Ecol. 2019, 137, 21–28. [Google Scholar] [CrossRef]
- Lobet, G.; Pagès, L.; Draye, X. A novel image-analysis toolbox enabling quantitative analysis of root system architecture. Plant Physiol. 2011, 157, 29–39. [Google Scholar] [CrossRef]
- Pansu, M.; Gautheyrou, J. Handbook of Soil Analysis: Mineralogical, Organic and Inorganic Methods; Springer: Berlin/Heidelberg, Germany, 2006; ISBN 3540312102. [Google Scholar]
- Weber, K.P.; Legge, R.L. One-dimensional metric for tracking bacterial community divergence using sole carbon source utilization patterns. J. Microbiol. Methods 2009, 79, 55–61. [Google Scholar] [CrossRef] [PubMed]
- Atlas, R.M. Handbook of Microbiological Media; CRC Press: Boca Raton, FL, USA, 2004. [Google Scholar]
- ISO-ISO 2293:1988-Meat and Meat Products—Enumeration of Micro-Organisms—Colony Count Technique at 30 Degrees C (Reference Method). Available online: https://www.iso.org/standard/7119.html (accessed on 7 July 2020).
- Cesana, B.M. What p-value must be used as the Statistical Significance Threshold? P<0.005, P<0.01, P<0.05 or no value at all? Biomed. J. Sci. Tech. Res. 2018, 6, 001–009. [Google Scholar] [CrossRef]

| Plant Species | Total Root Length (m) | Main Root Length (m) | Total Length of Lateral Roots (m) | Specific Root Length (m · g−1 FW) | Root Tips (Number) | Average Root Diameter (mm) | Total Root Surface Area (m2) |
|---|---|---|---|---|---|---|---|
| Broad bean | 24.59 ± 3.09 b | 0.17 ± 0.09 b | 24.41 ± 3.21 b | 0.45 ± 0.10 ab | 1151 ± 88 b | 3.24 ± 0.32 a | 0.25 ± 0.02 b |
| Pea | 15.24 ± 1.53 c | 0.20 ± 0.04 b | 15.04 ± 3.85 bc | 0.45 ± 0.03 b | 713 ± 34 c | 2.53 ± 0.15 b | 0.12 ± 0.03 c |
| Cabbage | 11.31 ± 3.00 cd | 0.55 ± 0.13 a | 10.76 ± 2.09 c | 0.58 ± 0.07 a | 529 ± 39 d | 2.07 ± 0.17 c | 0.07 ± 0.02 d |
| Fennel | 5.34 ± 0.79 d | 0.15 ± 0.05 b | 5.19 ± 1.77 d | 0.33 ± 0.04 c | 403 ± 12 e | 2.48 ± 0.20 b | 0.04 ± 0.01 e |
| Olive | 127.83 ± 20.11 a | 0.67 ± 0.16 a | 127.16 ± 26.79 a | 0.28 ± 0.08 c | 11965 ± 937 a | 5.05 ± 0.17 a | 6.78 ± 0.34 a |
| Plant Species | Green Tea Mass Loss (g) | Fraction of Remaining Green Tea (Xg) | Red Tea Mass Loss (g) | Fraction of Remaining Red Tea (Xr) | Stabilization Factor (S) | Decomposition Rate Constant (k) |
|---|---|---|---|---|---|---|
| Broad bean | 0.521 ± 0.035 b | 0.629 ± 0.025 b | 0.257 ± 0.062 b | 0.851 ± 0.039 b | 0.560 ± 0.029 b | 0.013 ± 0.007 b |
| Pea | 0.562 ± 0.087 b | 0.616 ± 0.040 b | 0.290 ± 0.035 ab | 0.824 ± 0.061 bc | 0.544 ± 0.048 b | 0.015 ± 0.007 b |
| Cabbage | 0.649 ± 0.102 a | 0.538 ± 0.040 c | 0.328 ± 0.029 a | 0.803 ± 0.020 c | 0.452 ± 0.066 c | 0.014 ± 0.005 b |
| Fennel | 0.542 ± 0.038 b | 0.621 ± 0.031 b | 0.326 ± 0.065 a | 0.806 ± 0.037 c | 0.549 ± 0.036 b | 0.029 ± 0.026 a |
| Olive | 0.395 ± 0,066 c | 0.719 ± 0.016 a | 0.172 ± 0.026 bc | 0.898 ± 0.019 b | 0.666 ± 0.019 a | 0.010 ± 0.003 bc |
| Control (no plants) | 0.381 ± 0.040 c | 0.726 ± 0.015 a | 0.102 ± 0.016 d | 0.938 ± 0.019 a | 0.675 ± 0.018 a | 0.005 ± 0.002 c |
| Green Tea | Red Tea | Soil | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Plant Species | LOC (g · kg−1) | LTN (g · kg−1) | LOC/LTN | LOC (g · kg−1) | LTN (g · kg−1) | LOC/LTN | SOC (g · kg−1) | STN (g · kg−1) | SOC/STN | pH |
| Before incubation/planting | 44.64 ± 2.03 a | 2.65 ± 0.43 b | 16.85 ± 1.35 a | 43.45 ± 3.54 a | 3.23 ± 0.20 a | 13.45 ± 1.92 bc | 38.54 ± 0.98 a | 3.20 ± 0.20 ab | 12.04 ± 0.87 a | 7.15 ± 0.11 a |
| Broad bean | 28.09 ± 4.21 b | 3.05 ± 0.04 a | 9.21 ± 1.90 c | 36.97 ± 1.21 b | 3.56 ± 0.43 a | 10.38 ± 0.23 c | 31.96 ± 3.42 c | 3.55 ± 0.20 a | 9.00 ± 1.12 d | 6.62 ± 0.09 b |
| Pea | 27.49 ± 6.78 b | 2.88 ± 0.14 a | 9.54 ± 0.43 c | 35.81 ± 0.73 b | 3.25 ± 0.02 a | 11.02 ± 2.34 c | 31.44 ± 4.53 c | 3.62 ± 0.53 a | 8.69 ± 0.86 d | 6.68 ± 0.09 b |
| Cabbage | 24.03 ± 2.61 c | 1.71 ± 0.04 c | 14.05 ± 2.11 ab | 34.90 ± 0244 b | 2.04 ± 0.04 c | 17.11 ± 2.86 a | 28.45 ± 2.90 d | 2.83 ± 0.38 b | 10.05 ± 0.05 c | 6.59 ± 0.12 b |
| Fennel | 27.70 ± 3.00 b | 1.80 ± 0.25 c | 15.39 ± 0.83 a | 35.01 ± 3.54 b | 2.21 ± 0.09 c | 15.84 ± 2.38 ab | 31.62 ± 0.77 c | 2.91 ± 0.19 c | 10.87 ± 2.45 c | 6.28 ± 0.04 c |
| Olive | 32.09 ± 2.39 b | 2.12 ± 0.28 b | 15.14 ± 0.54 a | 39.01 ± 3.63 b | 2.67 ± 0.29 b | 14.61 ± 1.55 b | 35.41 ± 1.41 b | 2.67 ± 0.36 c | 13.26 ± 2.32 a | 6.90 ± 0.14 ab |
| Control (no plants) | 32.41 ± 2.09 b | 2.50 ± 0.33 b | 12.96 ± 1.03 b | 40.76 ± 4.32 ab | 2.65 ± 0.17 b | 15.38 ± 2.45 ab | 35.69 ± 1.51 b | 3.09 ± 0.27 ab | 11.55 ±1.64 bc | 7.10 ± 0.05 a |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sofo, A.; Elshafie, H.S.; Camele, I. Structural and Functional Organization of the Root System: A Comparative Study on Five Plant Species. Plants 2020, 9, 1338. https://doi.org/10.3390/plants9101338
Sofo A, Elshafie HS, Camele I. Structural and Functional Organization of the Root System: A Comparative Study on Five Plant Species. Plants. 2020; 9(10):1338. https://doi.org/10.3390/plants9101338
Chicago/Turabian StyleSofo, Adriano, Hazem S. Elshafie, and Ippolito Camele. 2020. "Structural and Functional Organization of the Root System: A Comparative Study on Five Plant Species" Plants 9, no. 10: 1338. https://doi.org/10.3390/plants9101338
APA StyleSofo, A., Elshafie, H. S., & Camele, I. (2020). Structural and Functional Organization of the Root System: A Comparative Study on Five Plant Species. Plants, 9(10), 1338. https://doi.org/10.3390/plants9101338