Competitive Exclusion of Flavescence dorée Phytoplasma Strains in Catharanthus roseus Plants
Abstract
:1. Introduction
2. Results
2.1. Development of a Real Time PCR Protocol for Simultaneous Diagnosis and Quantification of FD-C and -D Strains
2.2. FD-C and FD-D Infections in Catharanthus roseus
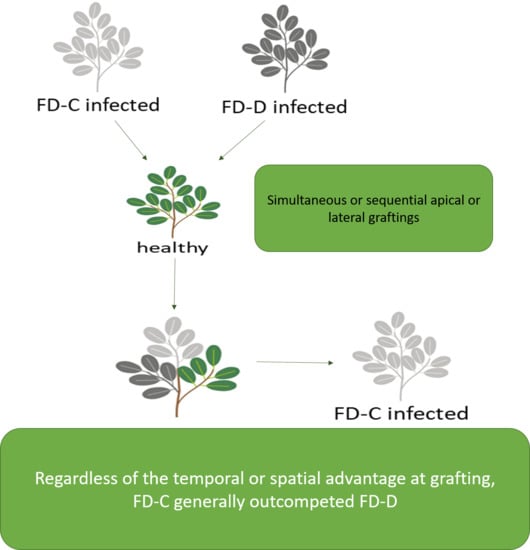
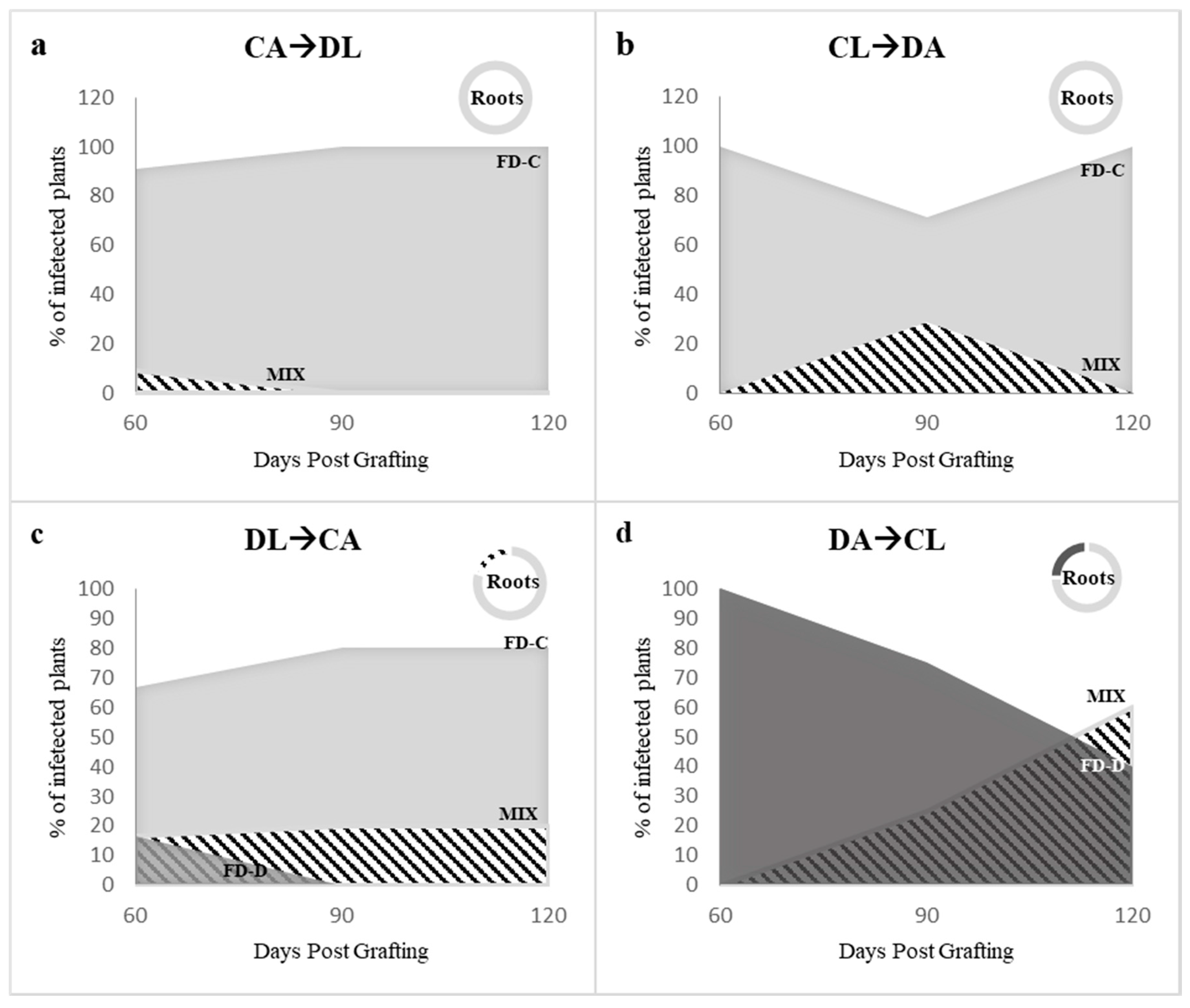
2.3. Effects of the Competition between FD-Cp and FD-Dp Strains in Catharanthus roseus Plants
3. Discussion
4. Materials and Methods
4.1. Phytoplasma Strains and Plant Hosts
4.2. Grafting for Systemic Single or Dual Infection of Catharantus roseus
4.2.1. Dynamics of FD-C and FD-D Single Infections in Catharanthus roseus
4.2.2. Dual Infection of FD-C and FD-D Phytoplasma Strains
4.3. Sampling and DNA Extraction
4.4. Development of a Strain-Specific Quantitative Assay for Flavescence dorée Phytoplasmas
4.5. Statistical Analyses
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Schvester, D.; Carle, P.; Moutous, G. Testing susceptibility of vine varieties to (Flavescence dorée) by means of inoculation by Scaphoideus littoralis Ball. Ann. Des. Epiphyt. 1967, 18, 143. [Google Scholar]
- Chuche, J.; Thiery, D. Biology and ecology of the Flavescence dorée vector Scaphoideus titanus: A review. Agron. Sustain. Dev. 2014, 34, 381–403. [Google Scholar] [CrossRef] [Green Version]
- Caudwell, A. Epidemiology and characterization of flavescence dorée (FD) and other grapevine yellows. Agronomie 1990, 10, 655–663. [Google Scholar] [CrossRef]
- Malembic-Maher, S.; Desqué, D.; Khalil, D.; Salar, P.; Bergey, B.; Danet, J.L.; Duret, S.; Dubrana-Ourabah, M.P.; Beven, L.; Ember, I.; et al. When a Palearctic bacterium meets a Nearctic insect vector: Genetic and ecological insights into the emergence of the grapevine Flavescence dorée epidemics in Europe. PLoS Pathog. 2020, 16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- EFSA PLH Panel (EFSA Panel on Plant Health); Jeger, M.; Bragard, C.; Caffier, D.; Candresse, T.; Chatzivassiliou, E.; Dehnen-Schmutz, K.; Gilioli, G.; Jaques Miret, J.A.; MacLeod, A.; et al. Scientific opinion on the risk to plant health of Flavescence dorée for the EU territory. EFSA J. 2016, 14. [Google Scholar] [CrossRef]
- Davis, R.E.; Dally, E.L. Revised subgroup classification of group 16SrV phytoplasmas and placement of flavescence dorée-associated phytoplasmas in two distinct subgroups. Plant Dis. 2001, 85, 790–797. [Google Scholar] [CrossRef] [Green Version]
- Martini, M.; Murari, E.; Mori, N.; Bertaccini, A. Identification and epidemic distribution of two flavescence dorée-related phytoplasmas in Veneto (Italy). Plant Dis. 1999, 83, 925–930. [Google Scholar] [CrossRef] [Green Version]
- Martini, M.; Botti, S.; Marcone, C.; Marzachı, C.; Casati, P.; Bianco, P.A.; Benedetti, R.; Bertaccini, A. Genetic variability among Flavescence dorée phytoplasmas from different origins in Italy and France. Mol. Cell. Probes 2002, 16, 197–208. [Google Scholar] [CrossRef]
- Lee, I.M.; Martini, M.; Marcone, C.; Zhu, S.F. Classification of phytoplasma strains in the elm yellows group (16SrV) and proposal of ‘Candidatus Phytoplasma ulmi’ for the phytoplasma associated with elm yellows. Int. J. Syst. Evol. Microbiol. 2004, 54, 337–347. [Google Scholar] [CrossRef] [Green Version]
- The Irpcm Phytoplasma/Spiroplasma Working Team-Phytoplasma Taxonomy Group. ‘Candidatus Phytoplasma’, a taxon for the wall-less, non-helical prokaryotes that colonize plant phloem and insects. Int. J. Syst. Evol. Microbiol. 2004, 54, 1243–1255. [Google Scholar] [CrossRef] [Green Version]
- Mori, N.; Bressan, A.; Martini, M.; Guadagnini, M.; Girolami, V.; Bertaccini, A. Experimental transmission by Scaphoideus titanus Ball of two Flavescence dorée-type phytoplasmas. Vitis 2002, 41, 99–102. [Google Scholar]
- Rossi, M.; Pegoraro, M.; Ripamonti, M.; Abbà, S.; Beal, D.; Giraudo, A.; Veratti, F.; Malembic-Maher, S.; Salar, P.; Bosco, D.; et al. Genetic diversity of Flavescence dorée phytoplasmas at the vineyard scale. Appl. Environ. Microbiol. 2019, 85. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wei, W.; Wu, W.; Davis, R.E.; Lee, I.-M.; Zhao, Y. Development of molecular markers and a diagnostic tool for investigation of coinfections by and interactions between potato purple top and potato witches’-broom phytoplasmas in tomato. Ann. Appl. Biol. 2016, 168, 133–141. [Google Scholar] [CrossRef]
- Plavec, J.; Budinšćak, Ž.; Križanac, I.; Škorić, D.; Foissac, X.; Šeruga Musić, M. Multilocus sequence typing reveals the presence of three distinct flavescence dorée phytoplasma genetic clusters in Croatian vineyards. Plant Pathol. 2019, 68, 18–30. [Google Scholar] [CrossRef]
- Zambon, Y.; Canel, A.; Bertaccini, A.; Contaldo, N. Molecular Diversity of Phytoplasmas Associated with Grapevine Yellows Disease in North-Eastern Italy. Phytopathology 2018, 108, 206–214. [Google Scholar] [CrossRef] [Green Version]
- Casati, P.; Jermini, M.; Quaglino, F.; Corbani, G.; Schaerer, S.; Passera, A.; Bianco, P.A.; Rigamonti, I.E. New insights on Flavescence dorée phytoplasma ecology in the vineyard agro-ecosystem in southern Switzerland. Ann. Appl. Biol. 2017, 171, 37–51. [Google Scholar] [CrossRef]
- Hardin, G. The competitive exclusion principle. Science 1960, 131, 1292–1297. [Google Scholar] [CrossRef] [Green Version]
- Salar, P.; Charenton, C.; Foissac, X.; Malembic-Maher, S. Multiplication kinetics of Flavescence dorée phytoplasma in broad bean. Effect of phytoplasma strain and temperature. Eur. J. Plant Pathol. 2013, 135, 371–381. [Google Scholar] [CrossRef]
- Roggia, C.; Caciagli, P.; Galetto, L.; Pacifico, D.; Veratti, F.; Bosco, D.; Marzachì, C. Flavescence dorée phytoplasma titre in field-infected B arbera and N ebbiolo grapevines. Plant Pathol. 2014, 63, 31–41. [Google Scholar] [CrossRef]
- Lessio, F.; Bocca, F.; Alma, A. Development, Spatial Distribution, and Presence on Grapevine of Nymphs of Orientus ishidae (Hemiptera: Cicadellidae), a New Vector of Flavescence Dorée Phytoplasmas. J. Econ. Entomol. 2019, 112, 2558–2564. [Google Scholar] [CrossRef]
- Strauss, G.; Reisenzein, H. First detection of Flavescence dorée phytoplasma in Phlogotettix cyclops (Hemiptera, Cicadellidae) and considerations on its possible role as vector in Austrian vineyards. IOBC-WPRS Bull. 2018, 139, 12–21. [Google Scholar]
- Angelini, E.; Squizzato, F.; Lucchetta, G.; Borgo, M. Detection of a phytoplasma associated with grapevine flavescence dorée in Clematis vitalba. Eur. J. Plant Pathol. 2004, 110, 193–201. [Google Scholar] [CrossRef]
- Hibbing, M.E.; Fuqua, C.; Parsek, M.R.; Peterson, S.B. Bacterial competition: Surviving and thriving in the microbial jungle. Nat. Rev. Microbiol. 2010, 8, 15–25. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alizon, S.; de Roode, J.C.; Michalakis, Y. Multiple infections and the evolution of virulence. Ecol. Lett. 2013, 16, 556–567. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tollenaere, C.; Susi, H.; Laine, A.-L. Evolutionary and epidemiological implications of multiple infection in plants. Trends Plant Sci. 2016, 21, 80–90. [Google Scholar] [CrossRef] [PubMed]
- Lherminier, J.; Courtois, M.; Caudwell, A. Determination of the distribution and multiplication sites of Flavescence Dorée mycoplasma-like organisms in the host plant Vicia faba by ELISA and immunocytochemistry. Physiol. Mol. Plant. Pathol. 1994, 45, 125–138. [Google Scholar] [CrossRef]
- Saracco, P.; Bosco, D.; Veratti, F.; Marzachi, C. Quantification over time of chrysanthemum yellows phytoplasma (16Sr-I) in leaves and roots of the host plant Chrysanthemum carinatum (Schousboe) following inoculation with its insect vector. Physiol. Mol. Plant Pathol. 2005, 67, 212–219. [Google Scholar] [CrossRef]
- Kuske, C.R.; Kirkpatrick, B.C. Phylogenetic relationships between the western aster yellows mycoplasmalike organism and other prokaryotes established by 16S rRNA gene sequence. Int. J. Syst. Bacteriol. 1992, 42, 226–233. [Google Scholar] [CrossRef] [Green Version]
- Read, A.F.; Taylor, L.H. The ecology of genetically diverse infections. Science 2001, 292, 1099–1102. [Google Scholar] [CrossRef] [Green Version]
- Mideo, N. Parasite adaptations to within-host competition. Trends Parasitol. 2009, 25, 261–268. [Google Scholar] [CrossRef]
- Bashey, F. Within-host competitive interactions as a mechanism for the maintenance of parasite diversity. Philos. Trans. R. Soc. B Biol. Sci. 2015, 370. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Margaria, P.; Abbà, S.; Palmano, S. Novel aspects of grapevine response to phytoplasma infection investigated by a proteomic and phospho-proteomic approach with data integration into functional networks. BMC Genom. 2013, 14, 1–15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jollard, C.; Foissac, X.; Desqué, D.; Garcion, C.; Razan, F.; Béven, L.; Eveillard, S. “Flavescence dorée” phytoplasma has multiple ftsH genes showing host dependent expression in both natural and experimental pathosystems. Phytopathog. Mollicutes 2019, 9, 117–118. [Google Scholar] [CrossRef]
- Abbà, S.; Galetto, L.; Carle, P.; Carrère, S.; Delledonne, M.; Foissac, X.; Palmano, S.; Veratti, F.; Marzachì, C. RNA-Seq profile of flavescence dorée phytoplasma in grapevine. BMC Genom. 2014, 15, 1088. [Google Scholar] [CrossRef] [Green Version]
- Lesniak, J.; Barton, W.A.; Nikolov, D.B. Structural and functional features of the Escherichia coli hydroperoxide resistance protein OsmC. Protein Sci. 2003, 12, 2838–2843. [Google Scholar] [CrossRef] [Green Version]
- Colonne, P.M.; Winchell, C.G.; Voth, D.E. Hijacking host cell highways: Manipulation of the host actin cytoskeleton by obligate intracellular bacterial pathogens. Front. Cell. Infect. Microbiol. 2016, 6, 107. [Google Scholar] [CrossRef] [Green Version]
- Buxa, S.V.; Degola, F.; Polizzotto, R.; De Marco, F.; Loschi, A.; Kogel, K.H.; di Toppi, L.S.; van Bel, A.J.; Musetti, R. Phytoplasma infection in tomato is associated with re-organization of plasma membrane, ER stacks, and actin filaments in sieve elements. Front. Plant Sci. 2015, 6, 650. [Google Scholar] [CrossRef] [Green Version]
- Boonrod, K.; Munteanu, B.; Jarausch, B.; Jarausch, W.; Krczal, G. An immunodominant membrane protein (Imp) of ‘Candidatus Phytoplasma mali’ binds to plant actin. Mol. Plant Microbe Interact. 2012, 25, 889–895. [Google Scholar] [CrossRef] [Green Version]
- Suzuki, S.; Oshima, K.; Kakizawa, S.; Arashida, R.; Jung, H.Y.; Yamaji, Y.; Nishigawa, H.; Ugaki, M.; Namba, S. Interaction between the membrane protein of a pathogen and insect microfilament complex determines insect-vector specificity. Proc. Natl. Acad. Sci. USA 2006, 103, 4252–4257. [Google Scholar] [CrossRef] [Green Version]
- Galetto, L.; Bosco, D.; Balestrini, R.; Genre, A.; Fletcher, J.; Marzachì, C. The major antigenic membrane protein of ‘Candidatus Phytoplasma asteris’ selectively interacts with ATP synthase and actin of leafhopper vectors. PLoS ONE 2011, 6, e22571. [Google Scholar] [CrossRef] [Green Version]
- Trivellone, V.; Ripamonti, M.; Angelini, E.; Filippin, L.; Rossi, M.; Marzachí, C.; Galetto, L. Evidence suggesting interactions between immunodominant membrane protein Imp of Flavescence dorée phytoplasma and protein extracts from distantly related insect species. J. Appl. Microbiol. 2019, 127, 1801–1813. [Google Scholar] [CrossRef] [PubMed]
- Galetto, L.; CNR, Institute for Sustainable Plant Protection, Torino, Italy. Personal communication, 2019.
- Filippin, L.; Trivellone, V.; Galetto, L.; Marzachì, C.; Elicio, V.; Angelini, E. Development of an anti-Imp serological assay for the detection of “flavescence dorée” phytoplasmas in grapevine, insect vectors and host plants. Phytopathog. Mollicutes 2019, 9, 75–76. [Google Scholar] [CrossRef]
- Schneider, B.; Sule, S.; Jelkmann, W.; Seemüller, E. Suppression of Aggressive Strains of ‘Candidatus Phytoplasma mali’ by Mild Strains in Catharanthus roseus and Nicotiana occidentalis and Indication of Similar Action in Apple Trees. Phytopathology 2014, 104, 453–461. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Haapalainen, M.; Mosorin, H.; Dorati, F.; Wu, R.F.; Roine, E.; Taira, S.; Nissinen, R.; Mattinen, L.; Jackson, R.; Pirhonen, M.; et al. Hcp2, a secreted protein of the phytopathogen Pseudomonas syringae pv. tomato DC3000, is required for fitness for competition against bacteria and yeasts. J. Bacteriol. 2012, 194, 4810–4822. [Google Scholar] [CrossRef] [PubMed]
- MacIntyre, D.L.; Miyata, S.T.; Kitaoka, M.; Pukatzki, S. The Vibrio cholerae type VI secretion system displays antimicrobial properties. Proc. Natl. Acad. Sci. USA 2010, 107, 19520–19524. [Google Scholar] [CrossRef] [Green Version]
- Russell, A.; Peterson, S.; Mougous, J. Type VI secretion system effectors: Poisons with a purpose. Nat. Rev. Microbiol. 2014, 12, 137–148. [Google Scholar] [CrossRef]
- Seemüller, E.; Zikeli, K.; Furch, A.C.; Wensing, A.; Jelkmann, W. Virulence of ‘Candidatus Phytoplasma mali’ strains is closely linked to conserved substitutions in AAA+ ATPase AP460 and their supposed effect on enzyme function. Eur. J. Plant Pathol. 2018, 150, 701–711. [Google Scholar] [CrossRef]
- Jollard, C.; Foissac, X.; Desqué, D.; Razan, F.; Garcion, C.; Beven, L.; Eveillard, S. Flavescence dorée phytoplasma has multiple ftsh genes that are differentially expressed in plants and insects. Int. J. Mol. Sci. 2020, 21, 150. [Google Scholar] [CrossRef] [Green Version]
- Hogenhout, S.A.; Oshima, K.; AMMAR, E.D.; Kakizawa, S.; Kingdom, H.N.; Namba, S. Phytoplasmas: Bacteria that manipulate plants and insects. Mol. Plant Pathol. 2008, 9, 403–423. [Google Scholar] [CrossRef]
- Carle, P.; Malembic-Maher, S.; Arricau-Bouvery, N.; Desque, D.; Eveillard, S.; Carrere, S.; Foissac, X. ‘Flavescence dorée’ phytoplasma genome: A metabolism oriented towards glycolysis and protein degradation. Bull. Insectol. 2011, 64, S13–S14. [Google Scholar]
- Firrao, G.; Martini, M.; Ermacora, P.; Loi, N.; Torelli, E.; Foissac, X.; Carle, P.; Kirkpatrick, B.C.; Liefting, L.; Schneider, B.; et al. Genome wide sequence analysis grants unbiased definition of species boundaries in “Candidatus Phytoplasma”. Syst. Appl. Microbiol. 2013, 36, 539–548. [Google Scholar] [CrossRef] [PubMed]
- Seemüller, E.; Kiss, E.; Sule, S.; Schneider, B. Multiple infection of apple trees by distinct strains of ‘Candidatus Phytoplasma mali’ and its pathological relevance. Phytopathology 2010, 100, 863–870. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pacifico, D.; Margaria, P.; Galetto, L.; Legovich, M.; Abbà, S.; Veratti, F.; Marzachì, C.; Palmano, S. Differential gene expression in two grapevine cultivars recovered from “flavescence dorée”. Microbiol. Res. 2019, 220, 72–82. [Google Scholar] [CrossRef] [PubMed]
- Pagliarani, C.; Gambino, G.; Ferrandino, A.; Chitarra, W.; Vrhovsek, U.; Cantu, D.; Palmano, S.; Marzachì, C.; Schubert, A. Molecular memory of Flavescence dorée phytoplasma in recovering grapevines. Hortic. Res. 2020, 7, 1–15. [Google Scholar] [CrossRef]
- Maggi, F.; Bosco, D.; Galetto, L.; Palmano, S.; Marzachì, C. Space-time point pattern analysis of flavescence dorée epidemic in a grapevine field: Disease progression and recovery. Front. Plant Sci. 2017, 7, 1987. [Google Scholar] [CrossRef] [Green Version]
- Galetto, L.; Miliordos, D.; Roggia, C.; Rashidi, M.; Sacco, D.; Marzachì, C.; Bosco, D. Acquisition capability of the grapevine Flavescence dorée by the leafhopper vector Scaphoideus titanus Ball correlates with phytoplasma titre in the source plant. J. Pest Sci. 2014, 87, 671–679. [Google Scholar] [CrossRef]
- Pelletier, C.; Salar, P.; Gillet, J.; Cloquemin, G.; Very, P.; Foissac, X.; Malembic-Maher, S. Triplex real-time PCR assay for sensitive and simultaneous detection of grapevine phytoplasmas of the 16SrV and 16SrXIIA groups with an endogenous analytical control. Vitis 2009, 48, 87–95. [Google Scholar] [CrossRef]
- Foissac, X.; INRAE, Univ. Bordeaux, UMR BFP, Villenave d’Ornon, France. Personal communication, 2013.
- Hammer, Ø.; Harper, D.A.T.; Ryan, P.D. PAST: Paleontological statistics software package for education and data analysis. Palaeontol. Electron. 2001, 4, 9. [Google Scholar]





| Locus | Chemistry | Name | Sequence (5′-3′) | Size (bp) | N° of SNP | TA (°C) | qPCR (MT-C; MT-D) | qPCR (R2; Efficiency) |
|---|---|---|---|---|---|---|---|---|
| nrdF | SYBR Green | nrdF_Real_F | AGATGACTTATTTCAATGGGGCG | 173 | 4 | 59 | 72.10 °C ± 0.14; | - |
| nrdF_Real_R | CAGAAGCGACCATTGCTTTCCA | 72.25 °C ± 0.07 | ||||||
| Taq Man | nrdF_F28 | TTTCACGGAGCTAATTGGAA | 94 | 2 | 60 | - | - | |
| nrdF_R121 | TGATATCTTCTGGACGCCA | |||||||
| nrdF -C | HEX-TGAAGATGAATATACCCAATATTTTTACGAACA-BHQ1 | - | - | - | - | 0.996; 99% | ||
| nrdF -D | FAM-TGAAGATGAATATACTCAATATTTTTATGAACA-BHQ1 | - | - | - | - | 0.999; 101% | ||
| malG | SYBR Green | malG_F | GCTTTCCGAGGCCAATTCCA | 267 | 6 | 59 | 75.70 °C ± 0.00; | - |
| malG_R | ATTCTGGCCAAGCATAAGCG | 76.05 °C ± 0.07 | ||||||
| contig 12 | SYBR Green | contig12Fw3 | AGTTTGATCCAGCTTGCGGA | 141 | 5 | 59 | 77.74 °C ± 0.05; | - |
| contig12Rv2 | CGGCAACTGTGTAAATCCGT | 78.08 °C ± 0.04 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rossi, M.; Vallino, M.; Galetto, L.; Marzachì, C. Competitive Exclusion of Flavescence dorée Phytoplasma Strains in Catharanthus roseus Plants. Plants 2020, 9, 1594. https://doi.org/10.3390/plants9111594
Rossi M, Vallino M, Galetto L, Marzachì C. Competitive Exclusion of Flavescence dorée Phytoplasma Strains in Catharanthus roseus Plants. Plants. 2020; 9(11):1594. https://doi.org/10.3390/plants9111594
Chicago/Turabian StyleRossi, Marika, Marta Vallino, Luciana Galetto, and Cristina Marzachì. 2020. "Competitive Exclusion of Flavescence dorée Phytoplasma Strains in Catharanthus roseus Plants" Plants 9, no. 11: 1594. https://doi.org/10.3390/plants9111594
APA StyleRossi, M., Vallino, M., Galetto, L., & Marzachì, C. (2020). Competitive Exclusion of Flavescence dorée Phytoplasma Strains in Catharanthus roseus Plants. Plants, 9(11), 1594. https://doi.org/10.3390/plants9111594