The Turnera Style S-Locus Gene TsBAHD Possesses Brassinosteroid-Inactivating Activity When Expressed in Arabidopsis thaliana
Abstract
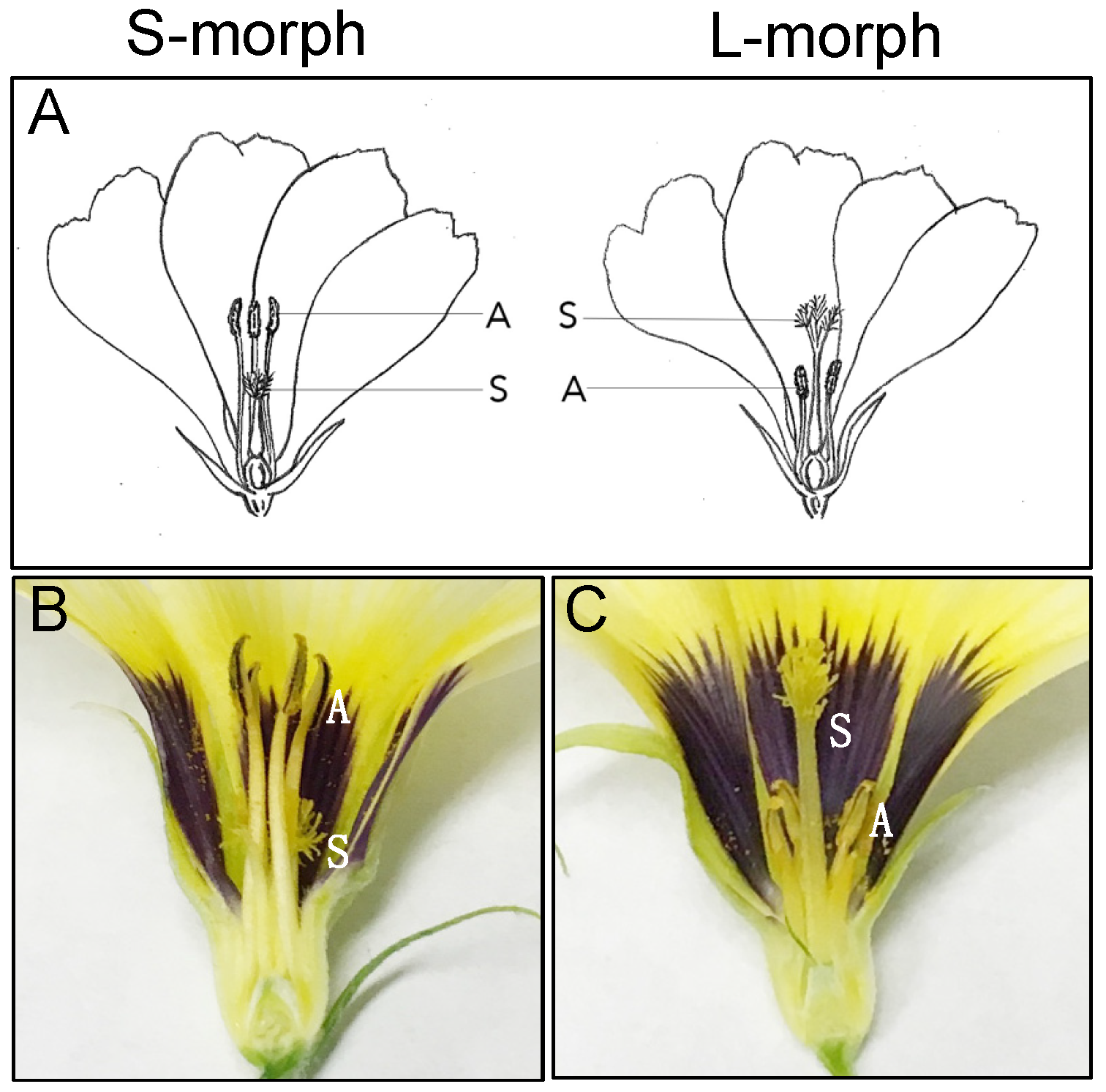
:1. Introduction
2. Results
2.1. Expression of TsBAHD in A. Thaliana Induces Dwarfism
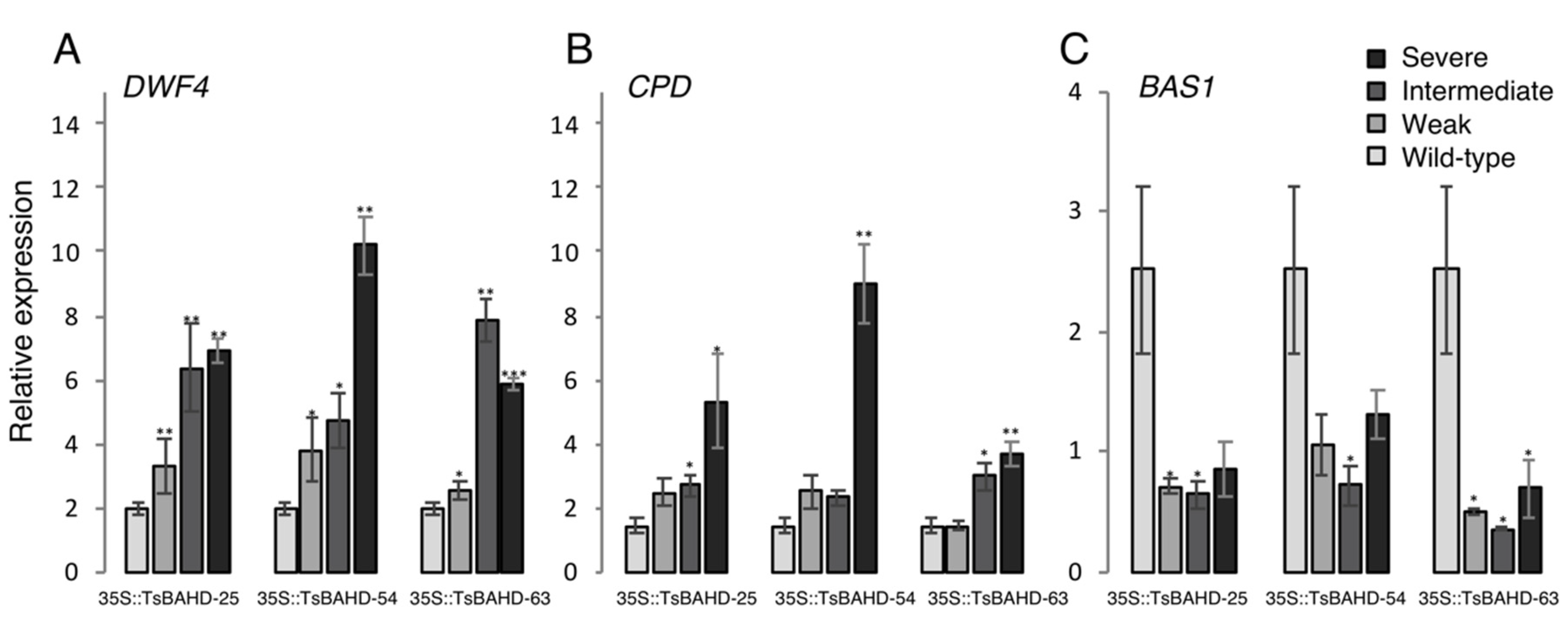
2.2. Assessment of Expression of BR Biosynthesis and Inactivating Genes
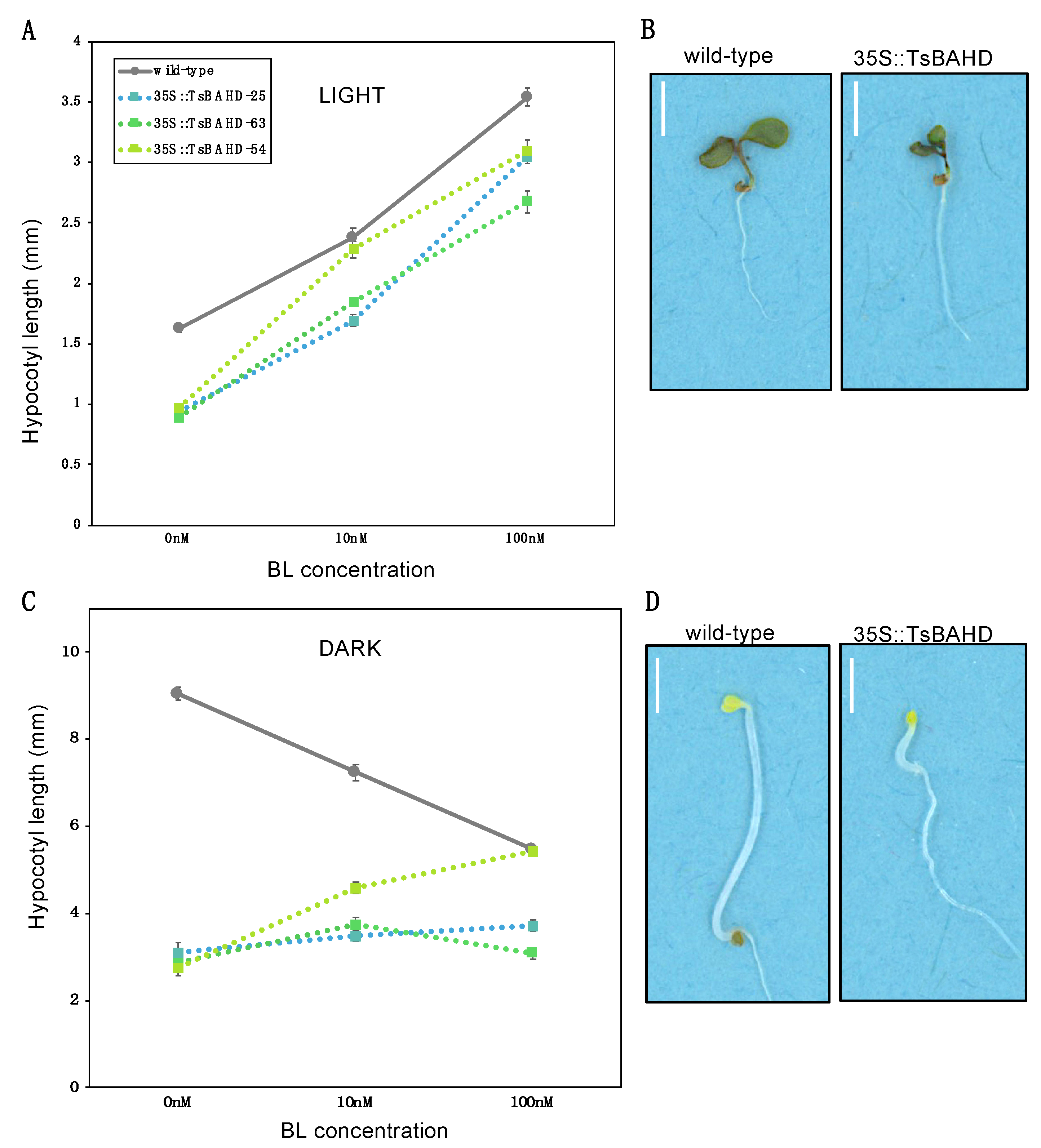
2.3. Response of TsBAHD Expressing Hypocotyls to Exogenous BR in Light and Dark
3. Discussion
4. Materials and Methods
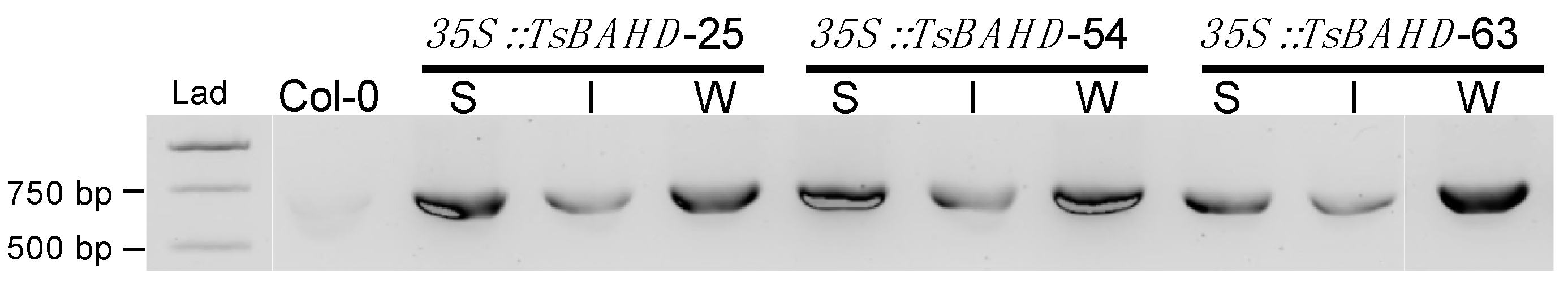
4.1. Plasmid Construction and Generation of Transgenic Lines
4.2. Plant Growth Conditions and Transformation Screening
4.3. RNA Extraction, Reverse Transcription, and RT-qPCR Analyses
4.4. BL Dose Response Growth and Hypocotyl Measurements
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Bateson, W.; Gregory, R.P. On the inheritance of heterostylism in Primula. Proc. R. Soc. Lond. Ser. B 1905, 76, 581–586. [Google Scholar] [CrossRef] [Green Version]
- Barrett, S.C.H.; Cruzan, M.B. Incompatibility in heterostylous plants. In Genetic Control of Self-Incompatibility and Reproductive Development in Flowering Plants; Williams, E.G., Ed.; Kluwer Academic Publishers: Dordrecht, The Netherlands, 1994; pp. 189–219. [Google Scholar] [CrossRef]
- Darwin, C. The Different Forms of Flowers on Plants of the Same Species; John Murray: London, UK, 1877. [Google Scholar]
- Mather, K. The genetical architecture of heterostyly in Primula sinensis. Evolution 1950, 4, 340–352. [Google Scholar] [CrossRef]
- Lewis, D.; Jones, D.A. The genetics of heterostyly. In Evolution and Function of Heterostyly; Barrett, S.C.H., Ed.; Springer: Berlin, Germany, 1992; pp. 129–150. [Google Scholar] [CrossRef]
- Barrett, S.C.H.; Shore, J.S. New insights on heterostyly: Comparative biology, ecology and genetics. In Self-Incompatibility in Flowering Plants–Evolution, Diversity and Mechanisms; Franklin-Tong, V.E., Ed.; Springer: Berlin, Germany, 2008; pp. 3–32. [Google Scholar]
- Kappel, C.; Huu, C.N.; Lenhard, M. A short story gets longer: Recent insights into the molecular basis of heterostyly. J. Exp. Bot. 2017, 68, 5719–5730. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yasui, Y.; Mori, M.; Aii, J.; Abe, T.; Masumoto, D.; Sato, S.; Hayashi, Y.; Ohnishi, O.; Ota, T. S-LOCUS EARLY FLOWERING 3 is exclusively present in the genomes of short-styled buckwheat plants that exhibit heteromorphic self-incompatibility. PLoS ONE 2012, 7, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Yasui, Y.; Hirakawa, H.; Ueno, M.; Matsui, K.; Katsube-Tanaka, T.; Yang, S.J.; Aii, J.; Sato, S.; Mori, M. Assembly of the draft genome of buckwheat and its applications in identifying agronomically useful genes. DNA Res. 2016, 23, 215–224. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Cocker, J.M.; Wright, J.; Webster, M.A.; McMullan, M.; Dyer, S.; Swarbreck, D.; Caccamo, M.; Oosterhout, C.V.; Gilmartin, P.M. Genetic architecture and evolution of the S locus supergene in Primula vulgaris. Nat. Plants 2016, 2, 16188. [Google Scholar] [CrossRef] [PubMed]
- Shore, J.S.; Hamam, H.J.; Chafe, P.D.J.; Labonne, J.D.J.; Henning, P.M.; McCubbin, A.G. The long and short of the S-locus in Turnera (Passifloraceae). New Phytol. 2019, 224, 1316–1329. [Google Scholar] [CrossRef]
- Nowak, M.D.; Russo, G.; Schlapbach, R.; Huu, C.N.; Lenhard, M.; Conti, E. The draft genome of Primula veris yields insights into the molecular basis of heterostyly. Genome Biol. 2015, 16, 12. [Google Scholar] [CrossRef] [Green Version]
- Huu, C.N.; Kappel, C.; Keller, B.; Sicard, A.; Takebayashi, Y.; Breuninger, H.; Nowak, M.D.; Bäurle, I.; Himmelbach, A.; Burkart, M.; et al. Presence versus absence of CYP734A50 underlies the style-length dimorphism in primroses. eLife 2016, 5, e17956. [Google Scholar] [CrossRef]
- Huu, C.N.; Keller, B.; Conti, E.; Kappel, C.; Lenhard, M. Supergene evolution via stepwise duplications and neofunctionalization of a floral-organ identity gene. Proc. Natl. Acad. Sci. USA 2020, 117, 23149–23157. [Google Scholar] [CrossRef]
- Burrows, B.A.; McCubbin, A.G. Sequencing the genomic regions flanking S-linked PvGLO sequences confirm the presence of two GLO loci, one of which lies adjacent to the style-length determinant gene CYP734A50. Plant Reprod. 2017, 30, 53–67. [Google Scholar] [CrossRef] [PubMed]
- Neff, M.M.; Nguyen, S.M.; Malancharuvil, E.J.; Fujioka, S.; Noguchi, T.; Seto, H.; Tsubuki, M.; Honda, T.; Takatsuto, S.; Yoshida, S.; et al. Bas1: A gene regulating brassinosteroid levels and light responsiveness in Arabidopsis. Proc. Natl. Acad. Sci. USA 1999, 96, 15316–15323. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheng, Y.; Dai, X.; Zhao, Y. Auxin biosynthesis by the YUCCA flavin monooxygenases controls formation of floral organs and vascular tissues in Arabidopsis. Gene Dev. 2006, 20, 1790–1799. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cecchetti, V.; Altamura, M.M.; Falasca, G.; Costantino, P.; Cardarelli, M. Auxin regulates Arabidopsis anther dehiscence, pollen maturation, and filament elongation. Plant Cell 2008, 20, 1760–1774. [Google Scholar] [CrossRef] [Green Version]
- Roh, H.; Jeong, C.W.; Fujioka, S.; Kim, Y.K.; Lee, S.; Ahn, J.H.; Choi, Y.D.; Lee, J.S. Genetic evidence for the reduction of brassinosteroid levels by a BAHD acyltransferase-like protein in Arabidopsis. Plant Physiol. 2012, 159, 696–709. [Google Scholar] [CrossRef] [Green Version]
- Henning, P.M.; Shore, J.S.; McCubbin, A.G. Transcriptome and Network Analyses of Heterostyly in Turnera subulata Provide Mechanistic Insights: Are S-Loci a Red-Light for Pistil Elongation? Plants 2020, 9, 713. [Google Scholar] [CrossRef]
- D’Auria, J.C. Acyltransferases in plants: A good time to be BAHD. Curr. Opin. Plant Biol. 2006, 9, 331–340. [Google Scholar] [CrossRef]
- St-Pierre, B.; Luca, V. The cell and developmental biology of alkaloid biosynthesis. Trends Plant Sci. 2000, 5, 168–173. [Google Scholar] [CrossRef]
- Beekwilder, J.; Alvarez-huerta, M.; Neef, E.; Verstappen, F.W.A.; Boumeester, H.J.; Aheroni, A. State of the Field: Functional Characterization of Enzymes Forming Volatile Esters from Strawberry and Banana. Plant Physiol. 2004, 135, 1865–1878. [Google Scholar] [CrossRef] [Green Version]
- Okada, T.; Hirai, M.Y.; Suzuki, H.; Yamazaki, M.; Saito, K. Molecular characterization of a novel quinolizidine alkaloid O-tigloyltransferase: cDNA cloning; catalytic activity of recombinant protein and expression analysis in Lupinus plants. Plant Cell Physiol. 2005, 46, 233–244. [Google Scholar] [CrossRef]
- Stewart, C.; Kang, B.C.; Liu, K.; Mazourek, M.; Moore, S.; Young, E.; Yoo, Y.; Kim, B.-D.; Paran, I.; Jahn, M.M. The Pun1 gene for pungency in pepper encodes a putative acyltransferase. Plant J. 2005, 42, 675–688. [Google Scholar] [CrossRef] [PubMed]
- Bishop, G.J.; Koncz, C. Brassinosteroids and plant steroid hormone signaling. Plant Cell 2002, 14, 97–111. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Symons, G.M.; Reid, J.B. Brassinosteroids do not undergo long-distance transport in pea. Implications for the regulation of endogenous brassinosteroid levels. Plant Physiol. 2004, 135, 2196–2206. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Choe, S.; Dilkes, B.P.; Fujioka, S.; Takatsuto, S.; Sakurai, A.; Feldmann, K.A. The DWF4 gene of Arabidopsis encodes a Cytochrome P450 that mediates multiple 22α-hydroxylation steps in brassinosteroid biosynthesis. Plant Cell 1998, 10, 231–243. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Nagpal, P.; Vitart, V.; McMorris, T.C.; Chory, J. A role for brassinosteroids in light-dependent development of Arabidopsis. Science 1996, 272, 398–401. [Google Scholar] [CrossRef]
- Szekeres, M.; Nemeth, K.; Kalman, Z.; Mathur, J.; Kauschmann, A.; Altmann, T.; Redei, G.P.; Nagy, F.; Schell, J.; Koncz, C. Brassinosteroids rescue the deficiency of CYP90, a cytochrome P450, controlling cell elongation and de-etiolation in Arabidopsis. Cell 1996, 85, 171–182. [Google Scholar] [CrossRef] [Green Version]
- Zhao, B.; Li, J. Regulation of Brassinosteroid Biosynthesis and Inactivation. J. Int. Plant Biol. 2012, 54, 746–759. [Google Scholar] [CrossRef]
- Tanaka, K.; Asami, T.; Yoshida, S.; Nakamura, Y.; Matsuo, T.; Okamoto, S. Brassinosteroid homeostasis in Arabidopsis is ensured by feedback expressions of multiple genes involved in its metabolism. Plant Physiol. 2005, 138, 1117–1125. [Google Scholar] [CrossRef] [Green Version]
- Turk, E.M.; Fujioka, S.; Seto, H.; Shimada, Y.; Takatsuto, S.; Yoshida, S.; Denzel, M.A.; Torres, Q.I.; Neff, M.M. CYP72B1 inactivates brassinosteroid hormones: An intersection between photomorphogenesis and plant steroid signal transduction. Plant Physiol. 2003, 133, 1643–1653. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Z.; Xu, L. Arabidopsis BRASSINOSTEROID INACTIVATOR2 is a typical BAHD acyltransferase involved in brassinosteroid homeostasis. J. Exp. Bot. 2018, 62, 1925–1941. [Google Scholar] [CrossRef]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koncz, C.; Schell, J. The promoter of TI-DNA gene5 controls the tissue-specific expression of chimeric genes carried by a novel type of Agrobacterium binary vector. Mol. Genet. Genom. 1986, 204, 383–396. [Google Scholar] [CrossRef]
- Choe, S.; Fujioka, S.; Noguchi, T.; Takatsuto, S.; Yoshida, S.; Feldmann, K.A. Overexpression of DWARF4 in the brassinosteroid biosynthetic pathway results in increased vegetative growth and seed yield in Arabidopsis. Plant J. 2001, 26, 573–582. [Google Scholar] [CrossRef] [PubMed]
- Song, L.; Zhou, X.Y.; Li, L.; Xue, L.J.; Yang, X.; Xue, H.W. Genome-wide analysis revealed the complex regulatory network of brassinosteroid effects in photomorphogenesis. Mol. Plant 2009, 2, 755–772. [Google Scholar] [CrossRef] [PubMed]
- Bancos, S.; Szatmári, A.-M.; Castle, J.; Kozma-Bognár, L.; Shibata, K.; Yokota, T.; Bishop, G.J.; Nagy, F.; Szekeres, M. Diurnal Regulation of the Brassinosteroid-Biosynthetic CPD Gene in Arabidopsis. Plant Physiol. 2006, 141, 299–309. [Google Scholar] [CrossRef] [Green Version]
- Chory, J.; Nagpal, P.; Peto, C.A. Phenotypic and genetic analysis of det2, a new mutant that affects light-regulated seedling development in Arabidopsis. Plant Cell 1991, 3, 445–459. [Google Scholar] [CrossRef]
- Choi, S.; Cho, Y.H.; Kim, K.; Matsui, M.; Son, S.-H.; Fujioka, S.; Hwang, I. BAT1, a putative acyltransferase, modulates brassinosteroid levels in Arabidopsis. Plant J. 2013, 73, 380–391. [Google Scholar] [CrossRef]
- Wang, M.; Liu, X.; Wang, R.; Wanchun, L.; Rodermel, S.; Yu, F. Overexpression of a putative Arabidopsis BAHD acyltransferase causes dwarfism that can be rescued by brassinosteroid. J. Exp. Bot. 2012, 63, 5787–5801. [Google Scholar] [CrossRef] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Jacques, C.N.; Hulbert, A.; Westenskow, S.; Neff, M.M. Production location of the gelling agent, Phytagel, has a significant impact on Arabidopsis thaliana seedling phenotypic analysis. PLoS ONE 2020, 15, e0228515. [Google Scholar] [CrossRef]
- Sandhu, K.S.; Koirala, P.S.; Neff, M.M. The ben1-1 brassinosteroid-catabolism mutation is unstable due to epigenetic modifications of the intronic T-DNA insertion. G3 2013, 4, 1587–1595. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abràmoff, M.D.; Magalhães, P.J.; Ram, S.J. Image processing with imageJ. Biophotonics Int. 2004, 11, 36–41. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Matzke, C.M.; Shore, J.S.; Neff, M.M.; McCubbin, A.G. The Turnera Style S-Locus Gene TsBAHD Possesses Brassinosteroid-Inactivating Activity When Expressed in Arabidopsis thaliana. Plants 2020, 9, 1566. https://doi.org/10.3390/plants9111566
Matzke CM, Shore JS, Neff MM, McCubbin AG. The Turnera Style S-Locus Gene TsBAHD Possesses Brassinosteroid-Inactivating Activity When Expressed in Arabidopsis thaliana. Plants. 2020; 9(11):1566. https://doi.org/10.3390/plants9111566
Chicago/Turabian StyleMatzke, Courtney M., Joel S. Shore, Michael M. Neff, and Andrew G. McCubbin. 2020. "The Turnera Style S-Locus Gene TsBAHD Possesses Brassinosteroid-Inactivating Activity When Expressed in Arabidopsis thaliana" Plants 9, no. 11: 1566. https://doi.org/10.3390/plants9111566





