Identification, Characterization, and Mutational Analysis of a Probable KEAP1 Ortholog in Rice (Oryza sativa L.)
Abstract
:1. Introduction
2. Results
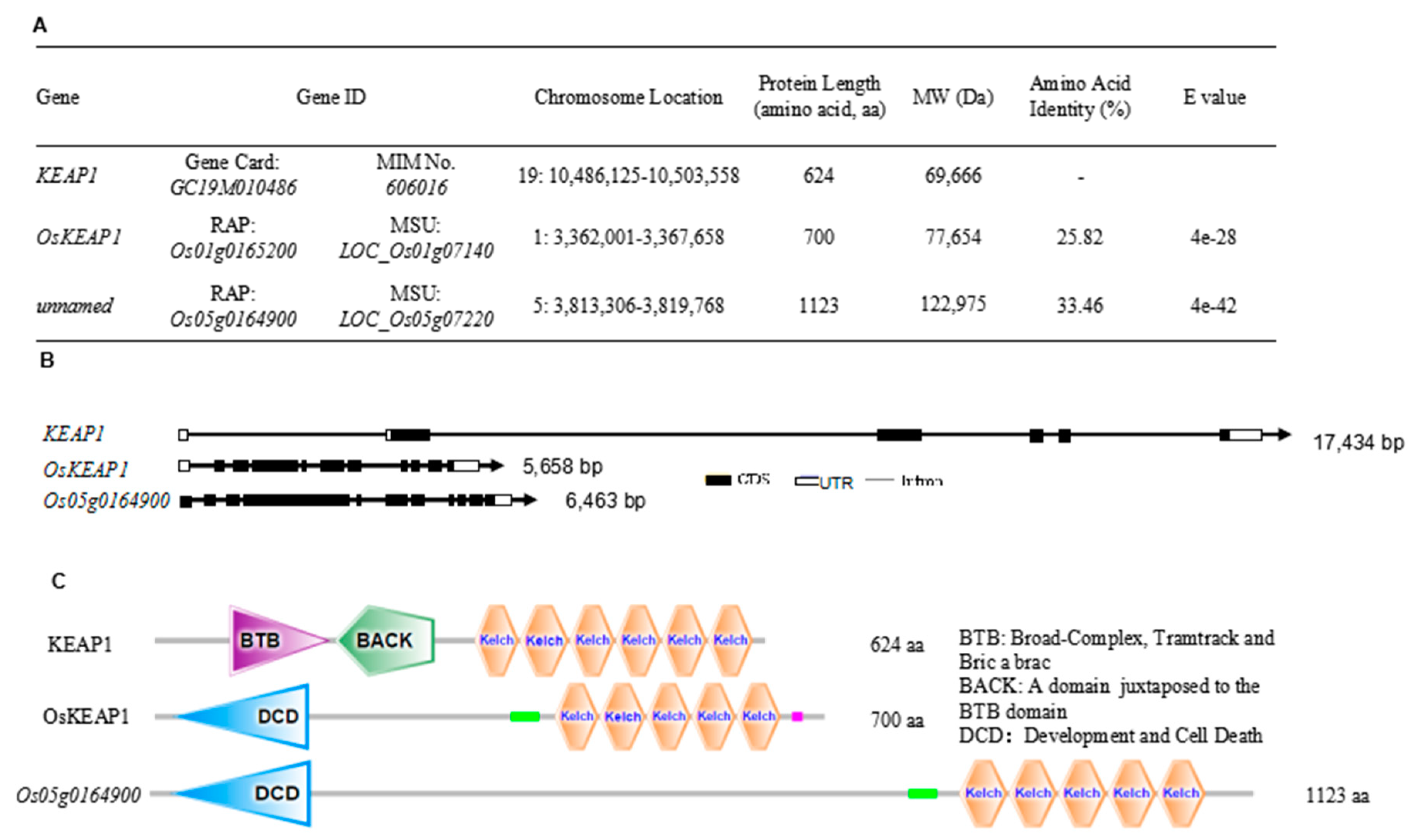
2.1. Identification and Characterization of the KEAP1 Ortholog in Rice
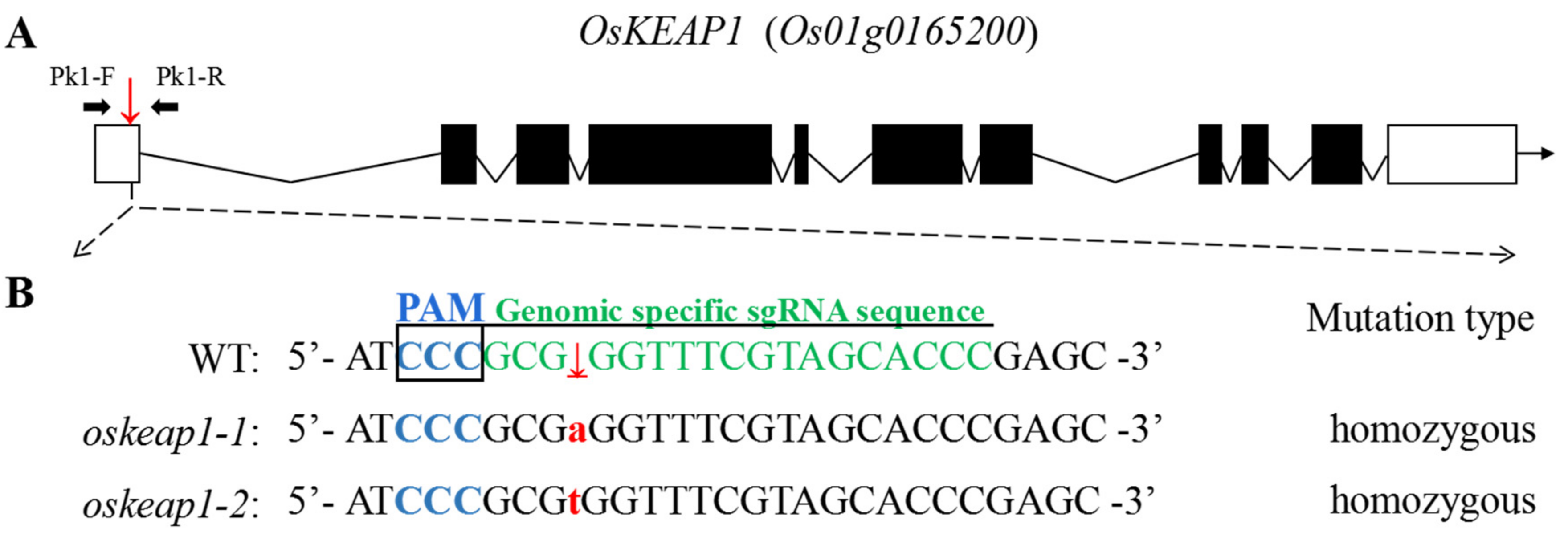
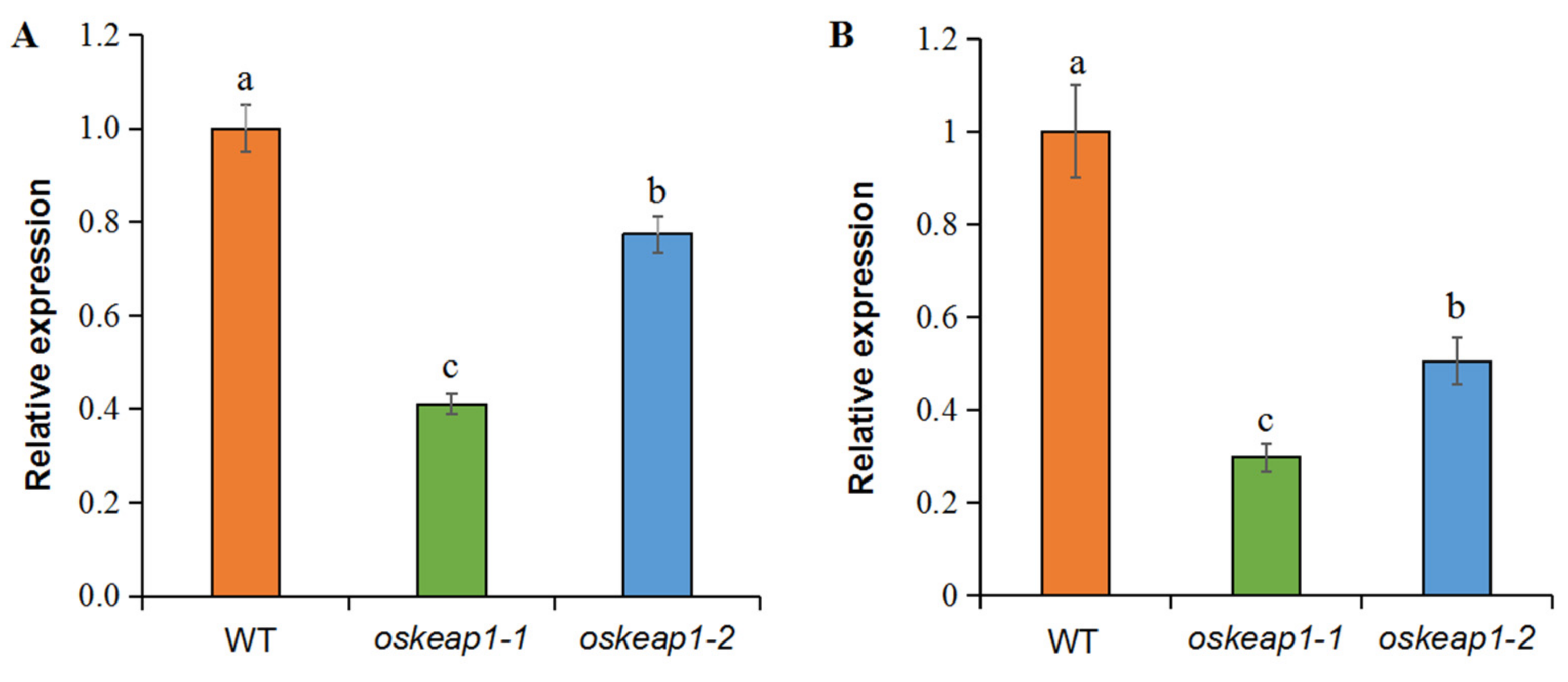
2.2. Targeted Mutagenesis of 5′ UTR Downregulated OsKEAP1 Expression
2.3. Impact of OsKEAP1 Down-Regulation on Plant Growth and Development
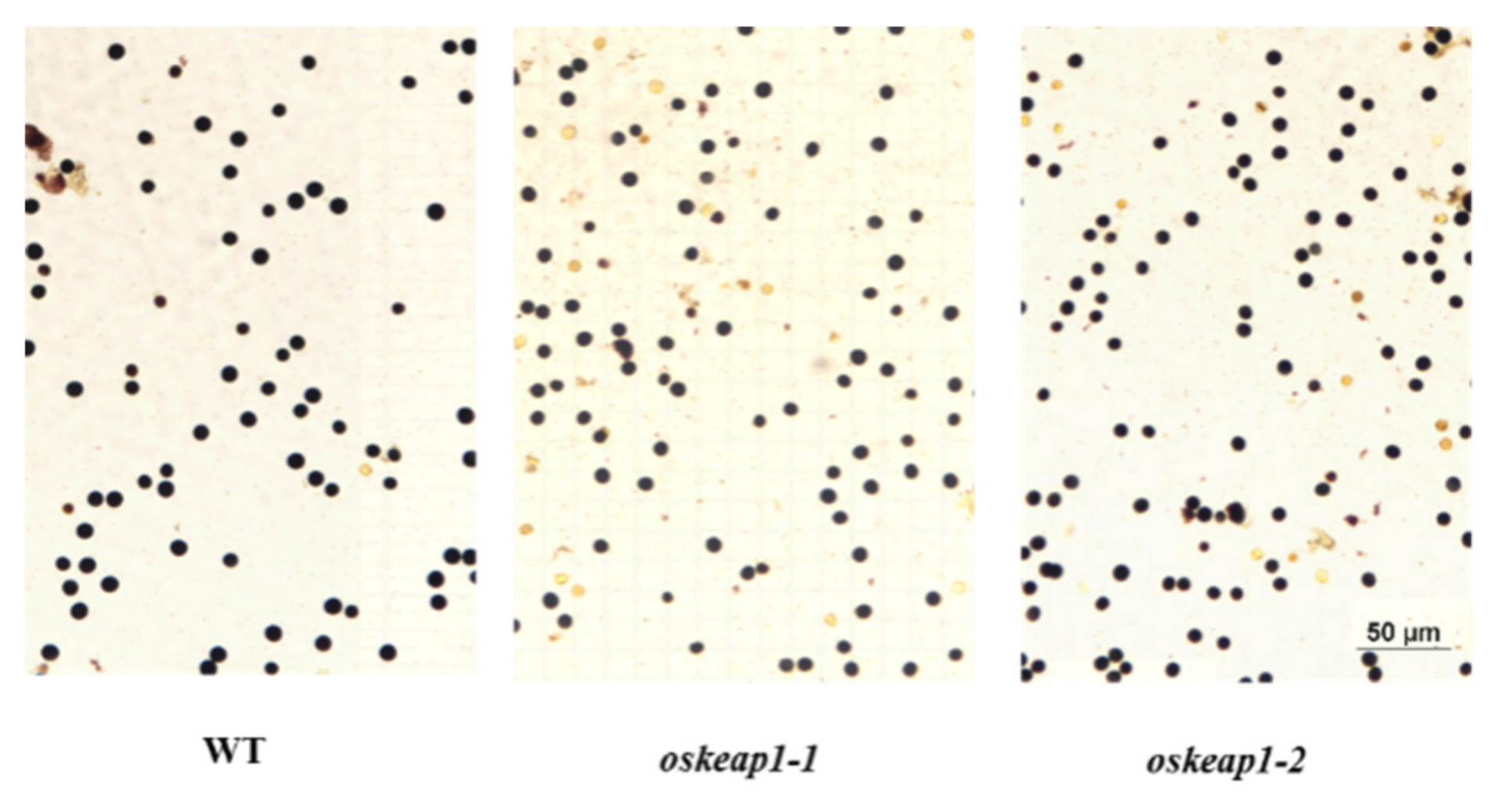
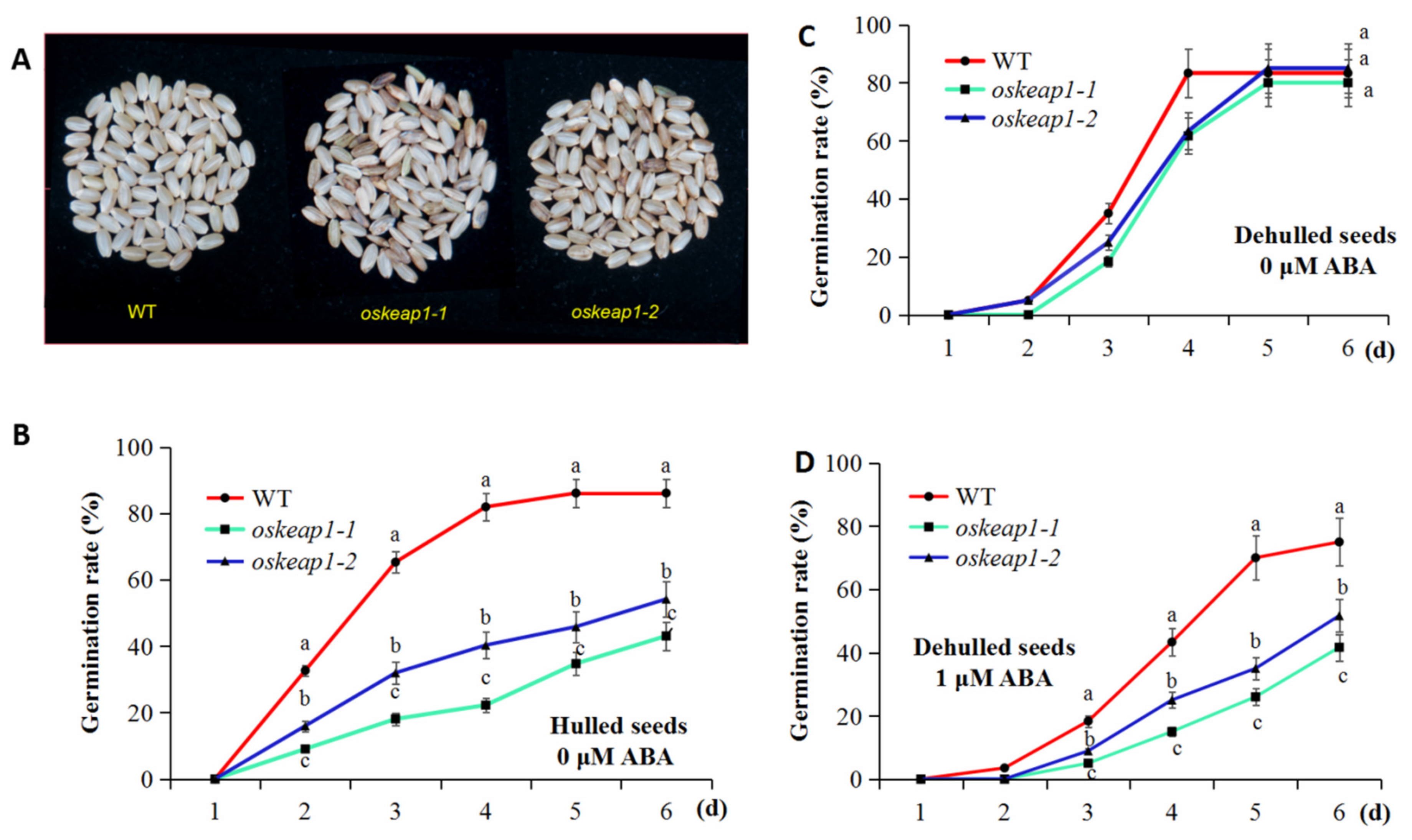
2.4. Impact of OsKEAP1 Downregulation on Seed Germination and Sensitivity to Abscisic Acid (ABA)
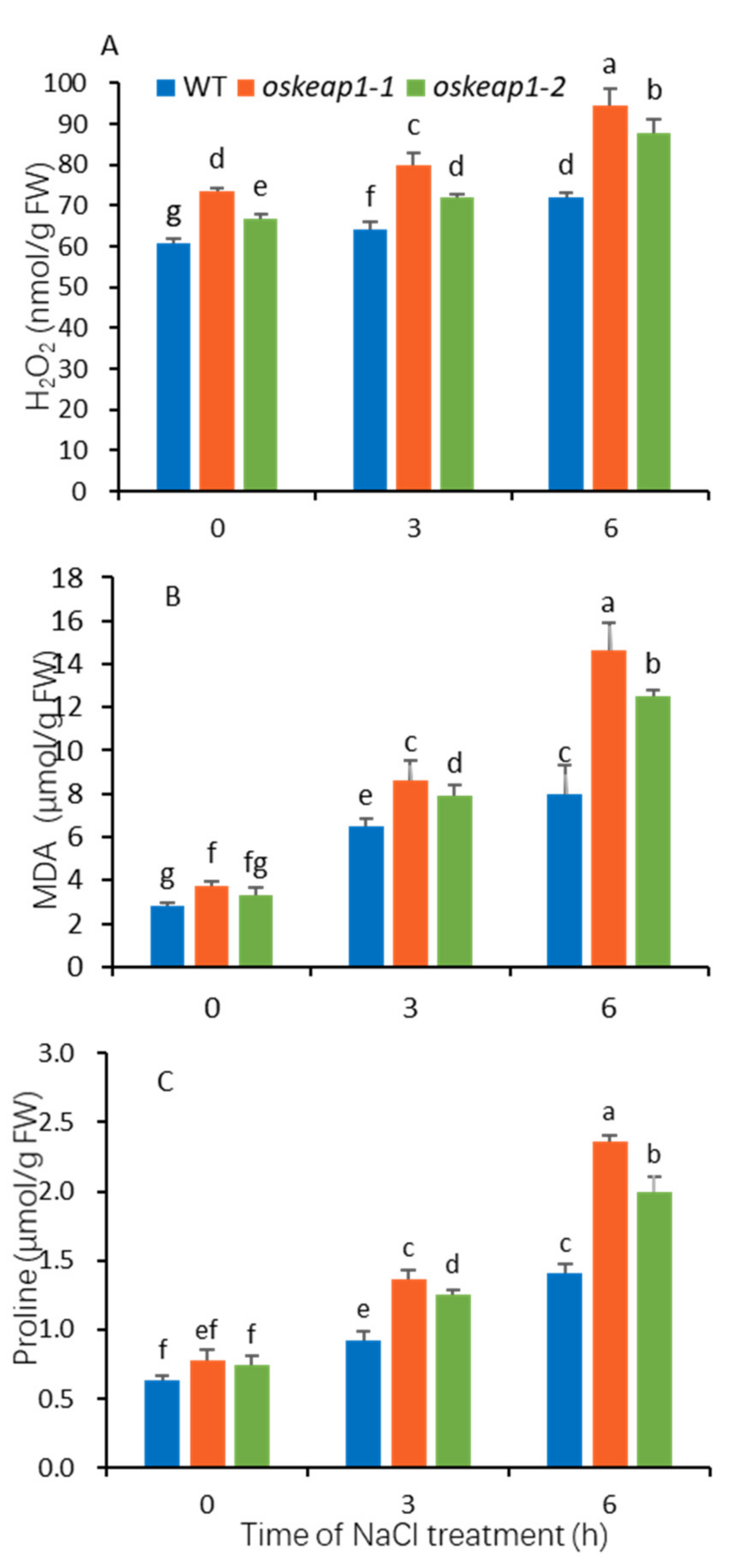
2.5. Impact of OsKEAP1 Downregulation on Redox Regulation
3. Discussion
4. Materials and Methods
4.1. In Silico Identification and Characterization of KEAP1 Orthologs in Plants
4.2. Construction of CRISPR/Cas9 Vector
4.3. Identification and Development of OsKEAP1 Mutants
4.4. Real-Time Quantitative-PCR and Subcellular Localization
4.5. Agronomic and Seed Germination Tests of OsKEAP1 Mutants
4.6. Seedling Growth
4.7. Measurement of H2O2, MDA and Proline Content
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Jain, A.; Lamark, T.; Sjøttem, E.; Larsen, K.B.; Awuh, J.A.; Øvervatn, A.; McMahon, M.; Hayes, J.D.; Johansen, T. p62/SQSTM1 is a target gene for transcription factor NRF2 and creates a positive feedback loop by inducing antioxidant response element-driven gene transcription. J. Biol. Chem. 2010, 285, 22576–22591. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fan, W.; Tang, Z.; Chen, D.; Moughon, D.; Ding, X.; Chen, S.; Zhu, M.; Zhong, Q. Keap1 facilitates p62-mediated ubiquitin aggregate clearance via autophagy. Autophagy. 2010, 6, 614–621. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lau, A.; Wang, X.-J.; Zhao, F.; Villeneuve, N.F.; Wu, T.; Jiang, T.; Sun, Z.; White, E.; Zhang, D.D. A Noncanonical mechanism of Nrf2 activation by autophagy deficiency: Direct interaction between Keap1 and p62. Mol. Cell. Biol. 2010, 30, 3275–3285. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Komatsu, M.; Kurokawa, H.; Waguri, S.; Taguchi, K.; Kobayashi, A.; Ichimura, Y.; Sou, Y.S.; Ueno, I.; Sakamoto, A.; Tong, K.I.; et al. The selective autophagy substrate p62 activates the stress responsive transcription factor Nrf2 through inactivation of Keap1. Nat. Cell Biol. 2010, 12, 213–223. [Google Scholar] [CrossRef] [PubMed]
- Leiser, S.F.; Miller, R.A. Nrf2 signaling, a mechanism for cellular stress resistance in long-lived mice. Mol. Cell. Biol. 2010, 30, 871–884. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Uruno, A.; Motohashi, H. The Keap1-Nrf2 system as an in vivo sensor for electrophiles. Nitric Oxide Biol. Chem. 2011, 25, 153–160. [Google Scholar] [CrossRef]
- Singh, A.; Misra, V.; Thimmulappa, R.; Lee, H.; Ames, S.; Hoque, M.; Herman, J.; Baylin, S.; Sidransky, D.; Gabrielson, E.; et al. Dysfunctional KEAP1–NRF2 interaction in non-small-cell lung cancer. PLoS Med. 2006, 3, e420. [Google Scholar] [CrossRef] [Green Version]
- Meng, H.; Guo, J.; Wang, H.; Yan, P.; Niu, X.; Zhang, J. Erythropoietin activates Keap1-Nrf2/ARE pathway in rat brain after ischemia. Int. J. Neurosci. 2014, 124, 362–368. [Google Scholar] [CrossRef]
- Cai, M.C.; Chen, M.; Ma, P.; Wu, J.; Lu, H.; Zhang, S.; Liu, J.; Zhao, X.; Zhuang, G.; Yu, Z.; et al. Clinicopathological, microenvironmental and genetic determinants of molecular subtypes in KEAP1/NRF2-mutant lung cancer. Int. J. Cancer 2019, 144, 788–801. [Google Scholar] [CrossRef] [Green Version]
- Gacesa, R.; Dunlap, W.C.; Barlow, D.J.; Laskowski, R.A.; Long, P.F. Rising levels of atmospheric oxygen and evolution of Nrf2. Sci. Rep. 2016, 6, 27740. [Google Scholar] [CrossRef] [Green Version]
- Gacesa, R.; Dunlap, W.C.; Long, P.F. Bioinformatics analyses provide insight into distant homology of the Keap1–Nrf2 pathway. Free Radic. Biol. Med. 2015, 88, 373–380. [Google Scholar] [CrossRef] [PubMed]
- Mhamdi, A.; Van Breusegem, F. Reactive oxygen species in plant development. Development 2018, 145, dev164376. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Noctor, G.; Reichheld, J.-P.; Foyer, C.H. ROS-related redox regulation and signaling in plants. Semin. Cell Dev. Biol. 2018, 80, 3–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, Y.; Ye, X.; Guo, R.; Huang, J.; Wang, W.; Tang, J.; Tan, L.; Zhu, J.K.; Chu, C.; Qian, Y. Genome-wide targeted mutagenesis in rice using the CRISPR/Cas9 system. Mol. Plant 2017, 10, 1242–1245. [Google Scholar] [CrossRef] [Green Version]
- Murashige, T.; Skoog, F. A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol. Plant. 1962, 15, 473–497. [Google Scholar] [CrossRef]
- Leppek, K.; Das, R.; Barna, M. Functional 5′ UTR mRNA structures in eukaryotic translation regulation and how to find them. Nat. Rev. Mol. Cell Biol. 2018, 19, 158–174. [Google Scholar] [CrossRef]
- Guo, Z.; Mo, Z. Keap1-Nrf2 signaling pathway in angiogenesis and vascular diseases. J. Tissue Eng. Regen. Med. 2020, 14, 869–883. [Google Scholar] [CrossRef]
- Watai, Y.; Kobayashi, A.; Nagase, H.; Mizukami, M.; McEvoy, J.; Singer, J.D.; Itoh, K.; Yamamoto, M. Subcellular localization and cytoplasmic complex status of endogenous Keap1. Genes Cells. 2007, 12, 1163–1178. [Google Scholar] [CrossRef]
- Tenhaken, R.; Doerks, T.; Bork, P. DCD—A novel plant specific domain in proteins involved in development and programmed cell death. BMC Bioinf. 2005, 6, 1–6. [Google Scholar] [CrossRef] [Green Version]
- Zou, M.; Guan, Y.; Ren, H.; Zhang, F.; Chen, F. A bZIP transcription factor, OsABI5, is involved in rice fertility and stress tolerance. Plant Mol. Biol. 2008, 66, 675–683. [Google Scholar] [CrossRef]
- Nakamura, S.; Lynch, T.J.; Finkelstein, R.R. Physical interactions between ABA response loci of Arabidopsis. Plant J. 2001, 26, 627–635. [Google Scholar] [CrossRef]
- NCBI. Available online: https://www.ncbi.nlm.nih.gov/ (accessed on 30 July 2016).
- Gramene. Available online: http://www.gramene.org/ (accessed on 23 August 2016).
- RAPDB. Available online: https://rapdb.dna.affrc.go.jp/index.html (accessed on 24 August 2016).
- OrthoDB. Available online: https://www.orthodb.org/ (accessed on 14 August 2016).
- UNAFold. Available online: http://unafold.rna.albany.edu/ (accessed on 14 August 2016).
- RiceXPro. Available online: http://ricexpro.dna.affrc.go.jp/GGEP/index.php/ (accessed on 14 August 2016).
- Skubacz, A.; Daszkowska-Golec, A.; Szarejko, I. The role and regulation of ABI5 (ABA-Insensitive 5) in plant development, abiotic stress responses and phytohormone crosstalk. Front. Plant Sci. 2016, 7, 1884. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, S.; Jiang, J.; Liu, Y.; Meng, J.; Xu, S.; Tan, Y.; Li, Y.; Shu, Q.; Huang, J. Characterization and evaluation of OsLCT1 and OsNramp5 mutants generated through CRISPR/Cas9-mediated mutagenesis for breeding low Cd rice. Rice Sci. 2019, 26, 88–97. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.L.; Huang, J.Z.; Chen, X.Y.; Tan, Y.Y.; Shu, Q.Y. Competitive amplification of differentially melting amplicons facilitates efficient genotyping of photoperiod- and temperature-sensitive genic male sterility in rice. Mol. Breed. 2014, 34, 1765–1776. [Google Scholar] [CrossRef]
- Li, S.; Liu, S.; Liu, Y.; Lu, H.; Tan, Y.; Huang, J.; Wei, P.; Shu, Q.Y. HRM-facilitated rapid identification and genotyping of mutations induced by CRISPR/ Cas9 mutagenesis in rice. Crop Breed. Appl. Biotechnol. 2018, 18, 184–191. [Google Scholar] [CrossRef] [Green Version]
- Basnet, R.; Zhang, J.; Hussain, N.; Shu, Q. Characterization and mutational analysis of a monogalactosyldiacylglycerol synthase gene OsMGD2 in rice. Front. Plant Sci. 2019, 10, 1–12. [Google Scholar] [CrossRef] [Green Version]
- He, F.; Chen, S.; Ning, Y.; Wang, G.-L. Rice (Oryza sativa) protoplast isolation and its application for transient expression analysis. Curr. Protoc. Plant Biol. 2016, 1, 373–383. [Google Scholar] [CrossRef]
- Lee, S.-K.; Eom, J.-S.; Hwang, S.-K.; Shin, D.; An, G.; Okita, T.W.; Jeon, J.-S. Plastidic phosphoglucomutase and ADP-glucose pyrophosphorylase mutants impair starch synthesis in rice pollen grains and cause male sterility. J. Exp. Bot. 2016, 67, 5557–5569. [Google Scholar] [CrossRef] [Green Version]
- Jiang, M.; Jiang, J.; Li, S.; Li, M.; Tan, Y.; Song, S.; Shu, Q.; Huang, J. Glutamate alleviates cadmium toxicity in rice via suppressing cadmium uptake and translocation. J. Hazard. Mater. 2020, 384, 121319. [Google Scholar] [CrossRef]
- He, J.; Duan, Y.; Hua, D.; Fan, G.; Wang, L.; Liu, Y.; Chen, Z.; Han, L.; Qu, L.-J.; Gong, Z. DEXH box RNA helicase–mediated mitochondrial reactive oxygen species production in arabidopsis mediates crosstalk between abscisic acid and auxin signaling. Plant Cell 2012, 24, 1815–1833. [Google Scholar] [CrossRef] [Green Version]



| Name of Primers | Sequences (5′-3′) | Remarks |
|---|---|---|
| KEAP1-F | ggcaGGGTGCTACGAAACCCGC | OsKEAP1 sgRNA synthesize |
| KEAP1-R | aaacGCGGGTTTCGTAGCACCC | |
| Pk1-F | GGTTGATCGATGCTTGCTGC | For oskeap1 mutant identification and high resolution melting (HRM) analysis |
| Pk1-R | CACCAATCGCGACCAAATCG | |
| KPqpcr-F | CAAGCACTGGCCAGCTTAAT | q-PCR analysis |
| KPqpcr-R | GATTAGCGCGAACAGGAGCA | |
| qKp2-F | GCAGCCCGTCTATGATGAACTCTC | |
| qKp2-R | AGAAACCCTCCCTTGGGTCATAC | |
| qCATA-F | TCCCAGTGTGATGAGTCGTTGG | |
| qCATA-R | TCTTCACATGCTTGGCTTCACG | |
| qCATB-F | TCCTACTGGTCGCAGTGTGATG | |
| qCATB-R | TTTCAGGTTGAGACGTGAAGCC | |
| KpGFP F | gaattcATGGGTGCTGGAAAGAAGACTCA | Complementary DNA amplification |
| KpGFP R | ggatccCAATGGCAACGGCGCATGC | |
| HYG-F | AGAAGAAGATGTTGGCGACCT | Hygromycin marker primer |
| HYG-R | GTCCTGCGGGTAAATAGCT |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, Y.-H.; Jiang, M.; Li, R.-Q.; Basnet, R.; Huang, J.-Z.; Song, S.-Y.; Shu, Q.-Y. Identification, Characterization, and Mutational Analysis of a Probable KEAP1 Ortholog in Rice (Oryza sativa L.). Plants 2020, 9, 1450. https://doi.org/10.3390/plants9111450
Liu Y-H, Jiang M, Li R-Q, Basnet R, Huang J-Z, Song S-Y, Shu Q-Y. Identification, Characterization, and Mutational Analysis of a Probable KEAP1 Ortholog in Rice (Oryza sativa L.). Plants. 2020; 9(11):1450. https://doi.org/10.3390/plants9111450
Chicago/Turabian StyleLiu, Yan-Hua, Meng Jiang, Rui-Qing Li, Rasbin Basnet, Jian-Zhong Huang, Shi-Yong Song, and Qing-Yao Shu. 2020. "Identification, Characterization, and Mutational Analysis of a Probable KEAP1 Ortholog in Rice (Oryza sativa L.)" Plants 9, no. 11: 1450. https://doi.org/10.3390/plants9111450





