Sugar and Hormone Dynamics and the Expression Profiles of SUT/SUC and SWEET Sugar Transporters during Flower Development in Petunia axillaris
Abstract
:1. Introduction
2. Results
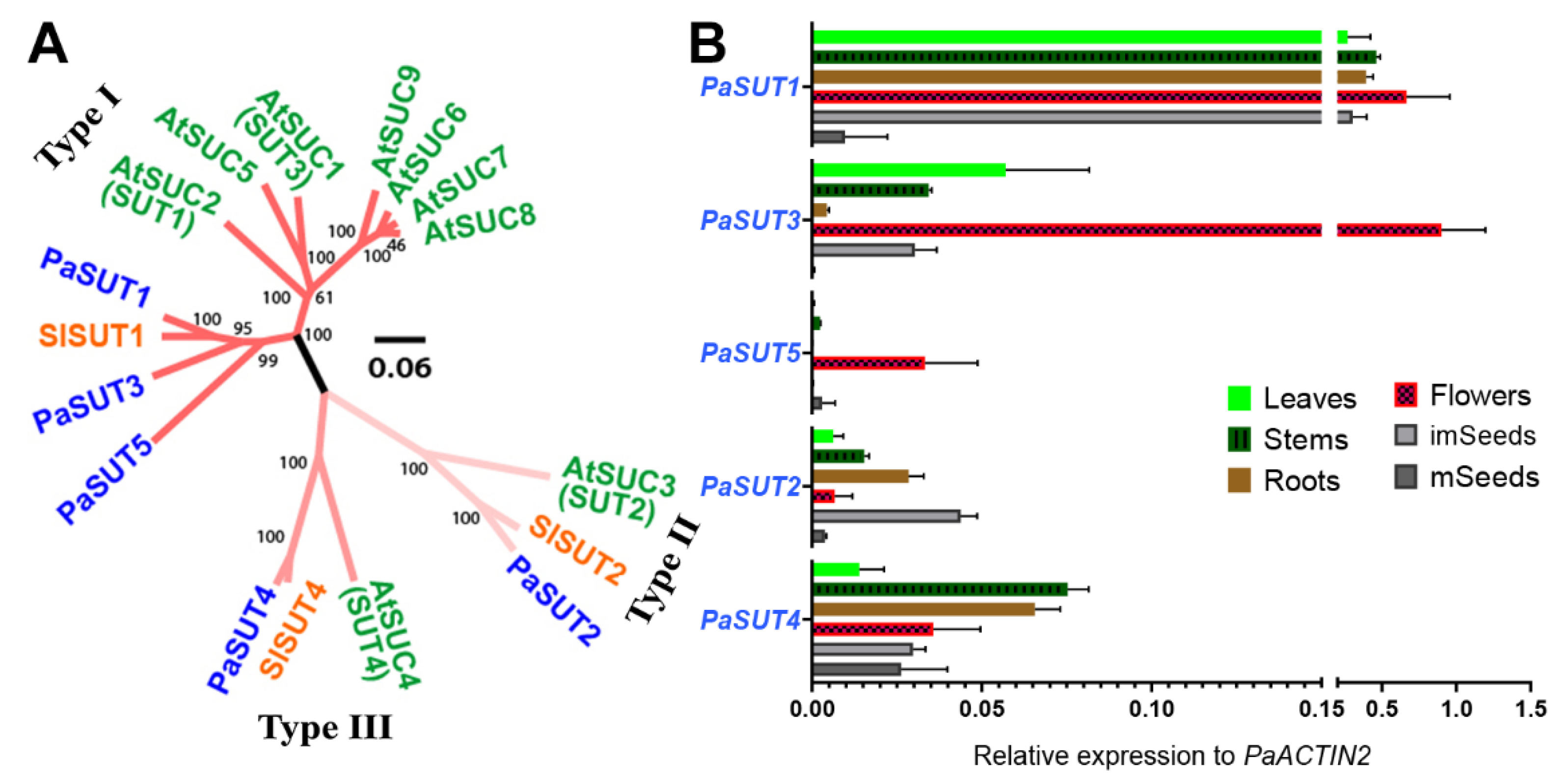
2.1. The Five-Gene Family of SUT/SUC Transporters in P. axillaris
2.2. The Genome of P. axillaris Encode 36 Genes for the SWEET Transporters with Disparate Tissue Expression Profile
2.3. Expression Profiles of PaSUTs and PaSWEETs during Flower Development
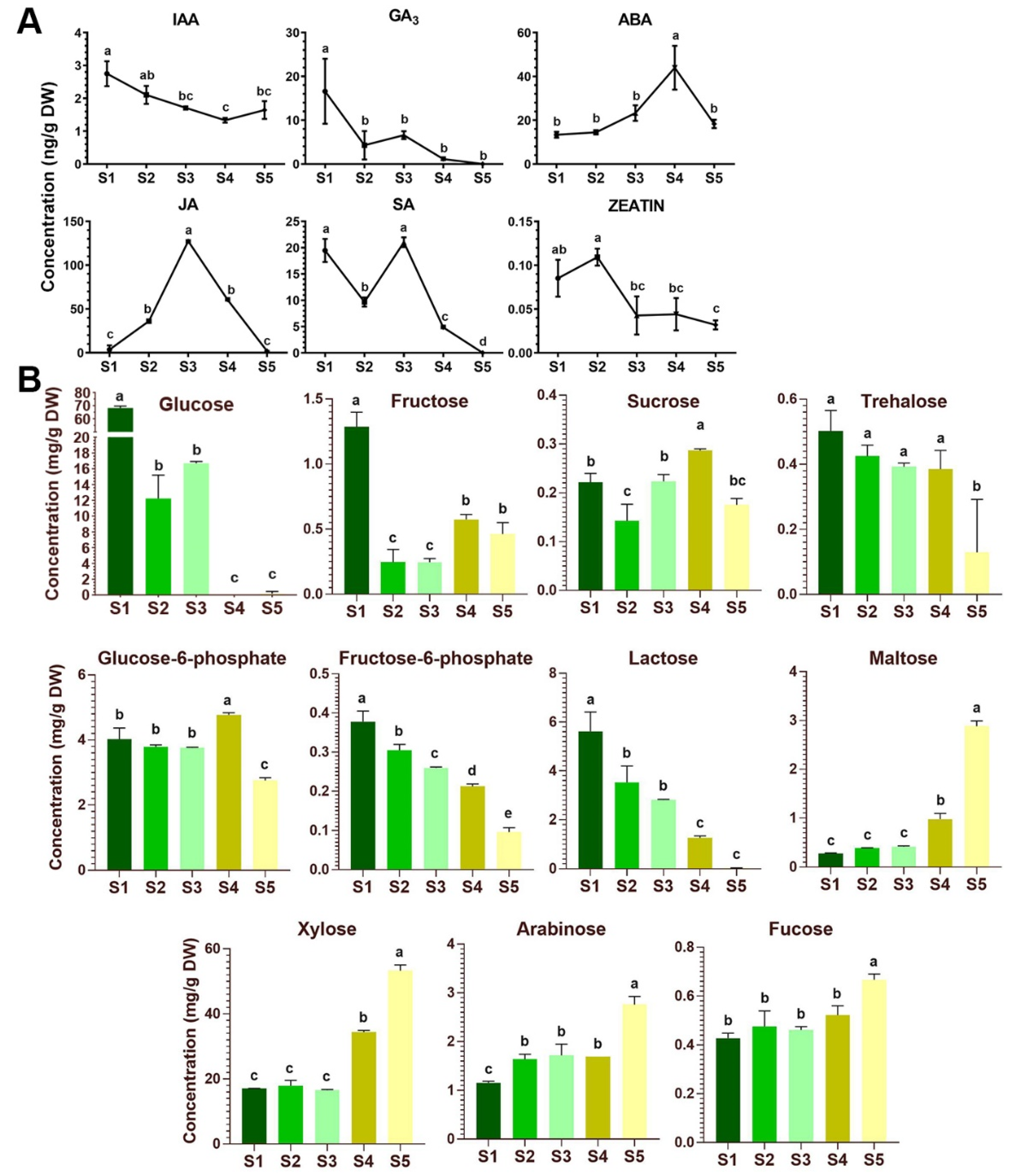
2.4. Phytohormone Dynamics and Sugar Contents during Flower Development
3. Discussion
4. Conclusions
5. Materials and Methods
5.1. Plant Materials, Growth Conditions and Sample Collections
5.2. Gene Identification, Sequence Analysis, Phylogenetics and Protein Subcellular Prediction
5.3. RNA Extraction and Quantitative Real-Time-PCR Analysis
5.4. Quantification of Phytohormones Using 6410 Triple Quad LCMS
5.5. Quantification of Sugars Using Gas Chromatography—Quadruple Time of Flight Mass Spectrometry (GC-Q ToF MS)
5.6. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Jian, H.; Lu, K.; Yang, B.; Wang, T.; Zhang, L.; Zhang, A.; Wang, J.; Liu, L.; Qu, C.; Li, J. Genome-Wide Analysis and Expression Profiling of the SUC and SWEET Gene Families of Sucrose Transporters in Oilseed Rape (Brassica napus L.). Front. Plant Sci. 2016, 7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kühn, C.; Grof, C.P.L. Sucrose transporters of higher plants. Curr. Opin. Plant Biol. 2010, 13, 287–297. [Google Scholar] [CrossRef] [PubMed]
- Peng, D.; Gu, X.; Xue, L.J.; Leebens-Mack, J.H.; Tsai, C.J. Bayesian phylogeny of sucrose transporters: Ancient origins, differential expansion and convergent evolution in monocots and dicots. Front. Plant Sci. 2014, 5, 615. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, L.Q.; Qu, X.Q.; Hou, B.H.; Sosso, D.; Osorio, S.; Fernie, A.R.; Frommer, W.B. Sucrose Efflux Mediated by SWEET Proteins as a Key Step for Phloem Transport. Science 2012, 335, 207–211. [Google Scholar] [CrossRef] [PubMed]
- Lemoine, R.; La Camera, S.; Atanassova, R.; Dédaldéchamp, F.; Allario, T.; Pourtau, N.; Bonnemain, J.L.; Laloi, M.; Coutos-Thévenot, P.; Maurousset, L.; et al. Source-to-sink transport of sugar and regulation by environmental factors. Front. Plant Sci. 2013, 4, 272. [Google Scholar] [CrossRef] [Green Version]
- Rennie, E.A.; Turgeon, R. A comprehensive picture of phloem loading strategies. Proc. Nat. Acad. Sci. USA 2009, 106, 14162–14167. [Google Scholar] [CrossRef] [Green Version]
- Eveland, A.L.; Jackson, D. Sugars, signalling, and plant development. J. Exp. Bot. 2012, 63, 3367–3377. [Google Scholar] [CrossRef] [Green Version]
- Matsoukas, I.G. Interplay between sugar and hormone signaling pathways modulate floral signal transduction. Front. Genet. 2014, 5, 218. [Google Scholar] [CrossRef] [Green Version]
- Julius, B.T.; Leach, K.A.; Tran, T.M.; Mertz, R.A.; Braun, D.M. Sugar Transporters in Plants: New Insights and Discoveries. Plant Cell Physiol. 2017, 58, 1442–1460. [Google Scholar] [CrossRef] [Green Version]
- Chen, L.Q.; Cheung, L.S.; Feng, L.; Tanner, W.; Frommer, W.B. Transport of Sugars. Annu. Rev. Biochem. 2015, 84, 865–894. [Google Scholar] [CrossRef] [PubMed]
- Büttner, M. The monosaccharide transporter(-like) gene family in Arabidopsis. FEBS Lett. 2007, 581, 2318–2324. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, L.Q. SWEET sugar transporters for phloem transport and pathogen nutrition. New Phytol. 2013, 201, 1150–1155. [Google Scholar] [CrossRef] [PubMed]
- Feng, L.; Frommer, W.B. Structure and function of SemiSWEET and SWEET sugar transporters. Trends Biochem. Sci. 2015, 40, 480–486. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reinders, A.; Sivitz, A.B.; Ward, J.M. Evolution of plant sucrose uptake transporters. Front. Plant Sci. 2012, 3, 22. [Google Scholar] [CrossRef] [Green Version]
- Gottwald, J.R.; Krysan, P.J.; Young, J.C.; Evert, R.F.; Sussman, M.R. Genetic evidence for the in planta role of phloem-specific plasma membrane sucrose transporters. Proc. Nat. Acad. Sci. USA 2000, 97, 13979–13984. [Google Scholar] [CrossRef] [Green Version]
- Riesmeier, J.; Willmitzer, L.; Frommer, W. Evidence for an essential role of the sucrose transporter in phloem loading and assimilate partitioning. EMBO J. 1994, 13, 1–7. [Google Scholar] [CrossRef]
- Sivitz, A.B.; Reinders, A.; Ward, J.M. Arabidopsis Sucrose Transporter AtSUC1 Is Important for Pollen Germination and Sucrose-Induced Anthocyanin Accumulation. Plant Physiol. 2008, 147, 92–100. [Google Scholar] [CrossRef]
- Slewinski, T.L.; Meeley, R.; Braun, D.M. Sucrose transporter1 functions in phloem loading in maize leaves. J. Exp. Bot. 2009, 60, 881–892. [Google Scholar] [CrossRef] [Green Version]
- Payyavula, R.S.; Tay, K.H.; Tsai, C.J.; Harding, S.A. The sucrose transporter family in Populus: The importance of a tonoplast PtaSUT4 to biomass and carbon partitioning. Plant J. 2011, 65, 757–770. [Google Scholar] [CrossRef]
- Schneider, S.; Hulpke, S.; Schulz, A.; Yaron, I.; Höll, J.; Imlau, A.; Schmitt, B.; Batz, S.; Wolf, S.; Hedrich, R.; et al. Vacuoles release sucrose via tonoplast-localised SUC4-type transporters. Plant Biol. 2011, 14, 325–336. [Google Scholar] [CrossRef]
- Barker, L.; Kühn, C.; Weise, A.; Schulz, A.; Gebhardt, C.; Hirner, B.; Hellmann, H.; Schulze, W.; Ward, J.M.; Frommer, W.B. SUT2, a Putative Sucrose Sensor in Sieve Elements. Plant Cell 2000, 12, 1153–1164. [Google Scholar] [CrossRef] [Green Version]
- Chincinska, I.; Gier, K.; Krügel, U.; Liesche, J.; He, H.; Grimm, B.; Harren, F.J.M.; Cristescu, S.M.; Kühn, C. Photoperiodic regulation of the sucrose transporter StSUT4 affects the expression of circadian-regulated genes and ethylene production. Front. Plant Sci. 2013, 4, 26. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chincinska, I.A.; Liesche, J.; Krügel, U.; Michalska, J.; Geigenberger, P.; Grimm, B.; Kühn, C. Sucrose Transporter StSUT4 from Potato Affects Flowering, Tuberization, and Shade Avoidance Response. Plant Physiol. 2007, 146, 515–528. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gamas, P. Use of a Subtractive Hybridization Approach to Identify NewMedicago truncatulaGenes Induced During Root Nodule Development. Mol. Plant-Microbe Interact. 1996, 9, 233–242. [Google Scholar] [CrossRef]
- Jaehme, M.; Guskov, A.; Slotboom, D.J. Crystal structure of the vitamin B3 transporter PnuC, a full-length SWEET homolog. Nat. Struct. Mol. Biol. 2014, 21, 1013–1015. [Google Scholar] [CrossRef] [PubMed]
- Tao, Y.; Cheung, L.S.; Li, S.; Eom, J.-S.; Chen, L.Q.; Xu, Y.; Perry, K.; Frommer, W.B.; Feng, L. Structure of a eukaryotic SWEET transporter in a homotrimeric complex. Nat. Cell Biol. 2015, 527, 259–263. [Google Scholar] [CrossRef] [Green Version]
- Chen, L.Q.; Hou, B.-H.; LaLonde, S.; Takanaga, H.; Hartung, M.L.; Qu, X.-Q.; Guo, W.-J.; Kim, J.-G.; Underwood, W.; Chaudhuri, B.; et al. Sugar transporters for intercellular exchange and nutrition of pathogens. Nat. Cell Biol. 2010, 468, 527–532. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, L.Q.; Lin, I.W.; Qu, X.Q.; Sosso, D.; McFarlane, H.E.; Londoño, A.; Samuels, A.L.; Frommer, W.B. A Cascade of Sequentially Expressed Sucrose Transporters in the Seed Coat and Endosperm Provides Nutrition for the Arabidopsis Embryo. Plant Cell 2015, 27, 607–619. [Google Scholar] [CrossRef] [Green Version]
- Jeena, G.S.; Kumar, S.; Shukla, R.K. Structure, evolution and diverse physiological roles of SWEET sugar transporters in plants. Plant Mol. Biol. 2019, 100, 351–365. [Google Scholar] [CrossRef]
- Yuan, M.; Wang, S. Rice MtN3/Saliva/SWEET Family Genes and Their Homologs in Cellular Organisms. Mol. Plant 2013, 6, 665–674. [Google Scholar] [CrossRef] [Green Version]
- Gautam, T.; Saripalli, G.; Gahlaut, V.; Kumar, A.; Sharma, P.K.; Balyan, H.S.; Gupta, P.K. Further studies on sugar transporter (SWEET) genes in wheat (Triticum aestivum L.). Mol. Biol. Rep. 2019, 46, 2327–2353. [Google Scholar] [CrossRef] [PubMed]
- Patil, G.B.; Valliyodan, B.; Deshmukh, R.; Prince, S.; Nicander, B.; Zhao, M.-Z.; Sonah, H.; Song, L.; Lin, L.; Chaudhary, J.; et al. Soybean (Glycine max) SWEET gene family: Insights through comparative genomics, transcriptome profiling and whole genome re-sequence analysis. BMC Genom. 2015, 16, 520. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mizuno, H.; Kasuga, S.; Kawahigashi, H. The sorghum SWEET gene family: Stem sucrose accumulation as revealed through transcriptome profiling. Biotechnol. Biofuels 2016, 9, 1–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chong, J.; Piron, M.-C.; Meyer, S.; Merdinoglu, D.; Bertsch, C.; Mestre, P. The SWEET family of sugar transporters in grapevine: VvSWEET4 is involved in the interaction with Botrytis cinerea. J. Exp. Bot. 2014, 65, 6589–6601. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wei, X.; Liu, F.; Chen, C.; Ma, F.; Li, M. The Malus domestica sugar transporter gene family: Identifications based on genome and expression profiling related to the accumulation of fruit sugars. Front. Plant Sci. 2014, 5, 569. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miao, H.; Sun, P.; Liu, Q.; Miao, Y.; Liu, J.; Zhang, K.; Hu, W.; Zhang, J.; Wang, J.; Wang, Z.; et al. Genome-wide analyses of SWEET family proteins reveal involvement in fruit development and abiotic/biotic stress responses in banana. Sci. Rep. 2017, 7, 1–15. [Google Scholar] [CrossRef] [Green Version]
- Zhang, W.; Wang, S.; Yu, F.; Tang, J.; Shan, X.; Bao, K.; Yu, L.; Wang, H.; Fei, Z.; Li, J. Genome-wide characterization and expression profiling of SWEET genes in cabbage (Brassica oleracea var. capitata L.) reveal their roles in chilling and clubroot disease responses. BMC Genom. 2019, 20, 93. [Google Scholar] [CrossRef]
- Feng, C.Y.; Han, J.X.; Han, X.X.; Jiang, J. Genome-wide identification, phylogeny, and expression analysis of the SWEET gene family in tomato. Gene 2015, 573, 261–272. [Google Scholar] [CrossRef]
- Li, M.; Xie, H.; He, M.; Su, W.; Yang, Y.; Wang, J.; Ye, G.; Zhou, Y. Genome-wide identification and expression analysis of the StSWEET family genes in potato (Solanum tuberosum L.). Genes Genom. 2019, 42, 135–153. [Google Scholar] [CrossRef]
- Eom, J.S.; Chen, L.Q.; Sosso, D.; Julius, B.T.; Lin, I.W.; Qu, X.Q.; Braun, D.M.; Frommer, W.B. SWEETs, transporters for intracellular and intercellular sugar translocation. Curr. Opin. Plant Biol. 2015, 25, 53–62. [Google Scholar] [CrossRef] [Green Version]
- Quirino, B.F.; Normanly, J.; Amasino, R.M. Diverse range of gene activity during Arabidopsis thaliana leaf senescence includes pathogen-independent induction of defense-related genes. Plant Mol. Biol. 1999, 40, 267–278. [Google Scholar] [CrossRef] [PubMed]
- Ge, Y.X.; Angenent, G.C.; Wittich, P.E.; Peters, J.; Franken, J.; Busscher, M.; Zhang, L.M.; Dahlhaus, E.; Kater, M.M.; Wullems, G.J.; et al. NEC1, a novel gene, highly expressed in nectary tissue of Petunia hybrida. Plant J. 2000, 24, 725–734. [Google Scholar] [CrossRef] [PubMed]
- Lin, I.W.; Sosso, D.; Chen, L.Q.; Gase, K.; Kim, S.G.; Kessler, D.; Klinkenberg, P.M.; Gorder, M.K.; Hou, B.H.; Qu, X.Q.; et al. Nectar secretion requires sucrose phosphate synthases and the sugar transporter SWEET9. Nat. Cell Biol. 2014, 508, 546–549. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.Y.; Huh, J.H.; Yu, Y.C.; Ho, L.H.; Chen, L.Q.; Tholl, D.; Frommer, W.B.; Guo, W.-J. The Arabidopsis vacuolar sugar transporter SWEET2 limits carbon sequestration from roots and restricts Pythium infection. Plant J. Cell Mol. Biol. 2015, 83, 1046–1058. [Google Scholar] [CrossRef] [Green Version]
- Cohn, M.; Bart, R.S.; Shybut, M.; Dahlbeck, D.; Gomez, M.; Morbitzer, R.; Hou, B.H.; Frommer, W.B.; Lahaye, T.; Staskawicz, B.J. Xanthomonas axonopodis virulence is promoted by a transcription activator like (TAL) effector mediated induction of a SWEET sugar transporter in cassava 3. Mol. Plant-Microbe Interact. MPMI 2014, 1186–1198. [Google Scholar] [CrossRef] [Green Version]
- Le Hir, R.; Spinner, L.; Klemens, P.A.; Chakraborti, D.; De Marco, F.; Vilaine, F.; Wolff, N.; Lemoine, R.; Porcheron, B.; Géry, C.; et al. Disruption of the Sugar Transporters AtSWEET11 and AtSWEET12 Affects Vascular Development and Freezing Tolerance in Arabidopsis. Mol. Plant 2015, 8, 1687–1690. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Y.; Wang, Y.; Zhang, H.; Zhang, Q.; Zhai, H.; Liu, Q.; He, S. The Plasma Membrane-Localized Sucrose Transporter IbSWEET10 Contributes to the Resistance of Sweet Potato to Fusarium oxysporum. Front. Plant Sci. 2017, 8, 197. [Google Scholar] [CrossRef] [Green Version]
- Zhao, D.; You, Y.; Fan, H.; Zhu, X.; Wang, Y.; Duan, Y.; Xuan, Y.H.; Chen, L. The Role of Sugar Transporter Genes during Early Infection by Root-Knot Nematodes. Int. J. Mol. Sci. 2018, 19, 302. [Google Scholar] [CrossRef] [Green Version]
- Yuan, K. Phytohormone signalling pathways interact with sugars during seed germination and seedling development. J. Exp. Bot. 2006, 57, 3359–3367. [Google Scholar] [CrossRef]
- Guan, Y.F.; Huang, X.Y.; Zhu, J.; Gao, J.F.; Zhang, H.X.; Yang, Z.N. RUPTURED POLLEN GRAIN1, a Member of the MtN3/saliva Gene Family, Is Crucial for Exine Pattern Formation and Cell Integrity of Microspores in Arabidopsis. Plant Physiol. 2008, 147, 852–863. [Google Scholar] [CrossRef] [Green Version]
- Sosso, D.; Luo, D.; Li, Q.-B.; Sasse, J.; Yang, J.; Gendrot, G.; Suzuki, M.E.; Koch, K.; Mccarty, D.R.; Chourey, P.S.; et al. Seed filling in domesticated maize and rice depends on SWEET-mediated hexose transport. Nat. Genet. 2015, 47, 1489–1493. [Google Scholar] [CrossRef] [PubMed]
- Sun, M.X.; Huang, X.Y.; Yang, J.; Guan, Y.F.; Yang, Z.N. Arabidopsis RPG1 is important for primexine deposition and functions redundantly with RPG2 for plant fertility at the late reproductive stage. Plant Reprod. 2013, 26, 83–91. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Luo, D.; Yang, B.; Frommer, W.B.; Eom, J.-S. SWEET11 and 15 as key players in seed filling in rice. New Phytol. 2018, 218, 604–615. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Z.; Zou, L.; Ren, C.; Ren, F.; Wang, Y.; Fan, P.; Li, S.; Liang, Z. VvSWEET10 Mediates Sugar Accumulation in Grapes. Genes 2019, 10, 255. [Google Scholar] [CrossRef] [Green Version]
- Durand, M.; Mainson, D.; Porcheron, B.; Maurousset, L.; Lemoine, R.; Pourtau, N. Carbon source–sink relationship in Arabidopsis thaliana: The role of sucrose transporters. Planta 2018, 247, 587–611. [Google Scholar] [CrossRef] [Green Version]
- Borghi, M.; Fernie, A.R. Floral Metabolism of Sugars and Amino Acids: Implications for Pollinators’ Preferences and Seed and Fruit Set. Plant Physiol. 2017, 175, 1510–1524. [Google Scholar] [CrossRef] [Green Version]
- Shan, H.; Cheng, J.; Zhang, R.; Yao, X.; Kong, H. Developmental mechanisms involved in the diversification of flowers. Nat. Plants 2019, 5, 917–923. [Google Scholar] [CrossRef]
- Vandenbussche, M.; Echambrier, P.; Bento, S.E.; Emorel, P. Petunia, Your Next Supermodel? Front. Plant Sci. 2016, 7, 72. [Google Scholar] [CrossRef]
- Bombarely, A.; Moser, M.; Amrad, A.; Bao, M.; Bapaume, L.; Barry, C.S.; Bliek, M.; Boersma, M.R.; Borghi, L.; Bruggmann, R.; et al. Insight into the evolution of the Solanaceae from the parental genomes of Petunia hybrida. Nat. Plants 2016, 2, 16074. [Google Scholar] [CrossRef] [Green Version]
- Solovyev, V.; Kosarev, P.; Seledsov, I.; Vorobyev, D. Automatic annotation of eukaryotic genes, pseudogenes and promoters. Genome Biol. 2006, 7 (Suppl. 1), 10.1–10.12. Available online: http://www.softberry.com/ (accessed on 1 July 2019). [CrossRef] [Green Version]
- Colquhoun, T.A.; Verdonk, J.C.; Schimmel, B.C.J.; Tieman, D.M.; Underwood, B.A.; Clark, D.G. Petunia floral volatile benzenoid/phenylpropanoid genes are regulated in a similar manner. Phytochemistry 2010, 71, 158–167. [Google Scholar] [CrossRef] [PubMed]
- Weiss, D.; Schönfeld, M.; Halevy, A.H. Photosynthetic Activities in the Petunia Corolla. Plant Physiol. 1988, 87, 666–670. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alvarez-Buylla, E.R.P.; Benítez, M.; Corvera-Poiré, A.; Cador, A.C.; De Folter, S.; De Buen, A.G.; Garay-Arroyo, A.; García-Ponce, B.; Jaimes-Miranda, F.; Pérez-Ruiz, R.V.; et al. Flower Development. Arab. Book 2010, 8, e0127. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, L.; Sui, X.; Lucas, W.J.; Li, Y.; Feng, S.; Ma, S.; Fan, J.; Gao, L.; Zhang, Z.-X. Down-regulation of the Sucrose Transporter CsSUT1 Causes Male Sterility by Altering Carbohydrate Supply. Plant Physiol. 2019, 180, 986–997. [Google Scholar] [CrossRef] [Green Version]
- Hackel, A.; Schauer, N.; Carrari, F.; Fernie, A.R.; Grimm, B.; Kühn, C. Sucrose transporter LeSUT1 and LeSUT2 inhibition affects tomato fruit development in different ways. Plant J. 2006, 45, 180–192. [Google Scholar] [CrossRef]
- Cheng, H. Gibberellin regulates Arabidopsis floral development via suppression of DELLA protein function. Development 2004, 131, 1055–1064. [Google Scholar] [CrossRef] [Green Version]
- Ravid, J.; Spitzer-Rimon, B.; Takebayashi, Y.; Seo, M.; Cna’Ani, A.; Aravena-Calvo, J.; Masci, T.; Farhi, M.; Vainstein, A. GA as a regulatory link between the showy floral traits color and scent. New Phytol. 2017, 215, 411–422. [Google Scholar] [CrossRef] [Green Version]
- Ishiguro, S.; Kawai-Oda, A.; Ueda, J.; Nishida, I.; Okada, K. The DEFECTIVE IN ANTHER DEHISCENCE1 Gene Encodes a Novel Phospholipase A1 Catalyzing the Initial Step of Jasmonic Acid Biosynthesis, Which Synchronizes Pollen Maturation, Anther Dehiscence, and Flower Opening in Arabidopsis. Plant Cell 2001, 13, 2191–2209. [Google Scholar] [CrossRef] [Green Version]
- Kumar, M.; Singh, V.P.; Arora, A.; Singh, N. The role of abscisic acid (ABA) in ethylene insensitive Gladiolus (Gladiolus grandiflora Hort.) flower senescence. Acta Physiol. Plant 2013, 36, 151–159. [Google Scholar] [CrossRef]
- Sekhar, K.C.; Sawhney, V. Role of ABA in stamen and pistil development in the normal and solanifolia mutant of tomato (Lycopersicon esculentum). Sex. Plant Reprod. 1991, 4, 279–283. [Google Scholar] [CrossRef]
- Browse, J. Jasmonate Passes Muster: A Receptor and Targets for the Defense Hormone. Annu. Rev. Plant Biol. 2009, 60, 183–205. [Google Scholar] [CrossRef] [PubMed]
- Smeekens, S.; Ma, J.; Hanson, J.; Rolland, F. Sugar signals and molecular networks controlling plant growth. Curr. Opin. Plant Biol. 2010, 13, 273–278. [Google Scholar] [CrossRef] [PubMed]
- Zeeman, S.C.; Kossmann, J.; Smith, A.M. Starch: Its Metabolism, Evolution, and Biotechnological Modification in Plants. Annu. Rev. Plant Biol. 2010, 61, 209–234. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ferrario, S.; Shchennikova, A.V.; Franken, J.; Immink, R.G.H.; Angenent, G.C. Control of Floral Meristem Determinacy in Petunia by MADS-Box Transcription Factors. Plant Physiol. 2006, 140, 890–898. [Google Scholar] [CrossRef] [Green Version]
- Preston, J.C.; Powers, B.; Kostyun, J.L.; Driscoll, H.; Zhang, F.; Zhong, J. Implications of region-specific gene expression for development of the partially fused petunia corolla. Plant J. 2019, 100, 158–175. [Google Scholar] [CrossRef]
- Ylstra, B.; Garrido, D.; Busscher, J.; Van Tunen, A.J. Hexose Transport in Growing Petunia Pollen Tubes and Characterization of a Pollen-Specific, Putative Monosaccharide Transporter. Plant Physiol. 1998, 118, 297–304. [Google Scholar] [CrossRef] [Green Version]
- Ge, Y.X.; Angenent, G.; Dahlhaus, E.; Franken, J.; Peters, J.; Wullens, G.; Creemers-Molenaar, J. Partial silencing of the NEC1 gene results in early opening of anthers in Petunia hybrida. Mol. Genet. Genom. 2001, 265, 414–423. [Google Scholar] [CrossRef]
- Amano, I.; Kitajima, S.; Suzuki, H.; Koeduka, T.; Shitan, N. Transcriptome analysis of Petunia axillaris flowers reveals genes involved in morphological differentiation and metabolite transport. PLoS ONE 2018, 13, e0198936. [Google Scholar] [CrossRef]
- Fernandez-Pozo, N.; Menda, N.; Edwards, J.D.; Saha, S.; Tecle, I.Y.; Strickler, S.R.; Bombarely, A.; Fisher-York, T.; Pujar, A.; Foerster, H.; et al. The Sol Genomics Network (SGN)—From genotype to phenotype to breeding. Nucleic Acids Res. 2015, 43, D1036–D1041. Available online: https://solgenomics.net/ (accessed on 1 October 2018). [CrossRef]
- Goodstein, D.M.; Shu, S.; Howson, R.; Neupane, R.; Hayes, R.D.; Fazo, J.; Mitros, T.; Dirks, W.; Hellsten, U.; Putnam, N.; et al. Phytozome: A comparative platform for green plant genomics. Nucleic Acids Res. 2012, 40, D1178–D1186. Available online: https://phytozome.jgi.doe.gov (accessed on 1 October 2018). [CrossRef]
- Hu, B.; Jin, J.; Guo, A.Y.; Zhang, H.; Luo, J.; Gao, G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 2015, 31, 1296–1297. Available online: http://gsds.cbi.pku.edu.cn (accessed on 1 January 2019). [CrossRef] [Green Version]
- Bailey, T.L.; Elkan, C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol. 1994, 2, 28–36. Available online: http://meme-suite.org/ (accessed on 1 January 2019). [PubMed]
- Chao, J.T.; Kong, Y.Z.; Wang, Q.; Sun, Y.H.; Gong, D.P.; Lv, J.; Liu, G.S. MapGene2Chrom, a tool to draw gene physical map based on Perl and SVG languages. Yi Chuan 2015, 37, 91–97. Available online: http://mg2c.iask.in/mg2c_v2.1/ (accessed on 1 January 2020). [CrossRef]
- Madeira, F.; Park, Y.M.; Lee, J.; Buso, N.; Gur, T.; Madhusoodanan, N.; Basutkar, P.; Tivey, A.R.N.; Potter, S.C.; Finn, R.D.; et al. The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 2019, 47, W636–W641. Available online: https://www.ebi.ac.uk/Tools/msa/muscle (accessed on 1 January 2019). [CrossRef] [PubMed] [Green Version]
- Artimo, P.; Jonnalagedda, M.; Arnold, K.; Baratin, D.; Csardi, G.; de Castro, E.; Duvaud, S.; Flegel, V.; Fortier, A.; Gasteiger, E.; et al. ExPASy: SIB bioinformatics resource portal. Nucleic Acids Res. 2012, 40, W597–W603. Available online: https://www.expasy.org/ (accessed on 1 January 2019). [CrossRef]
- Yu, C.S.; Cheng, C.W.; Su, W.C.; Chang, K.C.; Huang, S.W.; Hwang, J.K.; Lu, C.H. CELLO2GO: A Web Server for Protein subCELlular LOcalization Prediction with Functional Gene Ontology Annotation. PLoS ONE 2014, 9, e99368. [Google Scholar] [CrossRef] [Green Version]
- Savojardo, C.; Martelli, P.L.; Fariselli, P.; Profiti, G.; Casadio, R. BUSCA: An integrative web server to predict subcellular localization of proteins. Nucleic Acids Res. 2018, 46, W459–W466. [Google Scholar] [CrossRef]
- Horton, P.; Park, K.J.; Obayashi, T.; Fujita, N.; Harada, H.; Adams-Collier, C.; Nakai, K. WoLF PSORT: Protein localization predictor. Nucleic Acids Res. 2007, 35, W585–W587. [Google Scholar] [CrossRef] [Green Version]
- Untergasser, A.; Nijveen, H.; Rao, X.; Bisseling, T.; Geurts, R.; Leunissen, J.A.M. Primer3Plus, an enhanced web interface to Primer3. Nucleic Acids Res. 2007, 35, W71–W74. [Google Scholar] [CrossRef] [Green Version]



| Gene Mane | Locus ID | Gene Location | Gene Length (bp) | CDS Length (bp) | Peptide Residues | pI | |
|---|---|---|---|---|---|---|---|
| SUT/SUC Family | |||||||
| 1 | PaSUT1 | Peaxi162Scf00495g00036.1 | 354,487–359,931 | 5444 | 1548 | 516 | 5.0 |
| 2 | PaSUT2 | Peaxi162Scf00503g00419.1 | 445,731–452,274 | 6543 | 1680 | 560 | 5.0 |
| 3 | PaSUT3 | Peaxi162Scf00481g00028.1 | 281,749–287,120 | 5371 | 1521 | 507 | 5.0 |
| 4 | PaSUT4 | Peaxi162Scf00626g00416.1 | 429,871–441,778 | 11907 | 1524 | 508 | 5.0 |
| 5 | PaSUT5 | Peaxi162Scf00268g00137.1 | 1,330,847–1,335,013 | 4166 | 1575 | 525 | 5.0 |
| SWEET Family | |||||||
| 1 | PaSWEET1a | Peaxi162Scf00263g00161.1 | 1,672,743–1,674,573 | 1830 | 696 | 232 | 5.2 |
| 2 | PaSWEET1b | Peaxi162Scf00033g00610.1 | 626,372–627,611 | 1239 | 699 | 233 | 5.2 |
| 3 | PaSWEET1c | Peaxi162Scf00051g00211.1 | 208,784–211,388 | 2604 | 774 | 258 | 5.1 |
| 4 | PaSWEET1d | Peaxi162Scf00263g00156.1 | 1,501,206–1,504,332 | 3126 | 927 | 309 | 5.1 |
| 5 | PaSWEET1e | Peaxi162Scf00263g01423.1 | 1,475,112–1,477,300 | 2188 | 891 | 297 | 5.1 |
| 6 | PaSWEET2a | Peaxi162Scf00111g01241.1 | 1,263,409–1,265,523 | 2114 | 1194 | 398 | 5.1 |
| 7 | PaSWEET2b | Peaxi162Scf00899g00310.1 | 296,611–329,600 | 32,989 | 705 | 235 | 5.2 |
| 8 | PaSWEET2c | Peaxi162Scf01337g00018.1 | 11,448–14,668 | 3220 | 705 | 235 | 5.2 |
| 9 | PaSWEET3 | Peaxi162Scf00904g00131.1 | 169,563–171,965 | 2402 | 711 | 237 | 5.2 |
| 10 | PaSWEET4 * | Peaxi162Scf00102g01855.1 | 1,853,060–1,856,259 | 2021 | 750 | 249 | 5.2 |
| 11 | PaSWEET5a | Peaxi162Scf00689g00339.1 | 307,462–314,776 | 7314 | 1167 | 389 | 5.0 |
| 12 | PaSWEET5b | Peaxi162Scf00017g02829.1 | 2,794,729–2,795,924 | 1195 | 714 | 238 | 5.1 |
| 13 | PaSWEET6 | Peaxi162Scf00129g01027.1 | 1,045,179–1,046,437 | 1258 | 651 | 217 | 5.2 |
| 14 | PaSWEET7 | Peaxi162Scf01051g00019.1 | 196,117–196,821 | 704 | 705 | 235 | 5.1 |
| 15 | PaSWEET8 | Peaxi162Scf00074g00434.1 | 462,439–465,223 | 2784 | 612 | 204 | 5.2 |
| 16 | PaSWEET9a | Peaxi162Scf00386g00924.1 | 926,235–928,356 | 2121 | 792 | 264 | 5.2 |
| 17 | PaSWEET9b | Peaxi162Scf00028g00063.1 | 675,430–685,040 | 9610 | 685 | 228 | 5.2 |
| 18 | PaSWEET9c(NEC1) | Peaxi162Scf00303g00004.1 | 55,308–57,418 | 2110 | 843 | 281 | 5.2 |
| 19 | PaSWEET10a | Peaxi162Scf01039g00027.1 | 73,727–75,238 | 1511 | 852 | 284 | 5.1 |
| 20 | PaSWEET10b | Peaxi162Scf00067g00107.1 | 995,051–996,843 | 1792 | 816 | 272 | 5.1 |
| 21 | PaSWEET11a * | Peaxi162Scf00516g00125.1_126 | 165,635–167,699 | 1948 | 879 | 292 | 5.2 |
| 22 | PaSWEET11b | Peaxi162Scf00909g00223.1 | 308,317–310,150 | 1833 | 825 | 275 | 5.1 |
| 23 | PaSWEET12a | Peaxi162Scf00516g00033.1 | 55,540–65,536 | 9996 | 1599 | 533 | 5.0 |
| 24 | PaSWEET12b | Peaxi162Scf01064g00253.1 | 289,074–307,657 | 18,583 | 2304 | 768 | 4.9 |
| 25 | PaSWEET13a | Peaxi162Scf00502g00058.1 | 562,962–564,843 | 1881 | 750 | 250 | 5.2 |
| 26 | PaSWEET13b | Peaxi162Scf00358g01310.1 | 1,321,846–1,324,432 | 2586 | 867 | 289 | 5.1 |
| 27 | PaSWEET13c | Peaxi162Scf00516g00122.1 | 104,493–106,655 | 2162 | 852 | 284 | 5.1 |
| 28 | PaSWEET14a | Peaxi162Scf00516g00124.1 | 133,860–155,056 | 21,196 | 2355 | 785 | 4.9 |
| 29 | PaSWEET14b | Peaxi162Scf00647g00518.1 | 595,043–596,818 | 1775 | 984 | 328 | 5.1 |
| 30 | PaSWEET14c | Peaxi162Scf00516g00025.1 | 78,107–79,474 | 1367 | 717 | 239 | 5.2 |
| 31 | PaSWEET14d | Peaxi162Scf00516g00012.1 | 119,256–121,066 | 1810 | 690 | 230 | 5.2 |
| 32 | PaSWEET15a | Peaxi162Scf00128g01632.1 | 1,656,934–1,660,444 | 3510 | 966 | 322 | 5.1 |
| 33 | PaSWEET15b | Peaxi162Scf00255g00006.1 | 30,836–33,121 | 2285 | 816 | 272 | 5.1 |
| 34 | PaSWEET16a | Peaxi162Scf00471g01110.1 | 1,106,818–1,108,891 | 2073 | 618 | 206 | 5.2 |
| 35 | PaSWEET16b | Peaxi162Scf00037g01416.1 | 1,392,651–1,396,158 | 3507 | 723 | 241 | 5.2 |
| 36 | PaSWEET17 | Peaxi162Scf00037g01417.1 | 1,401,046–1,404,180 | 3134 | 894 | 298 | 5.1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Iftikhar, J.; Lyu, M.; Liu, Z.; Mehmood, N.; Munir, N.; Ahmed, M.A.A.; Batool, W.; Aslam, M.M.; Yuan, Y.; Wu, B. Sugar and Hormone Dynamics and the Expression Profiles of SUT/SUC and SWEET Sugar Transporters during Flower Development in Petunia axillaris. Plants 2020, 9, 1770. https://doi.org/10.3390/plants9121770
Iftikhar J, Lyu M, Liu Z, Mehmood N, Munir N, Ahmed MAA, Batool W, Aslam MM, Yuan Y, Wu B. Sugar and Hormone Dynamics and the Expression Profiles of SUT/SUC and SWEET Sugar Transporters during Flower Development in Petunia axillaris. Plants. 2020; 9(12):1770. https://doi.org/10.3390/plants9121770
Chicago/Turabian StyleIftikhar, Junaid, Meiling Lyu, Zhuoyi Liu, Nasir Mehmood, Nigarish Munir, Mohamed A. A. Ahmed, Wajjiha Batool, Mehtab Muhammad Aslam, Yuan Yuan, and Binghua Wu. 2020. "Sugar and Hormone Dynamics and the Expression Profiles of SUT/SUC and SWEET Sugar Transporters during Flower Development in Petunia axillaris" Plants 9, no. 12: 1770. https://doi.org/10.3390/plants9121770
APA StyleIftikhar, J., Lyu, M., Liu, Z., Mehmood, N., Munir, N., Ahmed, M. A. A., Batool, W., Aslam, M. M., Yuan, Y., & Wu, B. (2020). Sugar and Hormone Dynamics and the Expression Profiles of SUT/SUC and SWEET Sugar Transporters during Flower Development in Petunia axillaris. Plants, 9(12), 1770. https://doi.org/10.3390/plants9121770






