Differential Expression Profiling Reveals Stress-Induced Cell Fate Divergence in Soybean Microspores
Abstract
:1. Introduction
2. Results
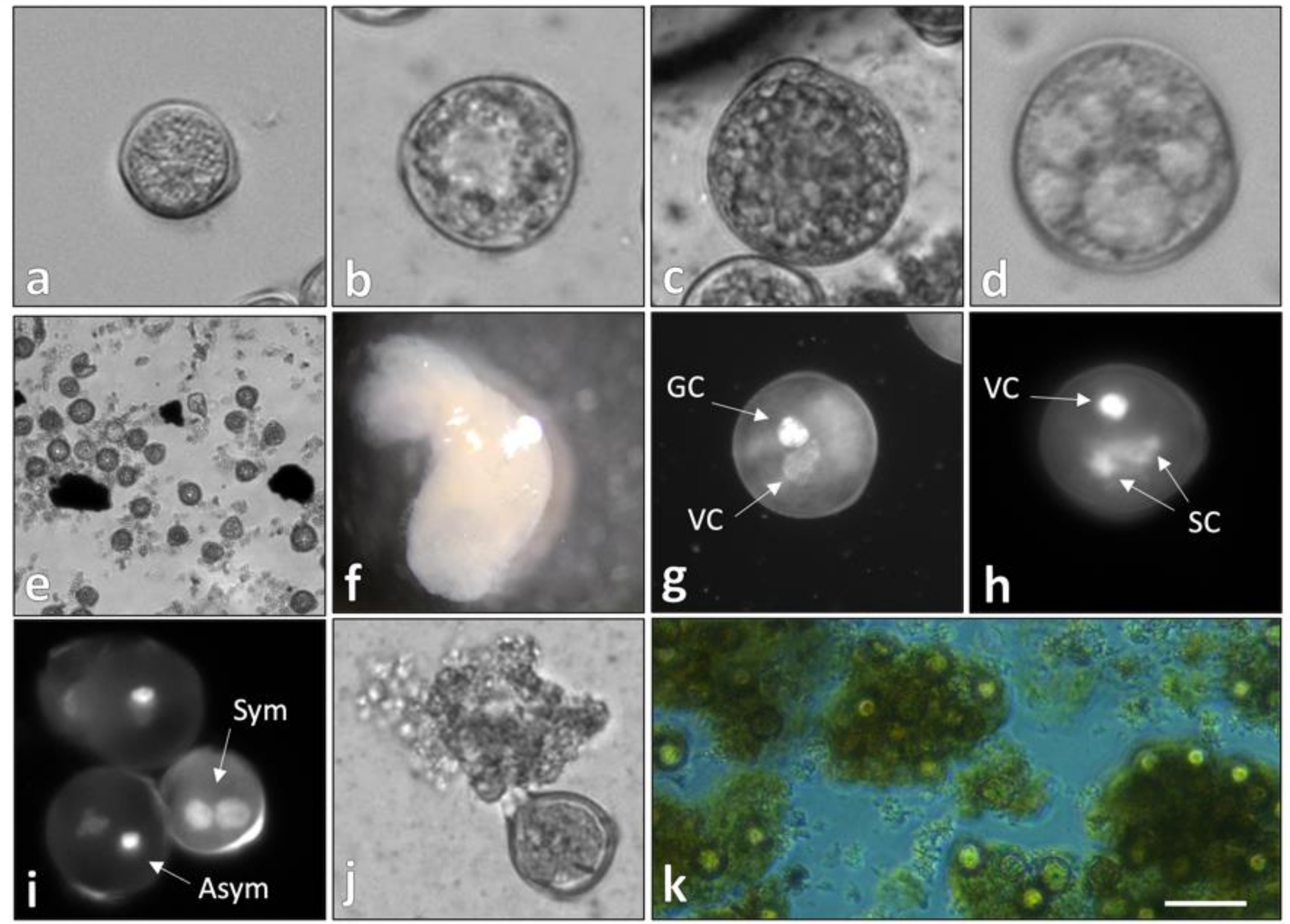
2.1. Temperature-Stressed Microspores Presented a Nongametophytic Morphology
2.2. Cold Treatment Elicited a Classic Stress Response
2.3. Microspore Totipotency Was Initiated by Suppression of the Gametophytic Program
2.4. Stressed Microspores Expressed Transcripts that Evidenced the Onset of Sporophytic Growth
3. Materials and Methods
3.1. Plant Materials
3.2. Floral Bud Selection and Microspore Isolation
3.3. Estimation of Microspore Density
3.4. Sample Preparation and Microscopy
3.5. RNA Isolation, Quantification, and Qualification
3.6. cDNA Library Preparation
3.7. Cluster Generation and Sequencing
3.8. Data Processing, Analyses, and Visualization
3.9. Data Availability
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations:
| ATP | adenosine triphosphate |
| DAPI | 4′6-diamidino-2-phenylindole |
| DEG | differentially expressed gene |
| EP3-3 | endochitinase protein3-3 |
| GO | gene ontology |
| HSP | heat shock protein |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| MCM | minichromosome maintenance protein |
| PBS | Phosphate-buffered saline |
| PCR | polymerase chain reaction |
| RIN | RNA integrity number |
| RNA | ribonucleic acid |
| RNA-Seq | RNA sequencing |
| RPKM | reads per kilobase of exon model per million mapped reads |
| Srna | small RNA |
References
- Seifert, F.; Bössow, S.; Kumlehn, J.; Gnad, H.; Scholten, S. Analysis of wheat microspore embryogenesis induction by transcriptome and small RNA sequencing using the highly responsive cultivar “Svilena”. BMC Plant Biol. 2016, 16, 97. [Google Scholar] [CrossRef]
- Seguí-Simarro, J.M. Androgenesis revisited. Bot. Rev. 2010, 76, 377–404. [Google Scholar] [CrossRef]
- Touraev, A.; Vicente, O.; Heberle-Bors, E. Initiation of microspore embryogenesis by stress. Trends Plant Sci. 1997, 2, 297–302. [Google Scholar] [CrossRef]
- Germanà, M.A. Gametic embryogenesis and haploid technology as valuable support to plant breeding. Plant Cell Rep. 2011, 30, 839–857. [Google Scholar] [CrossRef]
- Maluszynski, M.; Kasha, K.J.; Szarejko, I. Published Doubled Haploid Protocols in Plant Species. In Doubled Haploid Production in Crop Plants: A Manual; Maluszynski, M., Kasha, K.J., Forster, B.P., Szarejko, I., Eds.; Kluwer Academic Publishers: Dodrecht, The Netherlands, 2003; pp. 309–335. [Google Scholar] [CrossRef]
- Twell, D.; Park, S.K.; Lalanne, E. Asymmetric division and cell-fate determination in developing pollen. Trends Plant Sci. 1998, 3, 305–310. [Google Scholar] [CrossRef]
- Shariatpanahi, M.E.; Bal, U.; Heberle-Bors, E.; Touraev, A. Stresses applied for the reprogramming of plant microspores towards in vitro embryogenesis. Physiol. Plant 2006, 127, 519–534. [Google Scholar] [CrossRef]
- Touraev, A.; Indrianto, A.; Wratschko, I.; Vicente, O.; Heberle-Bors, E. Efficient microspore embryogenesis in wheat (Triticum aestivum L.) induced by starvation at high temperature. Sex. Plant Rep. 1996, 9, 209–215. [Google Scholar] [CrossRef]
- Maraschin, S.D.F.; De Priester, W.; Spaink, H.P.; Wang, M. Androgenic switch: An example of plant embryogenesis from the male gametophyte perspective. J. Exp. Bot. 2005, 56, 1711–1726. [Google Scholar] [CrossRef]
- Tang, X.C.; Sun, M.X. Exine-dehisced microspores: A novel model system for studying embryogenesis. Int. J. Plant Dev. Biol. 2007, 1, 28–33. [Google Scholar]
- Song, H.; Lou, Q.F.; Luo, X.D.; Wolukau, J.N.; Diao, W.P.; Qian, C.T.; Chen, J.F. Regeneration of doubled haploid plants by androgenesis of cucumber (Cucumis sativus L.). Plant Cell Tissue Organ. Cult. 2007, 90, 245–254. [Google Scholar] [CrossRef]
- Testillano, P.S. Microspore embryogenesis: Targeting the determinant factors of stress-induced cell reprogramming for crop improvement. J. Exp. Bot. 2019, 70, 2965–2978. [Google Scholar] [CrossRef]
- Pérez-Pérez, Y.O.; El-Tantawy, A.A.; González, M.S.; Risueno, M.C.; Testillano, P.S. Stress-induced microspore embryogenesis requires endogenous auxin synthesis and polar transport in barley. Front. Plant Sci. 2019, 10, 1200. [Google Scholar] [CrossRef] [PubMed]
- Rivas-Sendra, A.; Calabiug-Serna, A.; Seguí-Simarro, J.M. Dynamics of calcium during in vitro microspore embryogenesis and in vivo microspore development in Brassica napus and Solanum melongena. Front. Plant Sci. 2017, 7, 1177. [Google Scholar] [CrossRef]
- Pauls, K.P.; Chan, J.; Woronuk, G.; Schulze, D.; Brazolot, J. When microspores decide to become embryos—Cellular and molecular changes. Can. J. Bot. 2006, 84, 668–678. [Google Scholar] [CrossRef]
- Belanger, S.; Baldrich, P.; Lemay, M.A.; Marchand, S.; Esteves, P.; Meyers, B.C.; Belzile, F.C. The commitment of barley microspores into embryogenesis involves miRNA-directed regulation of members of the SPL, GRF and HD-ZIPIII transcription factor families. bioRxiv 2020, Preprint. [Google Scholar] [CrossRef]
- Solís, M.T.; El-Tantawy, A.A.; Cano, V.; Risueño, M.C.; Testillano, P.S. 5-azacytidine promotes microspore embryogenesis initiation by decreasing global DNA methylation, but prevents subsequent embryo development in rapeseed and barley. Front. Plant Sci. 2015, 6, 472. [Google Scholar] [CrossRef]
- Berenguer, E.; Bárány, I.; Solís, M.T.; Pérez-Pérez, Y.; Risueño, M.C.; Testillano, P.S. Inhibition of histone H3K9 methylation by BIX-01294 promotes stress-induced microspore totipotency and enhances embryogenesis initiation. Front. Plant Sci. 2017, 8, 1161. [Google Scholar] [CrossRef]
- Li, H.; Soriano, M.; Cordewener, J.; Muiño, J.M.; Riksen, T.; Fukuoka, H.; Angenent, G.C.; Boutilier, K. The histone deacetylase inhibitor trichostatin a promotes totipotency in the male gametophyte. Plant Cell 2014, 26, 195–209. [Google Scholar] [CrossRef]
- Malik, M.R.; Wang, F.; Dirpaul, J.M.; Zhou, N.; Polowick, P.L.; Ferrie, A.M.; Krochko, J.E. Transcript profiling and identification of molecular markers for early microspore embryogenesis in Brassica napus. Plant Phys. 2007, 144, 134–154. [Google Scholar] [CrossRef]
- Soriano, M.; Li, H.; Boutilier, K. Microspore embryogenesis: Establishment of embryo identity and pattern in culture. Plant Rep. 2013, 26, 181–196. [Google Scholar] [CrossRef]
- Bélanger, S.; Marchand, S.; Jacques, P.É.; Meyers, B.; Belzile, F. Differential expression profiling of microspores during the early stages of isolated microspore culture using the responsive barley cultivar gobernadora. G3 Genes Genomes Genet. 2018, 8, 1603–1614. [Google Scholar] [CrossRef]
- Corral-Martínez, P.; Siemons, C.; Horstman, A.; Angenent, G.C.; de Ruijter, N.; Boutilier, K. Live Imaging of embryogenic structures in Brassica napus microspore embryo cultures highlights the developmental plasticity of induced totipotent cells. Plant Rep. 2020, 1–16. [Google Scholar] [CrossRef]
- Lulsdorf, M.M.; Croser, J.S.; Ochatt, S. 11 Androgenesis and Doubled-Haploid Production in Food Legumes. Biol. Breed. Food Legumes 2011, 159, 159–177. [Google Scholar]
- Garda, M.; Hale, B.; Rao, N.; Lowe, M.; Bright, M.; Goodling, S.; Phillips, G.C. Soybean androgenesis I: Identification of pyramidal stressors in anther cultures that sustain cell divisions and putative embryo formation from isolated microspore cultures. Vitro Cell Dev. Biol. Plant 2020, 56, 415–429. [Google Scholar] [CrossRef]
- Nitsch, J.P.; Nitsch, C. Haploid plants from pollen grains. Science 1969, 163, 85–87. [Google Scholar] [CrossRef]
- Gamborg, O.L.; Miller, R.; Ojima, K. Nutrient requirements of suspension cultures of soybean root cells. Exp. Cell Res. 1968, 50, 151–158. [Google Scholar] [CrossRef]
- Lichter, R. Induction of haploid plants from isolated pollen of Brassica napus. Z Pflanzenphysiol. 1982, 105, 427–434. [Google Scholar] [CrossRef]
- Ranch, J.P. Plant regeneration from tissue cultures of soybean by somatic embryogenesis. Cell Cult. Somat. Cell Genet. Plants 1986, 97–110. [Google Scholar]
- Phillips, G.C.; Collins, G.B. In vitro tissue culture of selected legumes and plant regeneration from callus cultures of red clover. Crop Sci. 1979, 19, 59–64. [Google Scholar] [CrossRef]
- Hale, B.; Phipps, C.; Rao, N.; Kelley, C.; Phillips, G.C. Soybean androgenesis II: Characterization of early embryogenesis from isolated microspore cultures. Vitro Cell Dev. Biol. Plant 2020. (In Review) [Google Scholar]
- Ferrie, A.M.R.; Caswell, K.L. Isolated microspore culture techniques and recent progress for haploid and doubled haploid plant production. Plant Cell Tissue Organ. Cult. 2011, 104, 301–309. [Google Scholar] [CrossRef]
- Rodrigues, L.R.; Oliveira, J.M.S.; Mariath, J.E.; Bodanese-Zanettini, M.H. Histology of embryogenic responses in soybean anther culture. Plant Cell Tissue Organ. Cult. 2005, 80, 129–137. [Google Scholar] [CrossRef]
- Moraes, A.P.D.; Bonadese-Zanettini, M.H.; Callegari-Jacques, S.M.; Kaltchuk-Santos, E. Effect of temperature shock on soybean microspore embryogenesis. Braz. Arch. Biol. Technol. 2004, 47, 537–544. [Google Scholar] [CrossRef]
- Kaltchuk-Santos, E.; Mariath, J.E.; Mundstock, E.; Hu, C.Y.; Bodanese-Zanettini, M.H. Cytological analysis of early microspore divisions and embryo formation in cultured soybean anthers. Plant Cell Tissue Organ. Cult. 1997, 49, 107–115. [Google Scholar] [CrossRef]
- Kaltchuk-Santos, E.; Zanettini, M.H.B.; Mundstock, E. Pollen dimorphism in soybean. Protoplasma 1993, 174, 74–78. [Google Scholar] [CrossRef]
- Maraschin, S.D.F.; Caspers, M.; Potokina, E.; Wülfert, F.; Graner, A.; Spaink, H.P.; Wang, M. cDNA array analysis of stress-induced gene expression in barley androgenesis. Phys. Plant 2006, 127, 535–550. [Google Scholar] [CrossRef]
- Joosen, R.; Cordewener, J.; Supena, E.D.J.; Vorst, O.; Lammers, M.; Maliepaard, C.; Zeilmaker, T.; Miki, B.; America, T.; Custers, J.; et al. Combined transcriptome and proteome analysis identifies pathways and markers associated with the establishment of rapeseed microspore-derived embryo development. Plant Phys. 2007, 144, 155–172. [Google Scholar] [CrossRef]
- Muñoz-Amatriaín, M.; Svensson, J.T.; Castillo, A.M.; Close, T.J.; Vallés, M.P. Microspore embryogenesis: Assignment of genes to embryo formation and green vs. albino plant production. Funct. Int. Genom. 2009, 9, 311–323. [Google Scholar] [CrossRef]
- Seguí-Simarro, J.M.; Testillano, P.S.; Risueno, M.C. Hsp70 and Hsp90 change their expression and subcellular localization after microspore embryogenesis induction in Brassica napus L. J. Struct. Biol. 2003, 142, 379–391. [Google Scholar] [CrossRef]
- Bárány, I.; Testillano, P.S.; Mitykó, J.; Risueno, M.C. The switch of the microspore program in Capsicum involves HSP70 expression and leads to the production of haploid plants. Int. J. Dev. Biol. 2001, 45, 39–40. [Google Scholar]
- Zarsky, V.; Garrido, D.; Eller, N.; Tupy, J.; Vicente, O.; Schöffl, F.; Heberle-Bors, E. The expression of a small heat shock gene is activated during induction of tobacco pollen embryogenesis by starvation. Plant Cell Environ. 1995, 18, 139–147. [Google Scholar] [CrossRef]
- Telmer, C.A.; Newcomb, W.; Simmonds, D.H. Cellular changes during heat shock induction and embryo development of cultured microspores of Brassica napus cv. Topas. Protoplasma 1995, 185, 106–112. [Google Scholar] [CrossRef]
- Zhao, J.; Newcomb, W.; Simmonds, D. Heat-shock proteins 70 kDa and 19 kDa are not required for induction of embryogenesis of Brassica napus L. cv. Topas microspores. Plant Cell Phys. 2003, 44, 1417–1421. [Google Scholar] [CrossRef]
- Dubas, E.; Custers, J.; Kieft, H.; Wędzony, M.; van Lammeren, A.A. Characterization of polarity development through 2-and 3-D imaging during the initial phase of microspore embryogenesis in Brassica napus L. Protoplasma 2014, 251, 103–113. [Google Scholar] [CrossRef]
- Naya, L.; Paul, S.; Valdés-López, O.; Mendoza-Soto, A.B.; Nova-Franco, B.; Sosa-Valencia, G.; Reyes, J.L.; Hernández, G. Regulation of copper homeostasis and biotic interactions by microRNA 398b in common bean. PLoS ONE 2014, 9, e84416. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Gao, S.Y.; Liu, H.H.; Zhang, X.L.; Zeng, A.S.; Wang, J.J.; Hou, X.L.; Li, Y. cDNA-AFLP analysis of differentially expressed genes during microspore embryogenesis in non-heading Chinese cabbage. Vitro Cell Dev. Biol. Plant 2020, 56, 18–28. [Google Scholar] [CrossRef]
- Zondlo, S.C.; Irish, V.F. CYP78A5 encodes a cytochrome P450 that marks the shoot apical meristem boundary in Arabidopsis. Plant J. 1999, 19, 259–268. [Google Scholar] [CrossRef]
- Ehlting, J.; Sauveplane, V.; Olry, A.; Ginglinger, J.F.; Provart, N.J.; Werck-Reichhart, D. An extensive (co-) expression analysis tool for the cytochrome P450 superfamily in Arabidopsis thaliana. BMC Plant Biol. 2008, 8, 47. [Google Scholar] [CrossRef]
- McCormick, S. Male gametophyte development. Plant Cell 1993, 5, 1265–1275. [Google Scholar] [CrossRef]
- Datta, R.; Chourey, P.S.; Pring, D.R.; Tang, H.V. Gene-expression analysis of sucrose-starch metabolism during pollen maturation in cytoplasmic male-sterile and fertile lines in sorghum. Sex. Plant Rep. 2001, 14, 127–134. [Google Scholar] [CrossRef]
- Hosp, J.; Maraschin, S.F.D.; Touraev, A.; Boutilier, K. Functional genomics of microspore embryogenesis. Euphytica 2007, 158, 275–285. [Google Scholar] [CrossRef]
- van Hengel, A.J.; Guzzo, F.; van Kammen, A.; de Vries, S.C. Expression pattern of the carrot EP3 endochitinase genes in suspension cultures and in developing seeds. Plant Phys. 1993, 117, 43–53. [Google Scholar] [CrossRef]
- van Hengel, A.J.; Tadesse, Z.; Immerzeel, P.; Schols, H.; Van Kammen, A.B.; de Vries, S.C. N-acetylglucosamine and glucosamine-containing arabinogalactan proteins control somatic embryogenesis. Plant Phys. 2001, 125, 1880–1890. [Google Scholar] [CrossRef]
- Borderies, G.; Le Béchec, M.; Rossignol, M.; Lafitte, C.; Le Deunff, E.; Beckert, M.; Dumas, C.; Matthys-Rochon, E. Characterization of proteins secreted during maize microspore culture: Arabinogalactan proteins (AGPs) stimulate embryo development. Eur. J. Cell Biol. 2004, 83, 205–212. [Google Scholar] [CrossRef]
- Bárány, I.; Berenguer, E.; Solís, M.T.; Pérez-Pérez, Y.; Santamaría, M.E.; Crespo, J.L.; Risueño, M.C.; Díaz, I.; Testillano, P.S. Autophagy is activated and involved in cell death with participation of cathepsins during stress-induced microspore embryogenesis in barley. J. Exp. Bot. 2018, 69, 1387–1402. [Google Scholar] [CrossRef] [PubMed]
- Berenguer, E.; Minina, E.A.; Carneros, E.; Bárány, I.; Bozhkov, P.V.; Testillano, P.S. Suppression of Metacaspase-and Autophagy-Dependent Cell Death Improves Stress-Induced Microspore Embryogenesis in Brassica Napus. 2020. Available online: http://hdl.handle.net/10261/221508 (accessed on 21 October 2020).
- Bárány, I.; Fadón, B.; Risueño, M.C.; Testillano, P.S. Cell wall components and pectin esterification levels as markers of proliferation and differentiation events during pollen development and pollen embryogenesis in Capsicum annuum L. J. Exp. Bot. 2010, 61, 1159–1175. [Google Scholar] [CrossRef]
- Rodríguez-Sanz, H.; Manzanera, J.A.; Solís, M.T.; Gómez-Garay, A.; Pintos, B.; Risueño, M.C.; Testillano, P.S. Early markers are present in both embryogenesis pathways from microspores and immature zygotic embryos in cork oak, Quercus suber L. BMC Plant Biol. 2014, 14, 224. [Google Scholar] [CrossRef]
- El-Tantawy, A.A.; Solís, M.T.; Da Costa, M.L.; Coimbra, S.; Risueño, M.C.; Testillano, P.S. Arabinogalactan protein profiles and distribution patterns during microspore embryogenesis and pollen development in Brassica napus. Plant Rep. 2013, 26, 231–243. [Google Scholar] [CrossRef]
- Lee, J.Y.; Lee, D.H. Use of serial analysis of gene expression technology to reveal changes in gene expression in Arabidopsis pollen undergoing cold stress. Plant Phys. 2003, 132, 517–529. [Google Scholar] [CrossRef]
- Honys, D.; Twell, D. Comparative analysis of the Arabidopsis pollen transcriptome. Plant Phys. 2003, 132, 640–652. [Google Scholar] [CrossRef]
- Thibaud-Nissen, F.; Shealy, R.T.; Khanna, A.; Vodkin, L.O. Clustering of microarray data reveals transcript patterns associated with somatic embryogenesis in soybean. Plant Phys. 2003, 132, 18–136. [Google Scholar] [CrossRef]
- Chong, J.P.; Hayashi, M.K.; Simon, M.N.; Xu, R.M.; Stillman, B. A double-hexamer archaeal minichromosome maintenance protein is an ATP-dependent DNA helicase. Proc. Nat. Acad. Sci. USA 2000, 97, 1530–1535. [Google Scholar] [CrossRef]
- Ni, D.A.; Sozzani, R.; Blanchet, S.; Domenichini, S.; Reuzeau, C.; Cella, R.; Bergounioux, C.; Raynaud, C. The Arabidopsis MCM2 gene is essential to embryo development and its over-expression alters root meristem function. New Phytol. 2009, 184, 311–322. [Google Scholar] [CrossRef]
- Greenway, M.B.; Phillips, I.C.; Lloyd, M.N.; Hubstenberger, J.F.; Phillips, G.C. A nutrient medium for diverse applications and tissue growth of plant species in vitro. Vitro Cell Dev. Biol. Plant 2012, 48, 403–410. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Shi, W. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- R Core Team. R: A language and environment for statistical computing. R Found. Stat. Comp. 2017. Available online: https://www.R-project.org/ (accessed on 15 June 2020).
- Yu, G. clusterProfiler: Universal enrichment tool for functional and comparative study. BioRxiv 2018, 256784. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef]
- Wickham, H. Ggplot2: Elegant Graphics for Data Analysis; Springer: Berlin/Heidelberg, Germany, 2016; ISBN 978-3-319-24277-4. [Google Scholar]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hale, B.; Phipps, C.; Rao, N.; Wijeratne, A.; Phillips, G.C. Differential Expression Profiling Reveals Stress-Induced Cell Fate Divergence in Soybean Microspores. Plants 2020, 9, 1510. https://doi.org/10.3390/plants9111510
Hale B, Phipps C, Rao N, Wijeratne A, Phillips GC. Differential Expression Profiling Reveals Stress-Induced Cell Fate Divergence in Soybean Microspores. Plants. 2020; 9(11):1510. https://doi.org/10.3390/plants9111510
Chicago/Turabian StyleHale, Brett, Callie Phipps, Naina Rao, Asela Wijeratne, and Gregory C. Phillips. 2020. "Differential Expression Profiling Reveals Stress-Induced Cell Fate Divergence in Soybean Microspores" Plants 9, no. 11: 1510. https://doi.org/10.3390/plants9111510





