Physiological and Anatomical Differences and Differentially Expressed Genes Reveal Yellow Leaf Coloration in Shumard Oak
Abstract
1. Introduction
2. Results
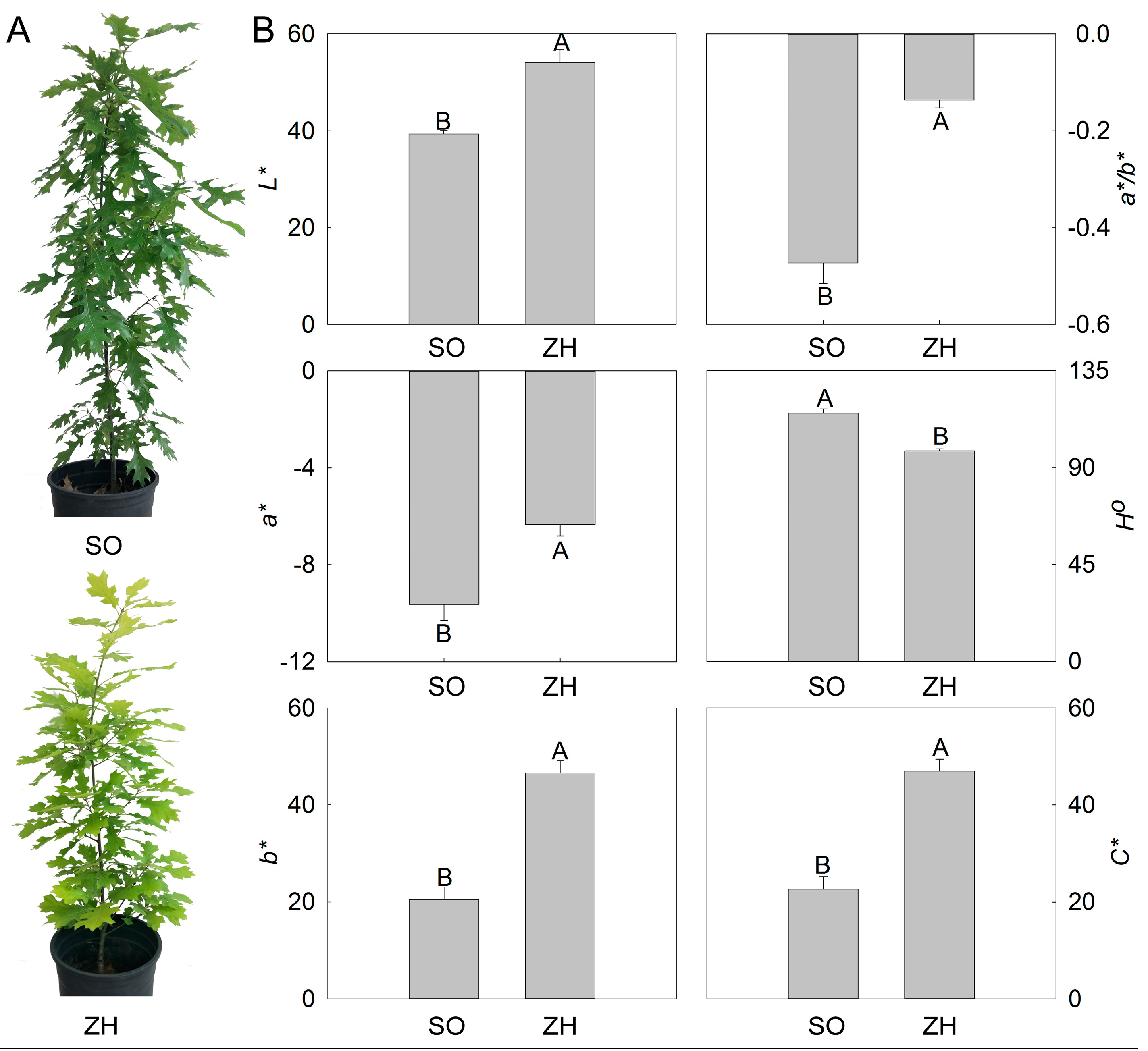
2.1. Color Indices
2.2. SPAD Value and Pigment Content
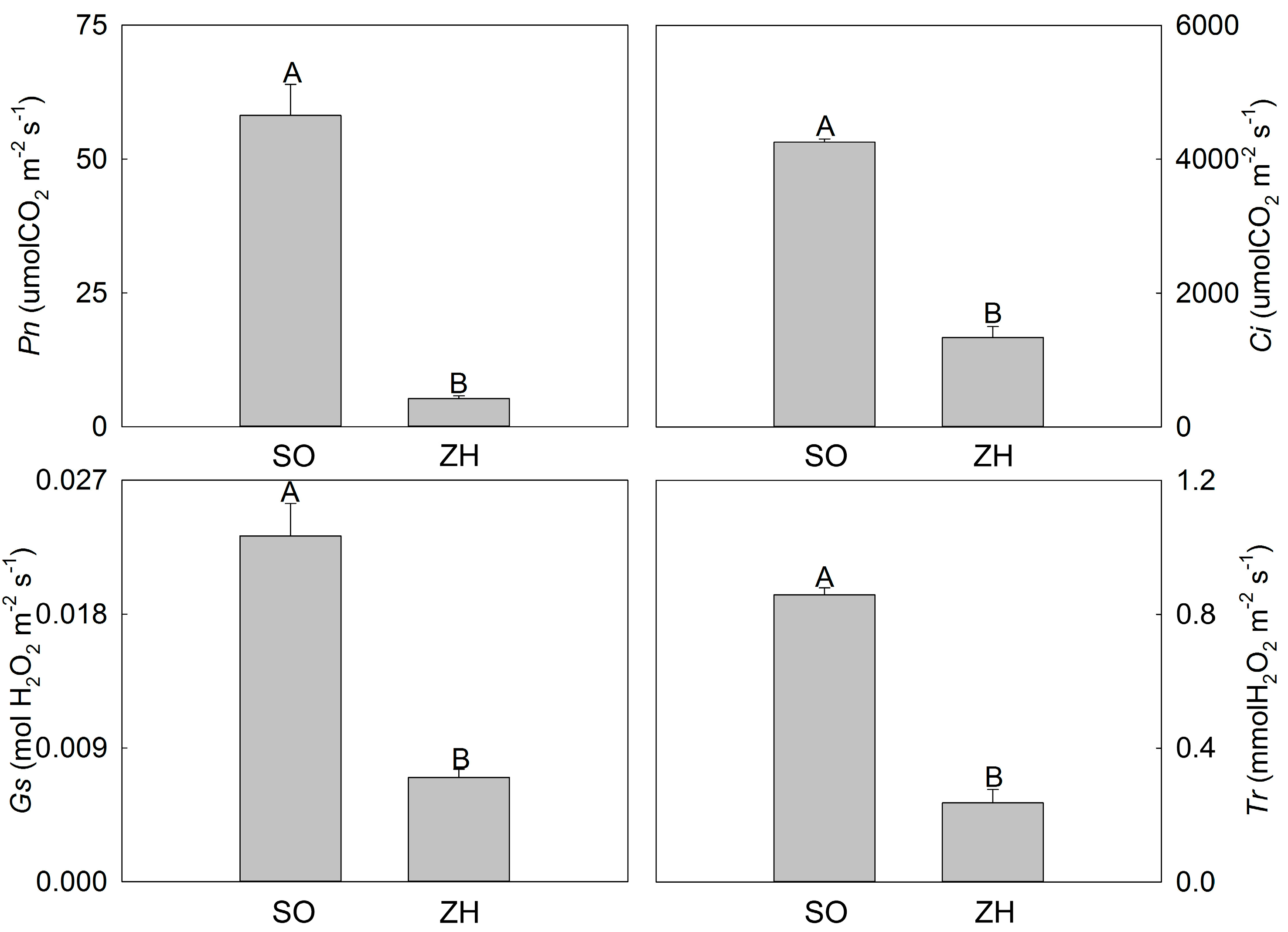
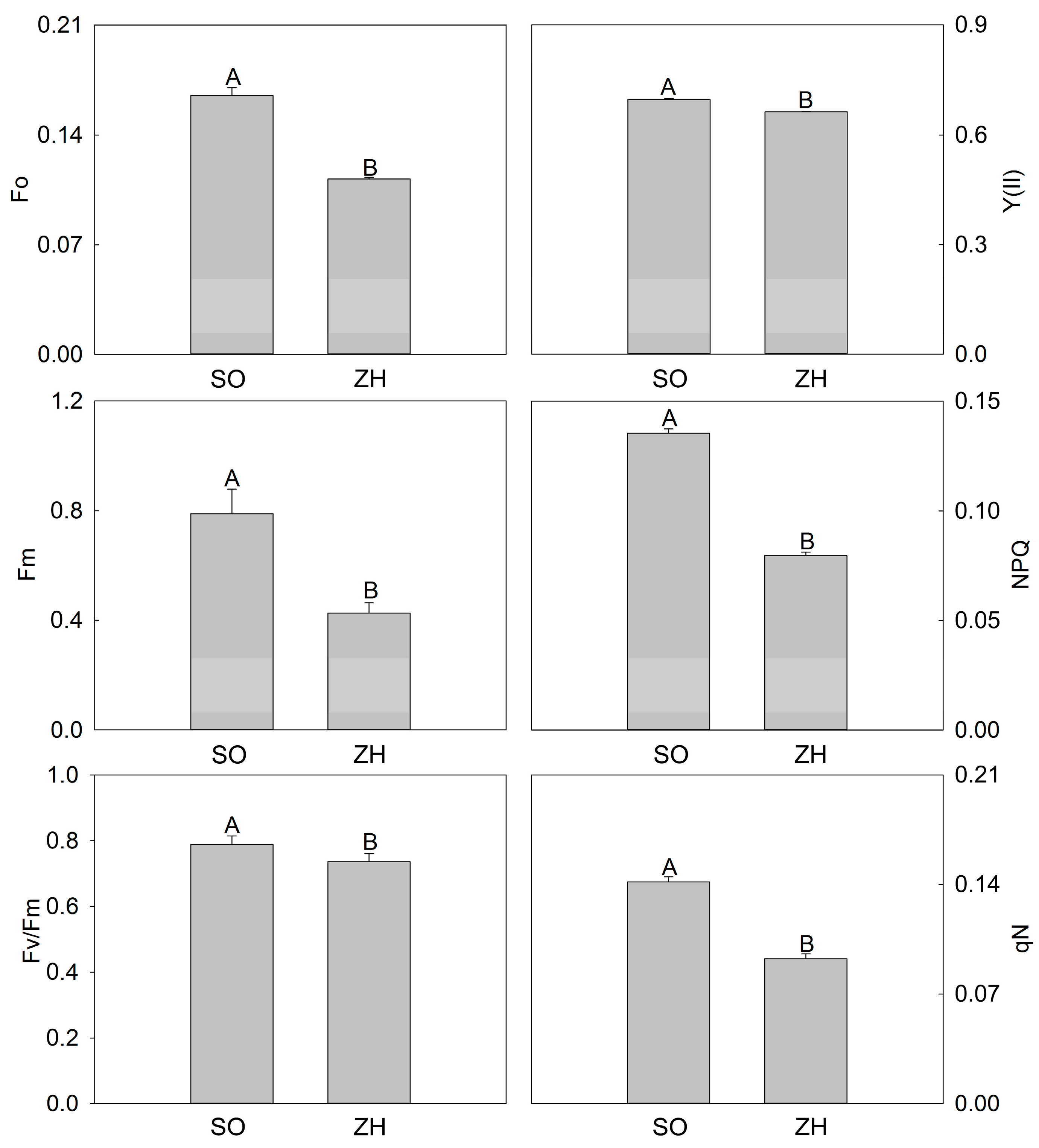
2.3. Photosynthetic Characteristics and Chlorophyll Fluorescence Parameters
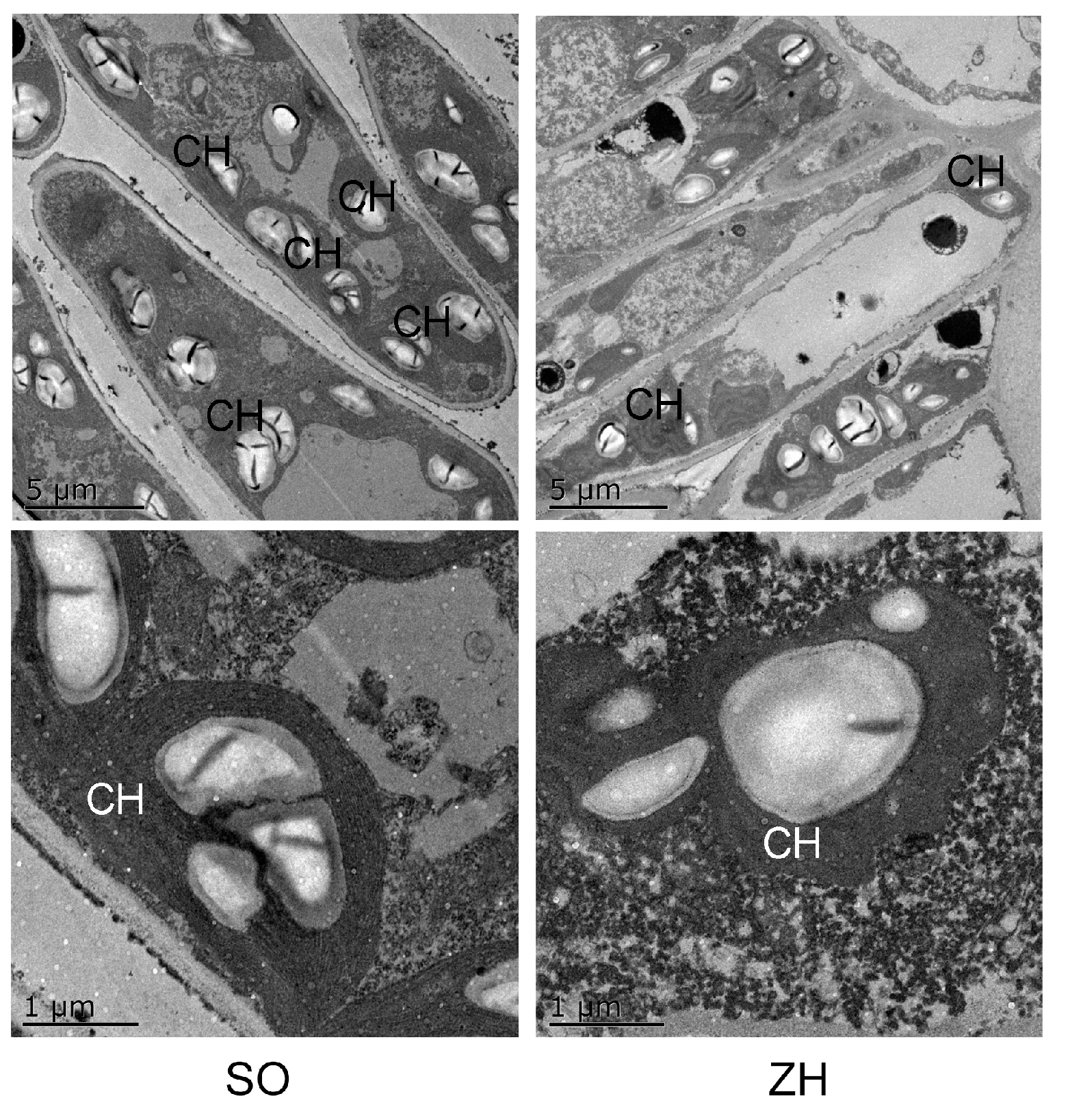
2.4. Anatomical Structures
2.5. Transcriptome Sequencing Results and Quality Assessment
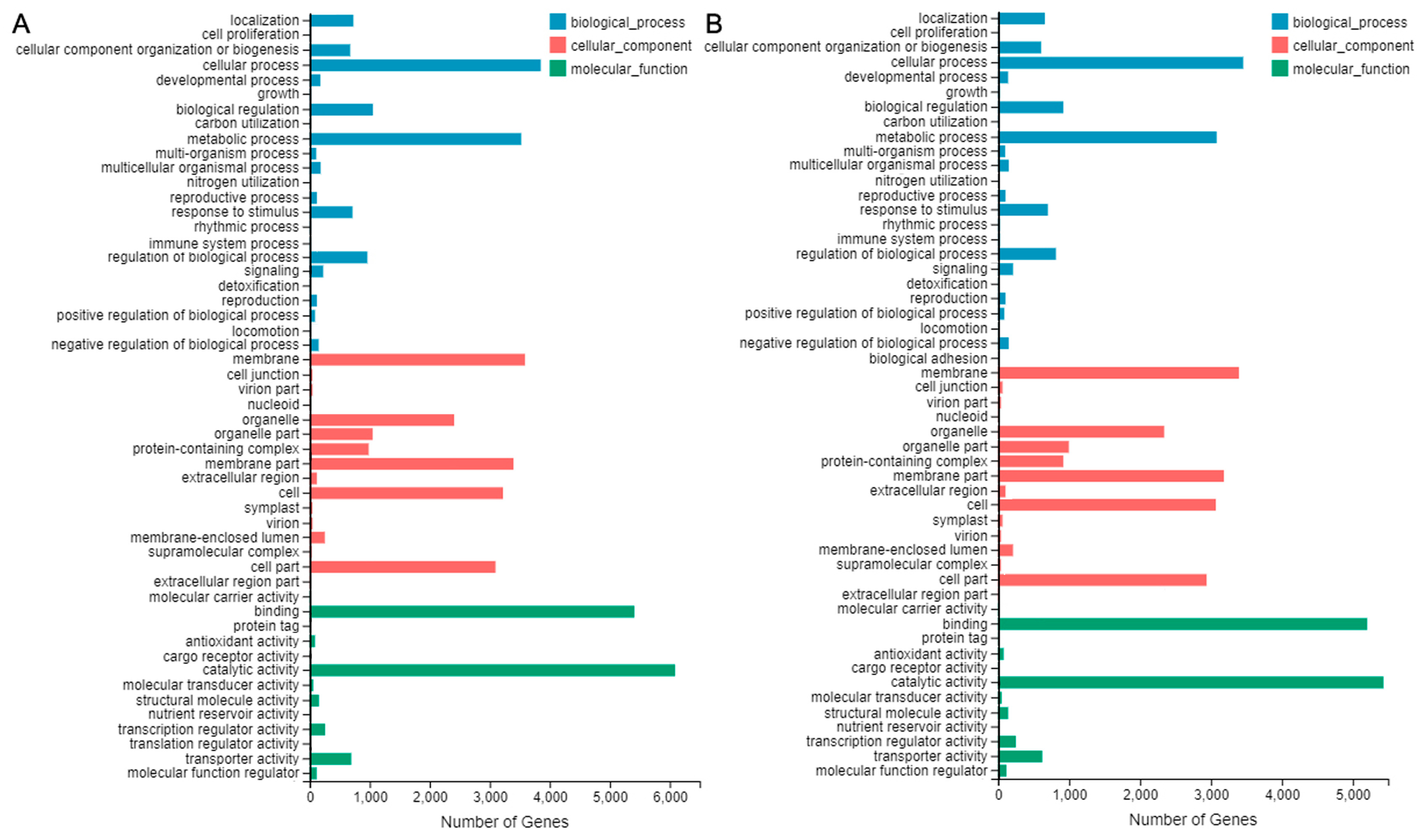
2.6. Functional Annotation for Unigenes
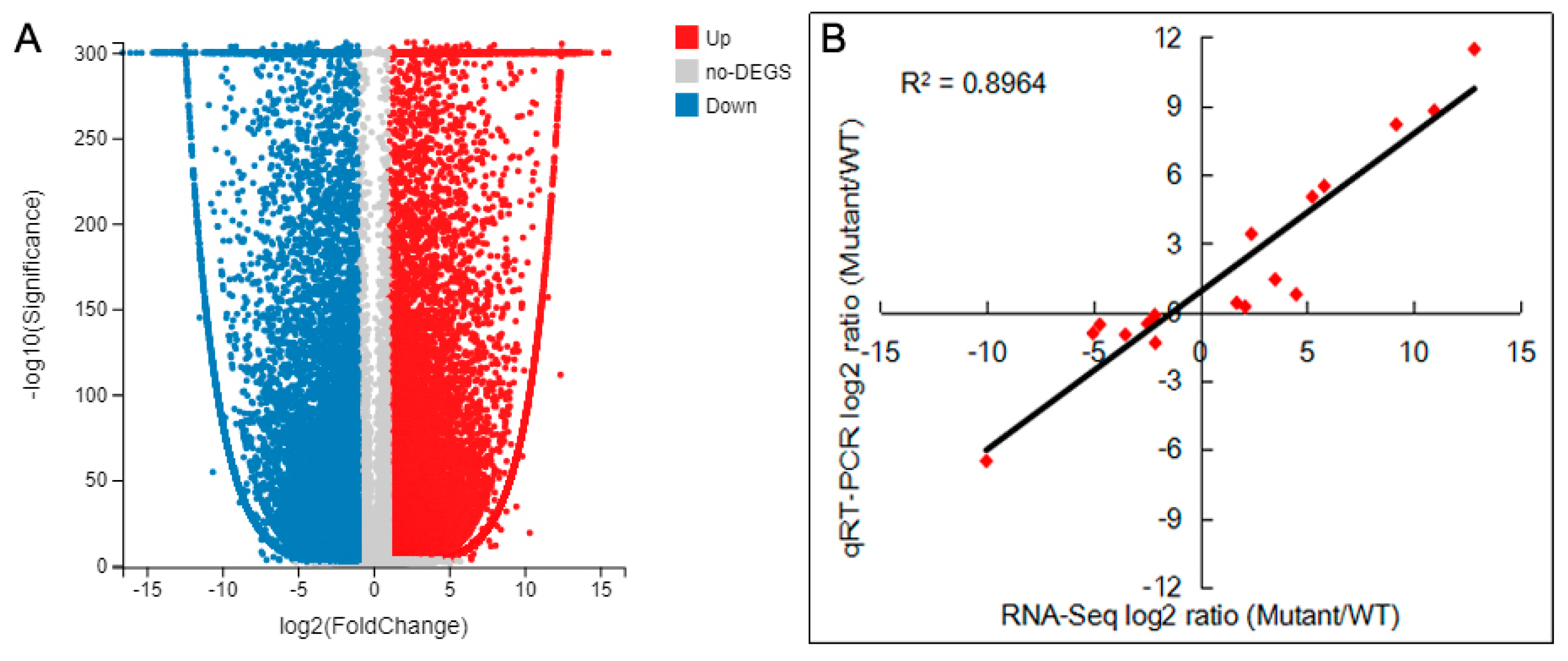
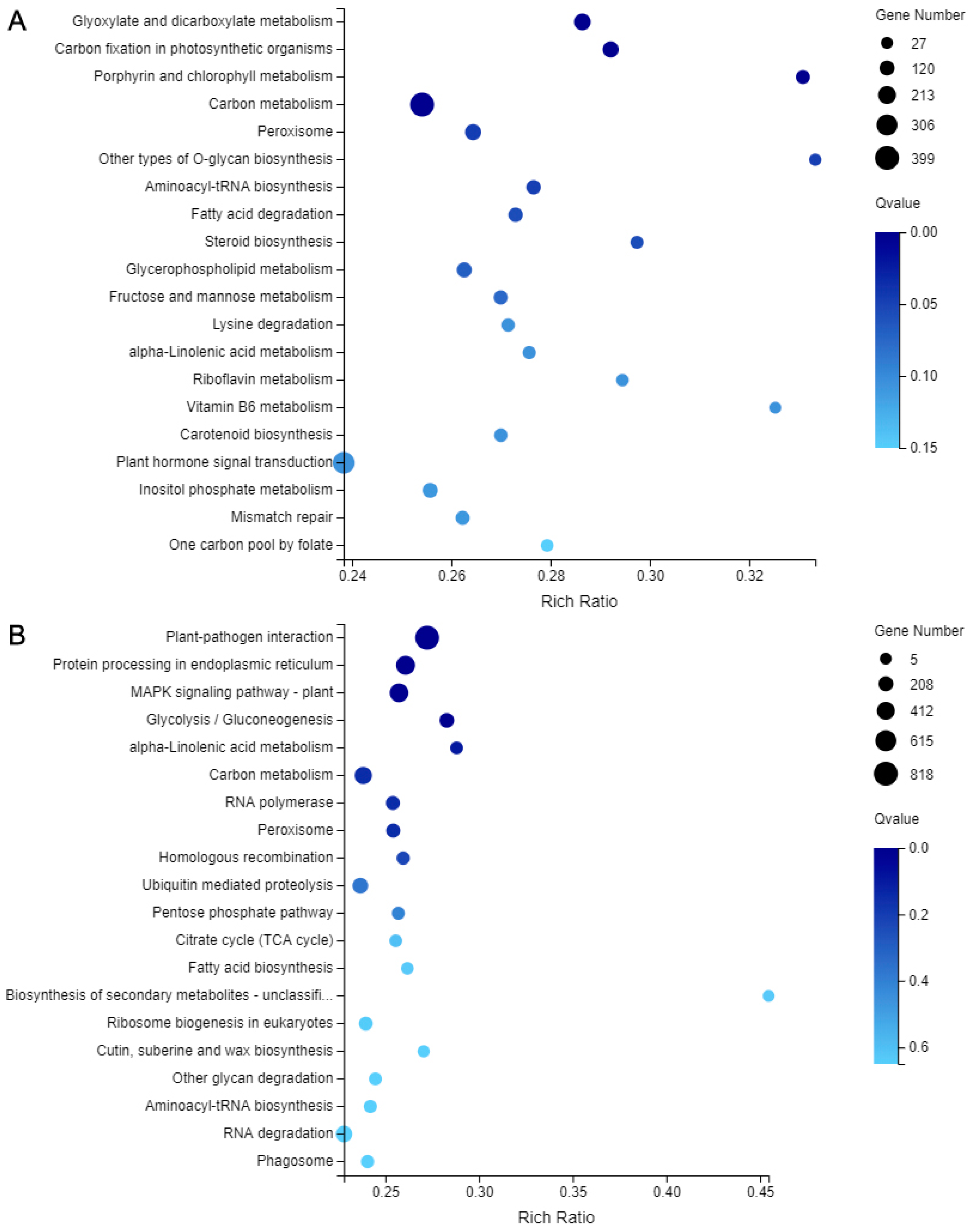
2.7. DEGs Analysis and Verification
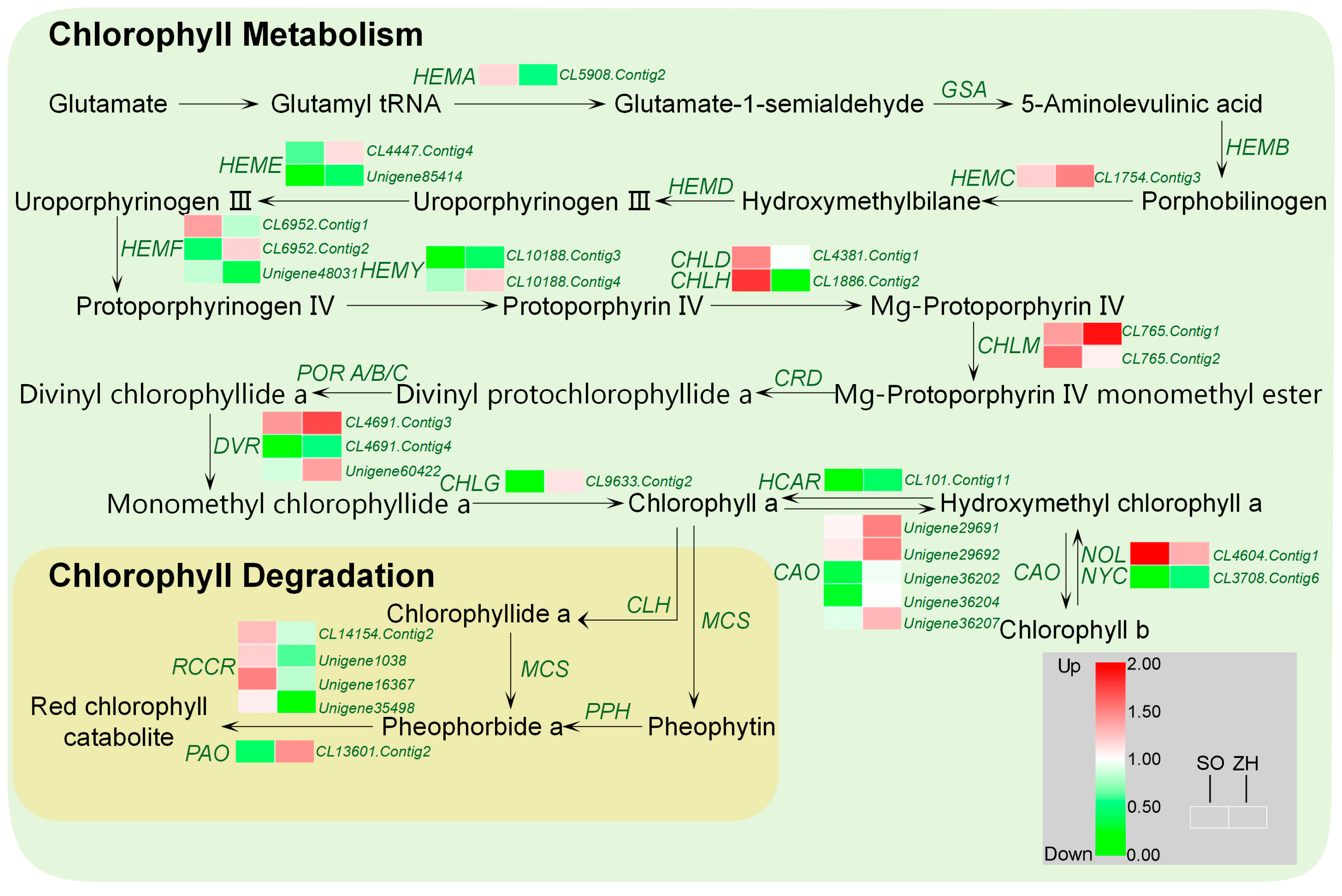
2.8. Chlorophyll and Carotenoid Biosynthetic Genes Involved in Leaf Coloration
3. Discussion
4. Materials and Methods
4.1. Plant Materials
4.2. Color Indices Measurement
4.3. SPAD Value and Pigment Content Measurement
4.4. Photosynthetic Characteristics and Chlorophyll Fluorescence Parameters Measurement
4.5. Anatomy Observation
4.6. RNA-seq and Data Analysis
4.7. Gene Expression Analysis
4.8. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Chen, Z.; Lu, X.Y.; Xuan, Y.; Tang, F.; Wang, J.J.; Shi, D.; Fu, S.L.; Ren, J. Transcriptome analysis based on a combination of sequencing platforms provides insights into leaf pigmentation in Acer rubrum. BMC Plant Biol. 2019, 19, 240. [Google Scholar] [CrossRef]
- Li, W.X.; Yang, S.B.; Lu, Z.G.; He, Z.C.; Ye, Y.L.; Zhao, B.B.; Wang, L.; Jin, B. Cytological, physiological, and transcriptomic analyses of golden leaf coloration in Ginkgo biloba L. Hortic. Res. 2018, 5, 12. [Google Scholar] [CrossRef] [PubMed]
- Lev-Yadun, S.; Yamazaki, K.; Holopainen, J.K.; Sinkkonen, A. Spring versus autumn leaf colours: Evidence for different selective agents and evolution in various species and floras. Flora-Morphology, Distribution. Funct. Ecol. 2012, 207, 80–85. [Google Scholar]
- Sinkkonen, A.; Somerkoski, E.; Paaso, U.; Holopainen, J.K.; Rousi, M.; Mikola, J. Genotypic variation in yellow autumn leaf colours explains aphid load in silver birch. New Phytol. 2012, 195, 461–469. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.X.; Chen, X.X.; Xu, B.; Li, Y.X.; Ma, Y.H.; Wang, G.D. Phenotype and transcriptome analysis reveals chloroplast development and pigment biosynthesis together influenced the leaf color formation in mutants of Anthurium andraeanum ‘Sonate’. Front. Plant Sci. 2015, 6, 139–155. [Google Scholar] [CrossRef] [PubMed]
- Gu, C.; Liao, L.; Zhou, H.; Wang, L.; Deng, X.B.; Han, Y.P. Constitutive activation of an anthocyanin regulatory gene pcMYB10.6 is related to red coloration in purple-foliage plum. PLoS ONE 2015, 10, 81–88. [Google Scholar] [CrossRef]
- Chang, Q.S.; Zhang, L.X.; Hou, X.G.; Wang, Z.; Wang, N.; Gong, M.G.; Zhang, Q.M.; Chen, H.; Shi, Z.Q.; Den, C.C. The anatomical, physiological, and molecular analysis of a chlorophyll-deficient mutant in tree peony (Paeonia suffruticosa). Photosynthetica 2019, 57, 724–730. [Google Scholar] [CrossRef]
- Li, Y.; Zhang, Z.; Wang, P.; Wang, S.; Ma, L.L.; Li, L.F.; Yang, R.T.; Ma, Y.Z.; Wang, Q. Comprehensive transcriptome analysis discovers novel candidate genes related to leaf color in a Lagerstroemia indica yellow leaf mutant. Genes Genom. 2015, 37, 851–863. [Google Scholar] [CrossRef]
- Zhu, G.F.; Yang, F.X.; Shi, S.; Li, D.M.; Wang, Z.; Liu, H.L.; Huang, D.; Wang, C.Y. Transcriptome characterization of Cymbidium sinense ‘Dharma’ using 454 pyrosequencing and its application in the identification of genes associated with leaf color variation. PLoS ONE 2015, 10, e0128592. [Google Scholar] [CrossRef]
- Nagata, N.; Tanaka, R.; Satoh, S.; Tanaka, A. Identification of a vinyl reductase gene for chlorophyll synthesis in Arabidopsis thaliana and implications for the evolution of prochlorococcus species. Plant Cell 2005, 17, 233–240. [Google Scholar] [CrossRef]
- Adhikari, N.D.; Froehlich, J.E.; Strand, D.D.; Buck, S.M.; Kramer, D.M.; Larkin, R.M. GUN4-porphyrin complexes bind the ChlH/GUN5 subunit of Mg-chelatase and promote chlorophyll biosynthesis in Arabidopsis. Plant Cell 2011, 23, 1449–1467. [Google Scholar] [CrossRef] [PubMed]
- Niu, Y.; Chen, G.; Peng, D.L.; Song, B.; Yang, Y.; Li, Z.M.; Sun, H. Grey leaves in an alpine plant: A cryptic colouration to avoid attack. New Phytol. 2014, 203, 953–963. [Google Scholar] [CrossRef] [PubMed]
- Whiston, J. A chlorophyll deficient rice mutant with impaired chlorophyllide esterification in chlorophyll biosynthesis. Plant Physiol. 2007, 145, 29–40. [Google Scholar]
- Sato, Y.; Morita, R.; Nishimura, M.; Yamaguchi, H.; Kusaba, M. Mendel’s gene cotyledon gene encodes a positive regulator of the chlorophyll-degrading pathway. Proc. Natl. Acad. Sci. USA 2007, 104, 14169–14174. [Google Scholar] [CrossRef] [PubMed]
- Cazzonelli, C.I.; Pogson, B.J. Source to sink: Regulation of carotenoid biosynthesis in plants. Trends Plant Sci. 2010, 15, 266–274. [Google Scholar] [CrossRef]
- Zhu, C.; Yamamura, S.; Koiwa, H.; Nishihara, M.; Sandmann, G. cDNA cloning and expression of carotenogenic genes during flower development in Gentiana lutea. Plant Mol. Biol. 2002, 48, 277–285. [Google Scholar] [CrossRef]
- Zhang, J.C.; Zhou, W.J.; Xu, Q.; Tao, N.G.; Ye, J.L.; Guo, F.; Xu, J.; Deng, X.X. Two lycopene β-cyclases genes from sweet orange (Citrus sinensis L. Osbeck) encode enzymes with different functional efficiency during the conversion of lycopene-to-provitamin A. J. Integr. Agric. 2013, 12, 1731–1747. [Google Scholar] [CrossRef]
- Kato, M.; Matsumoto, H.; Ikoma, Y.; Kuniga, T.; Nakajima, N.; Yoshida, T.; Yano, M. Accumulation of carotenoids and expression of carotenoid biosynthetic genes and carotenoid cleavage dioxygenase genes during fruit maturation in the juice sacs of ‘Tamami,’ ‘Kiyomi’ Tangor, and ‘Wilking’ Mandarin. J. Jpn. Soc. Hortic. Sci. 2007, 76, 103–111. [Google Scholar] [CrossRef]
- Bai, Y.H.; Pattanaik, S.; Patra, B.; Werkman, J.R.; Xie, C.H.; Yuan, L. Flavonoid-related basic helix-loop-helix regulators, NtAn1a and NtAn1b, of tobacco have originated from two ancestors and are functionally active. Planta 2011, 234, 363. [Google Scholar] [CrossRef]
- Gao, H.J.; Xu, J.; Liu, X.; Liu, B.Z.; Deng, X.X. Light effect on carotenoids production and expression of carotenogenesis genes in citrus callus of four genotypes. Acta Physiol. Plant. 2011, 33, 2485–2492. [Google Scholar] [CrossRef]
- Zhang, Q.; Zhang, M.; Ding, Y.; Zhou, P.; Fang, Y.M. Composition of photosynthetic pigments and photosynthetic characteristics in green and yellow sectors of the variegated Aucuba japonica ‘Variegata’ leaves. Flora 2018, 240, 25–33. [Google Scholar] [CrossRef]
- Aluru, M.R.; Yu, F.; Fu, A.; Rodermel, S. Arabidopsis variegation mutants: New insights into chloroplast biogenesis. J. Exp. Bot. 2006, 57, 1871–1881. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; Liu, Z.Y.; Shan, X.F.; Li, C.Y.; Tang, X.Y.; Chi, M.Y.; Feng, H. Physiological properties and chlorophyll biosynthesis in a Pak-choi (Brassica rapa L. ssp. chinensis) yellow leaf mutant, pylm. Acta Physiol. Plant. 2017, 39, 22. [Google Scholar] [CrossRef]
- Zhu, L.X.; Zeng, X.H.; Chen, Y.L.; Yang, Z.H.; Qi, L.P.; Pu, Y.Y.; Yi, B.; Wen, J.; Ma, C.Z.; Shen, J.X.; et al. Genetic characterisation and fine mapping of a chlorophyll-deficient mutant (BnaC.ygl) in Brassica napus. Mol. Breed. 2014, 34, 603–614. [Google Scholar] [CrossRef]
- Siefermann-Harms, D.; Boxler-Baldoma, C.; Wilpert, K.; Heumanna, H. The rapid yellowing of spruce at a mountain site in the Central Black Forest (Germany). Combined effects of Mg deficiency and ozone on biochemical, physiological and structural properties of the chloroplasts. J. Plant Physiol. 2004, 161, 423–443. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Fan, Y.; Jiao, Y.; Wu, J.; Zhang, Z.; Yu, X.; Ma, Y. Transcriptome profiling of yellow leafy head development during the heading stage in Chinese cabbage (Brassica rapa subsp. pekinensis). Physiol. Plant. 2019, 165, 800–813. [Google Scholar] [CrossRef]
- Crisosto, C.H.; Garner, D.; Crisosto, G.M.; Sibbett, S.; Day, K.R. Late harvest and delayed cooling induce internal browning of ‘Ya Li’ and ‘Seuri’ Chinese pears. HortScience 1994, 29, 667–670. [Google Scholar] [CrossRef]
- Intelmann, D.; Jaros, D.; Rohm, H. Identification of color optima of commercial tomato catsup. Eur. Food Res. Technol. 2005, 22, 662–666. [Google Scholar] [CrossRef]
- Zhao, C.L.; Guo, H.C. Research advances in the subcellular organization of the enzymes catalyzing anthocyanins bioynthesis in higher plants. Acta Bot. Boreal. 2007, 27, 1695–1701. [Google Scholar]
- Li, Y.H.; Wang, B.H.; Dai, Z.Y.; Li, A.H.; Liu, G.Q.; Zuo, S.M.; Zhang, H.X.; Pan, B. Morphological structure and genetic mapping of new leaf-color mutant gene in rice (Oryza sativa). Rice Sci. 2011, 25, 587–593. [Google Scholar] [CrossRef]
- Maekawa, S.; Takabayashi, A.; Reyes, T.H.; Yamamoto, H.; Tanaka, A.; Sato, T.; Yamaguchi, J. Pale-green phenotype of atl31atl6 double mutant leaves is caused by disruption of 5-aminolevulinic acid biosynthesis in Arabidopsis thaliana. PLoS ONE 2015, 10, e0117662. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.M.; Zhang, X.; Wang, J.L.; Wan, J.M. Leaf chloroplast ultrastructure and photosynthetic properties of a chlorophyll-deficient mutant of rice. Photosynthetica 2014, 52, 217–222. [Google Scholar] [CrossRef]
- Chang, Q.S.; Chen, S.M.; Chen, Y.; Deng, Y.M.; Chen, F.D.; Zhang, F.; Wang, S.W. Anatomical and physiological differences and differentially expressed genes between the green and yellow leaf tissue in a variegated chrysanthemum variety. Mol. Biotechnol. 2013, 54, 393–411. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.Y.; Xia, X.W.; Fang, W.; Fu, Y.; An, M.M.; Zhou, M.B. Identification of genes involved in spontaneous leaf color variation in Pseudosasa japonica. Genet. Mol. Res. 2015, 14, 11827–11840. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Gerstein, M.; Snyder, M. RNA-Seq: A revolutionary tool for transcriptomics. Nat. Rev. Genet. 2009, 10, 57–63. [Google Scholar] [CrossRef]
- Wang, J.Y.; Turner, N.C.; Liu, Y.X.; Siddique, K.H.M.; Xiong, Y.C. Effects of drought stress on morphological, physiological and biochemical characteristics of wheat species differing in ploidy level. Funct. Plant Biol. 2017, 44, 219–234. [Google Scholar] [CrossRef]
- Li, G.L.; Wu, H.X.; Sun, Y.Q.; Zhang, S.Y. Response of chlorophyll fluorescence parameters to drought stress in sugar beet seedlings. Russ. J. Plant Physiol. 2013, 60, 337–342. [Google Scholar] [CrossRef]
- Zhang, H.T.; Li, J.J.; Yoo, J.H.; Yoo, S.C.; Cho, S.H.; Koh, H.J.; Seo, H.S.; Paek, N.C. Rice Chlorina-1 and Chlorina-9 encode ChlD and ChlI subunits of Mg-chelatase, a key enzyme for chlorophyll synthesis and chloroplast development. Plant Mol. Biol. 2006, 62, 325–337. [Google Scholar] [CrossRef]
- Li, X.; Kanakala, S.; He, Y.H.; Zhong, X.L.; Yu, S.M.; Li, R.X.; Sun, L.X.; Ma, J. Physiological characterization and comparative transcriptome analysis of white and green leaves of Ananas comosus var. Bracteatus. PLoS ONE 2017, 12, e0169838. [Google Scholar] [CrossRef]
- Mochizuki, N.; Brusslan, J.A.; Larkin, R.; Nagatani, A.; Chory, J. Arabidopsis genomes uncoupled5 (GUN5) mutant reveals the involvement of Mg-chelatase H subunit in plastid-to-nucleus signal transduction. Proc. Natl. Acad. Sci. USA 2001, 98, 2053–2058. [Google Scholar] [CrossRef]
- Kumar, A.M.; Söll, D. Antisense HEMA1 RNA expression inhibits heme and chlorophyll biosynthesis in Arabidopsis. Plant Physiol. 2000, 122, 49–56. [Google Scholar] [CrossRef] [PubMed]
- Song, L.; Ma, Q.P.; Zou, Z.W.; Sun, K.; Yao, Y.T.; Tao, J.H.; Kaleri, N.A.; Li, X.H. Molecular link between leaf coloration and gene expression of flavonoid and carotenoid biosynthesis in Camellia sinensis cultivar ‘Huangjinya’. Front. Plant Sci. 2017, 8, 803. [Google Scholar] [CrossRef] [PubMed]
- Yuan, H.; Zhang, J.X.; Nageswaran, D.; Li, L. Carotenoid metabolism and regulation in horticultural crops. Hortic. Res. 2015, 2, 15036. [Google Scholar] [CrossRef] [PubMed]
- Voss, D.H. Relating colourimeter measurement of plant colour to the Royal Horticultural Society colour chart. Hortscience 1992, 27, 1256–1260. [Google Scholar] [CrossRef]
- Zhao, D.Q.; Zhou, C.H.; Tao, J. Carotenoid accumulation and carotenogenic genes expression during two types of persimmon fruit (Diospyros kaki L.) development. Plant Mol. Biol. Rep. 2011, 29, 646–654. [Google Scholar] [CrossRef]
- Zou, Q. (Ed.) Plant Physiology Experimental Guidance; China Agricultural Press: Beijing, China, 2000. [Google Scholar]
- Genty, B.; Briantais, J.M.; Baker, N.R. The relationship between the quantum yield of photosynthetic electron transport and quenching of chlorophyll fluorescence. Biochim. Biophys. Acta 1989, 990, 87–92. [Google Scholar] [CrossRef]
- Kooten, O.V.; Snel, J.F.H. The use of chlorophyll fluorescence nomenclature in plant stress physiology. Photosynth. Res. 1990, 25, 147–150. [Google Scholar] [CrossRef]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.D.; et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef]
- Mortazavi, A.; Williams, B.A.; Mccue, K.; Schaeffer, L.; Wold, B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 2008, 5, 621–628. [Google Scholar] [CrossRef]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 2008, 36, 1101–1108. [Google Scholar] [CrossRef]



| Sample | Total Raw Reads (M) | Total Clean Bases (Gb) | GC (%) | Q30 (%) | Adapter (%) | Low Quality (%) |
|---|---|---|---|---|---|---|
| SO-1 | 52.42 | 7.46 | 40.40 | 93.57 | 0.63 | 4.49 |
| SO-2 | 50.78 | 7.28 | 40.40 | 95.23 | 0.56 | 3.89 |
| SO-3 | 50.78 | 7.24 | 40.47 | 93.66 | 0.46 | 4.42 |
| ZH-1 | 50.78 | 7.25 | 40.45 | 93.73 | 0.52 | 4.24 |
| ZH-2 | 50.78 | 7.29 | 40.44 | 95.25 | 0.57 | 3.73 |
| ZH-3 | 50.78 | 7.24 | 40.55 | 93.61 | 0.59 | 4.32 |
| Average | 51.05 | 7.29 | 40.45 | 94.18 | 0.56 | 4.18 |
| No. | Pathway Name | DEGs Num. | Pathway ID |
|---|---|---|---|
| 1 | Carbon metabolism | 773 (5.58%) | ko01200 |
| 2 | Plant-pathogen interaction | 1419 (10.25%) | ko04626 |
| 3 | Porphyrin and chlorophyll metabolism | 152 (1.10%) | ko00860 |
| 4 | Peroxisome | 306 (2.21%) | ko04146 |
| 5 | alpha-Linolenic acid metabolism | 137 (0.99%) | ko00592 |
| 6 | Carbon fixation in photosynthetic organisms | 274 (1.98%) | ko00710 |
| 7 | Mitogen-activated protein kinase (MAPK) signaling pathway - plan | 851 (6.15%) | ko04016 |
| 8 | Aminoacyl-tRNA biosynthesis | 195 (1.41%) | ko00970 |
| 9 | Glycolysis / Gluconeogenesis | 334 (2.41%) | ko00010 |
| 10 | Homologous recombination | 197 (1.42%) | ko03440 |
| 11 | Carotenoid biosynthesis | 153 (1.11%) | ko00906 |
| 12 | Plant hormone signal transduction | 645 (4.66%) | ko04075 |
| 13 | Steroid biosynthesis | 103 (0.74%) | ko00100 |
| 14 | Fatty acid degradation | 189 (1.37%) | ko00071 |
| 15 | Glyoxylate and dicarboxylate metabolism | 292 (2.11%) | ko00630 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dong, X.; Huang, L.; Chen, Q.; Lv, Y.; Sun, H.; Liang, Z. Physiological and Anatomical Differences and Differentially Expressed Genes Reveal Yellow Leaf Coloration in Shumard Oak. Plants 2020, 9, 169. https://doi.org/10.3390/plants9020169
Dong X, Huang L, Chen Q, Lv Y, Sun H, Liang Z. Physiological and Anatomical Differences and Differentially Expressed Genes Reveal Yellow Leaf Coloration in Shumard Oak. Plants. 2020; 9(2):169. https://doi.org/10.3390/plants9020169
Chicago/Turabian StyleDong, Xiaoyun, Libin Huang, Qingsheng Chen, Yunzhou Lv, Hainan Sun, and Zhenhai Liang. 2020. "Physiological and Anatomical Differences and Differentially Expressed Genes Reveal Yellow Leaf Coloration in Shumard Oak" Plants 9, no. 2: 169. https://doi.org/10.3390/plants9020169
APA StyleDong, X., Huang, L., Chen, Q., Lv, Y., Sun, H., & Liang, Z. (2020). Physiological and Anatomical Differences and Differentially Expressed Genes Reveal Yellow Leaf Coloration in Shumard Oak. Plants, 9(2), 169. https://doi.org/10.3390/plants9020169




