Abstract
Although it is well established that flavonoid synthesis is induced in diverse plant species during nematode parasitism, little is known about the regulation of genes controlling flavonol biosynthesis during the plant–nematode interaction. In this study, expression of the Arabidopsis thaliana flavonol-specific transcription factor, AtMYB12, the flavonol synthase genes, AtFLS1, 2, 3, 4, and 5, and the gene encoding the central flavonoid enzyme, chalcone synthase (AtCHS), were examined in plant roots during infection by Heterodera schachtii (sugar beet cyst) and Meloidogyne incognita (root-knot) nematodes. These experiments showed that AtMYB12 was transiently upregulated at 9 dpi in syncytia associated with sugar beet cyst nematode infection and that an Atmyb12-deficient line was less susceptible to the parasite. This suggests that, rather than contributing to plant defense, this gene is essential for productive infection. However, the AtCHS and AtFLS1 genes, which are controlled by AtMYB12, did not exhibit a similar transient increase, but rather were expressly downregulated in syncytia relative to adjacent uninfected root tissue. Genetic analyses further indicated that AtFLS1 contributes to plant defense against Cyst nematode infection, while other AtFLS gene family members do not, consistent with prior reports that these other genes encode little or no enzyme activity. Together, these findings indicate a role of AtMyb12 in promoting the early stages of Cyst nematode infection, while flavonols produced through the action of AtFLS1 are essential for plant defense. On the other hand, a transient induction of AtMYB12 was not observed in galls produced during root-knot nematode infection, but this gene was instead substantially downregulated, starting at the 9 dpi sampling point, as were AtCHS and AtFLS1. In addition, both the AtMYB12- and AtFLS1-deficient lines were more susceptible to infection by this parasite. There was again little evidence for contributions from the other AtFLS gene family members, although an AtFLS5-deficient line appeared to be somewhat more susceptible to infection. Taken together, this study shows that sugar-beet cyst and root-knot nematodes modulate differently the genes involved in flavonol biosynthesis in order to successfully infect host roots and that AtFLS1 may be involved in the plant basal defense response against nematode infection.
1. Introduction
Plant parasitic nematodes are the causal agents of an estimated USD 100 billion in crop losses to the world-wide agro-economy annually, with Cyst (Heterodera and Globodera spp.) and root-knot (Meloidogyne spp.) nematodes causing the most extensive damage [1]. Plant infection by these nematodes involves the penetration of second-stage juveniles (J2) into host roots and migration through the root and subsequent intracellular migration (for cyst nematodes) or intercellular migration between the cortical cells to the vascular cylinder [2] until the favored site for feeding site induction is reached [3]. The nematodes then secrete an array of effectors into host cells through their stylet (a protrusible hollow mouth spear) to initiate the formation of specialized feeding structures known as syncytia for cyst nematodes and giant cells for root-knot nematodes [4,5,6]. These plant feeding cells, which are characterized by dense cytoplasm, enlarged multiple nuclei, a fragmented central vacuole and proliferation of organelles [3,5] act as the permanent source of nutrients for further nematode development. The feeding sites formed by cyst and root-knot nematodes have similarities in final phenotype but differ in ontogeny. Cell expansion with karyokinesis uncoupled from cytokinesis characterizes giant cells formed around the root-knot nematode head, while coordinated dissolution of walls adjacent to an initial syncytial cell give rise to a syncytium induced by cyst nematode [3]. The majority of effectors reported from cyst and root-knot nematodes differ, consistent with the differences observed in the parasitic process [6]. The secretion of nematode effectors is accompanied by an extensive alteration of gene expression in parasitized plant cells and roots including genes related to defense responses, cell-wall modifications, metabolic and signaling pathways, phytohormone balance, and the phenylpropanoid pathway [6,7,8,9,10,11,12]. Cyst and root knot nematodes induce the expression of different genes in Arabidopsis thaliana roots [7,12]. This is likely to be a reflection of the different modes of parasitism between cyst and root-knot nematodes as reflected in feeding cell ontogeny.
Flavonoids are natural products found in all plant species that are produced as part of the phenylpropanoid pathway [13]. In addition to ubiquitous pigments that play key roles in plant reproduction, seed dispersal, and UV protection [14,15], flavonoids are essential as chemical cues and defense molecules in interactions with microbes and insects [16,17,18,19,20]. There is also growing evidence that flavonoids contribute to defense against parasitic nematodes, although this remains poorly understood [21,22,23]. Flavonols represent the most abundant class of flavonoids and are also believed to be among the most ancient [24]. They possess potent free radical scavenging activity [25] and can have insecticidal properties [26]. In fact, soybean (Glycine max) genotype PI 227687, which accumulates the flavonol rutin, has been used widely in breeding programs as a source of insect resistance [27]. Flavonols are also involved in modulating auxin transport and signaling [28,29]. Several studies support a role of auxin in nematode feeding site formation of both cyst and root-knot nematode [30,31,32]. Moreover, flavonols have been shown to have a direct effect on chemotaxis, motility, and egg hatching of many nematode species [33]. Flavonols are synthesized from dihydroflavonols by a 2-oxoglutarate-dependent dioxygenase enzyme, flavonol synthase (FLS). Arabidopsis contains six FLS genes (AtFLS1 to AtFLS6) located at three sites on chromosome 5 [34]. AtFLS1, 3, and 5 encode full-length proteins while AtFLS2, 4, 6 are considered pseudo-genes unlikely to contribute to flavonol synthase activity [34]. AtFLS1 has been shown to have substantial flavonol synthase activity, both in vivo and in vitro [34,35], while AtFLS2 exhibits a relatively small amount of activity [36] and AtFLS5 no detectable activity under any conditions tested to date.
The R2R3-MYB family of transcription factors participates in a variety of cellular processes, including development [37,38,39], signal transduction [40,41], cell division [42], secondary metabolism [43,44,45], and plant disease resistance [46]. Among members of the R2R3-MYB family in Arabidopsis, AtMYB12 is a key regulator of FLS and chalcone synthase (CHS) gene expression [47,48,49]. CHS is the first enzyme in the flavonoid branch of the phenylpropanoid pathway and is encoded by the single-copy AtCHS gene in Arabidopsis [50]. Expression of AtCHS and AtFLS1 is induced by overexpression of AtMYB12 and substantially reduced in an Atmyb12 T-DNA knockout, with corresponding changes in flavonol levels in Arabidopsis seedlings [48].
In the study presented here, quantitative real-time PCR was used to analyze the expression patterns of AtMYB12 and the flavonoid biosynthetic genes, AtCHS and AtFLS1-5 during Arabidopsis parasitism by the beet cyst nematode, Heterodera schachtii and the southern root-knot nematode, Meloidogyne incognita. Histochemical analyses were used to examine the cell-type expression of these genes in nematode-infected root tissues. In addition, the effects on nematode parasitism of AtMYB12 overexpression and null mutations in Atmyb12 and five Atfls genes were examined. Our hypothesis is that sugar beet cyst and root knot nematodes modulate the expression of genes involved in the biosynthesis of flavonols in order to successfully infect host roots.
2. Results
2.1. AtMYB12 Is Upregulated in Syncytia and Downregulated in Galls
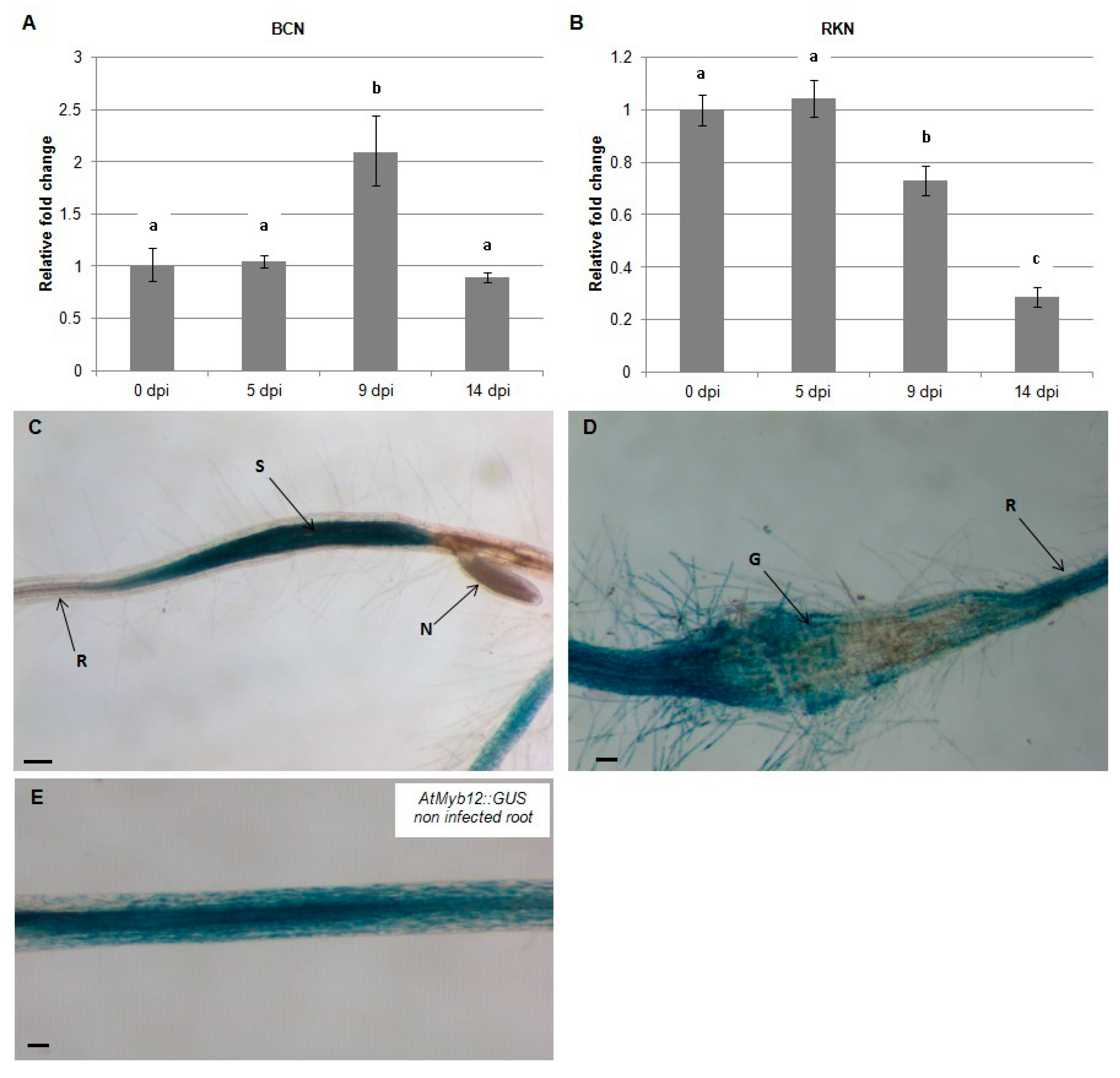
To test the effect of nematode infection on the flavonol transcriptional regulator, AtMYB12, quantitative real-time PCR (qRT-PCR) was used to quantify mRNA levels in Arabidopsis roots at 0, 5, 9, and 14 dpi with H. schachtii and M. incognita. During infection by H. schachtii, expression of AtMYB12 increased at -9 dpi and returned to preinfection level by 14 dpi (Figure 1A). In contrast, during infection with M. incognita, expression of AtMYB12 decreased over this time course, first detected at 9 dpi and continuing at 14 dpi (Figure 1B). These results indicate that AtMYB12 is affected differently during H. schachtii and M. incognita infection.
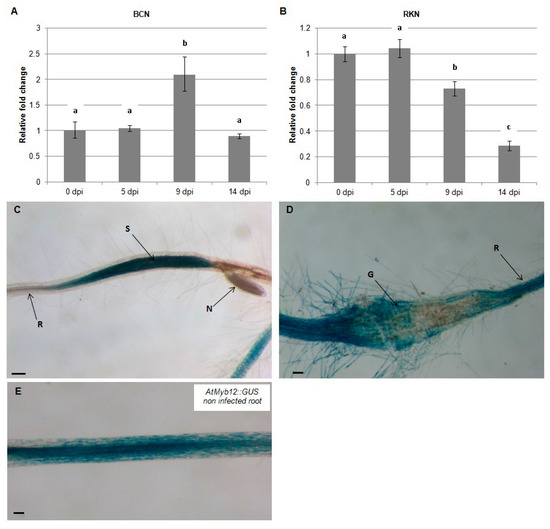
Figure 1.
Temporal pattern of Arabidopsis AtMYB12 gene expression during nematode infection. AtMYB12 expression was quantified by qRT-PCR in roots of wild-type Arabidopsis plants at 0, 5, 9 and 14 dpi with the beet-cyst nematode, H. schachtii (BCN) (A) or the southern root-knot nematode M. incognita (RKN) (B). Histochemical localization of GUS activity directed by AtMYB12p::GUS fusion in H. schachtii-induced feeding sites (C) and in M. incognita-formed galls (D) at 9 dpi as compared to non-infected roots (E). G, gall; N, nematode; S, syncytium; R, Arabidopsis root. Quantitative expression analyses were normalized to the Arabidopsis Actin8 gene and presented relative to uninfected control tissue (baseline set at 1.0). Values are means ± SE (3 technical replicates) with different letters indicating significant differences between time points p < 0.05 (One-way ANOVA test). Similar results were obtained from two independent biological replicates. Scale bar = 200 μm (C), and 100 μm (D,E).
To localize the expression of AtMYB12 in nematode-infected roots, AtMYB12p::GUS plants were infected with H. schachtii and M. incognita and GUS expression was examined by histochemical staining at 9 dpi. The results of this experiment indicate that expression of AtMYB12 is upregulated in syncytia formed by H. schachtii (Figure 1C) but strongly downregulated in galls generated by M. incognita (Figure 1D). These results further support the hypothesis that AtMYB12 mediates different responses to H. schachtii and M. incognita infection.
2.2. AtMYB12 Expression Promotes Sugar Beet Cyst Infection and May Contribute to Basal Defense Response to Root-Knot Nematode Infection
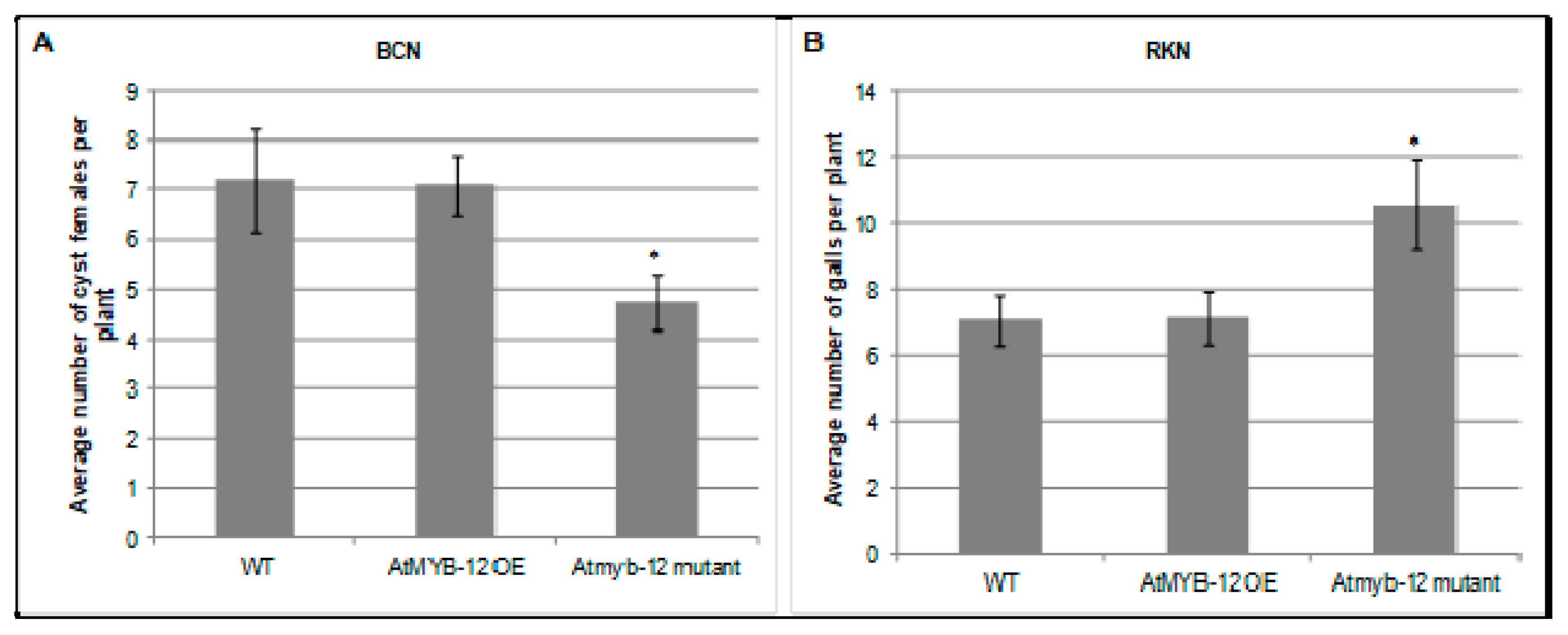
To assess the roles of AtMYB12 in nematode infection, Arabidopsis plants that either ectopically overexpress AtMYB12 or carry a T-DNA insertion in the Atmyb12 coding region were challenged with H. schachtii or M. incognita in six-well plates containing agar-based plant growth medium and the numbers of developing H. schachtii cyst females or M. incognita root galls in wild-type and mutant plants were counted 3–4 weeks post infection. No difference was observed in the number of H. schachtii-formed cysts or M. incognita-formed galls between plants over-expressing AtMYB12 and wild-type Arabidopsis plants. However, the Atmyb12 T-DNA mutant line was less susceptible to H. schachtii than were wild type plants, suggesting that the presence of AtMYB12 may promote H. schachtii parasitism (Figure 2). In contrast, Atmyb12 T-DNA mutant plants were more susceptible to M. incognita than wild-type plants, suggesting that AtMYB12 may contribute to basal defenses against M. incognita. These findings further suggest that Myb12 upregulation in syncytia and downregulation in galls is induced by the nematodes in order to promote infection.
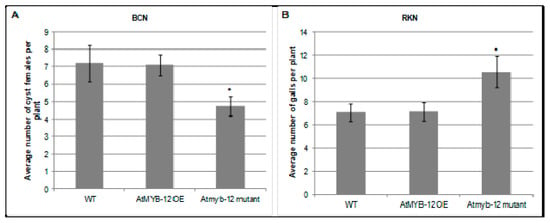
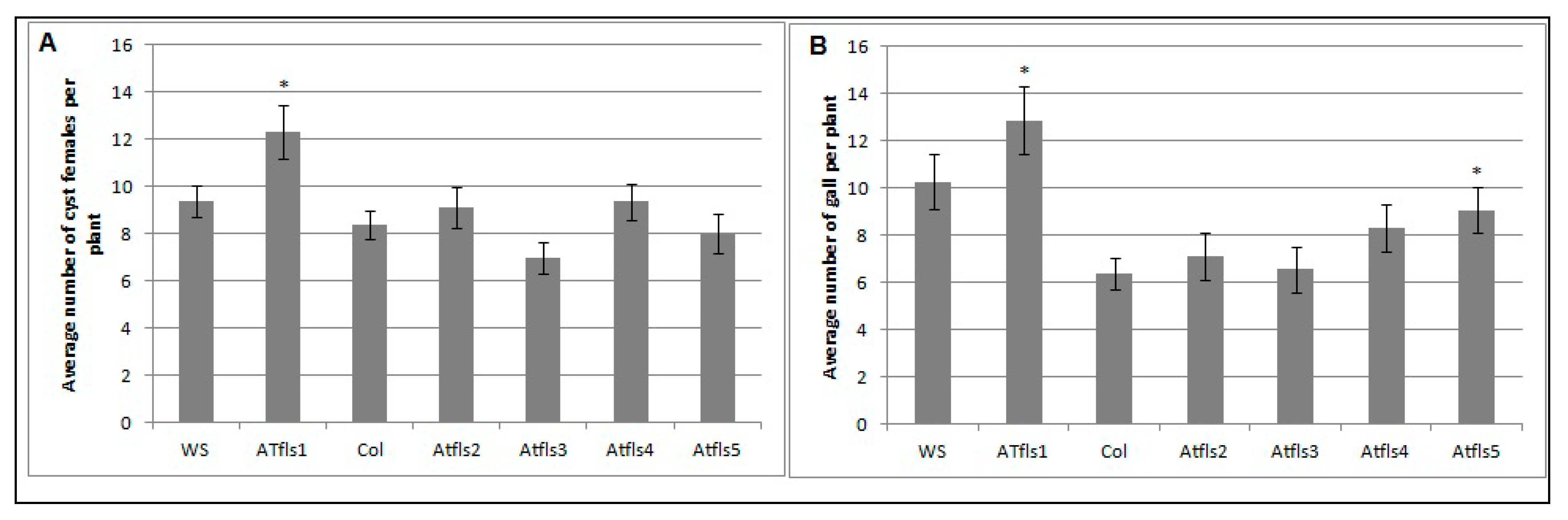
Figure 2.
Role of AtMYB12 in H. schachtii (A) and M. incognita (B) infection. Homozygous lines overexpressing AtMYB12, Atmyb12 knock-out, and wild-type Col-0 (WT) plants were planted on modified Knop’s medium and 10 days-old seedlings were inoculated with approximately 60 surface sterilized J2 of either H. schachtii or M. incognita. Three weeks after inoculation, the number of cyst female nematodes (for suagr beet cyst nematode) per root system or galls (for southern root-knot nematode) were counted. Data are presented as means ± standard errors. Mean values significantly different from wild type (Col 0) as determined by t test (p < 0.05) are denoted by asterisks. Similar results were obtained from a second, independent biological replicate.
2.3. Expression of AtCHS and AtFLS1 Is Downregulated in Both Syncytia and Root Galls
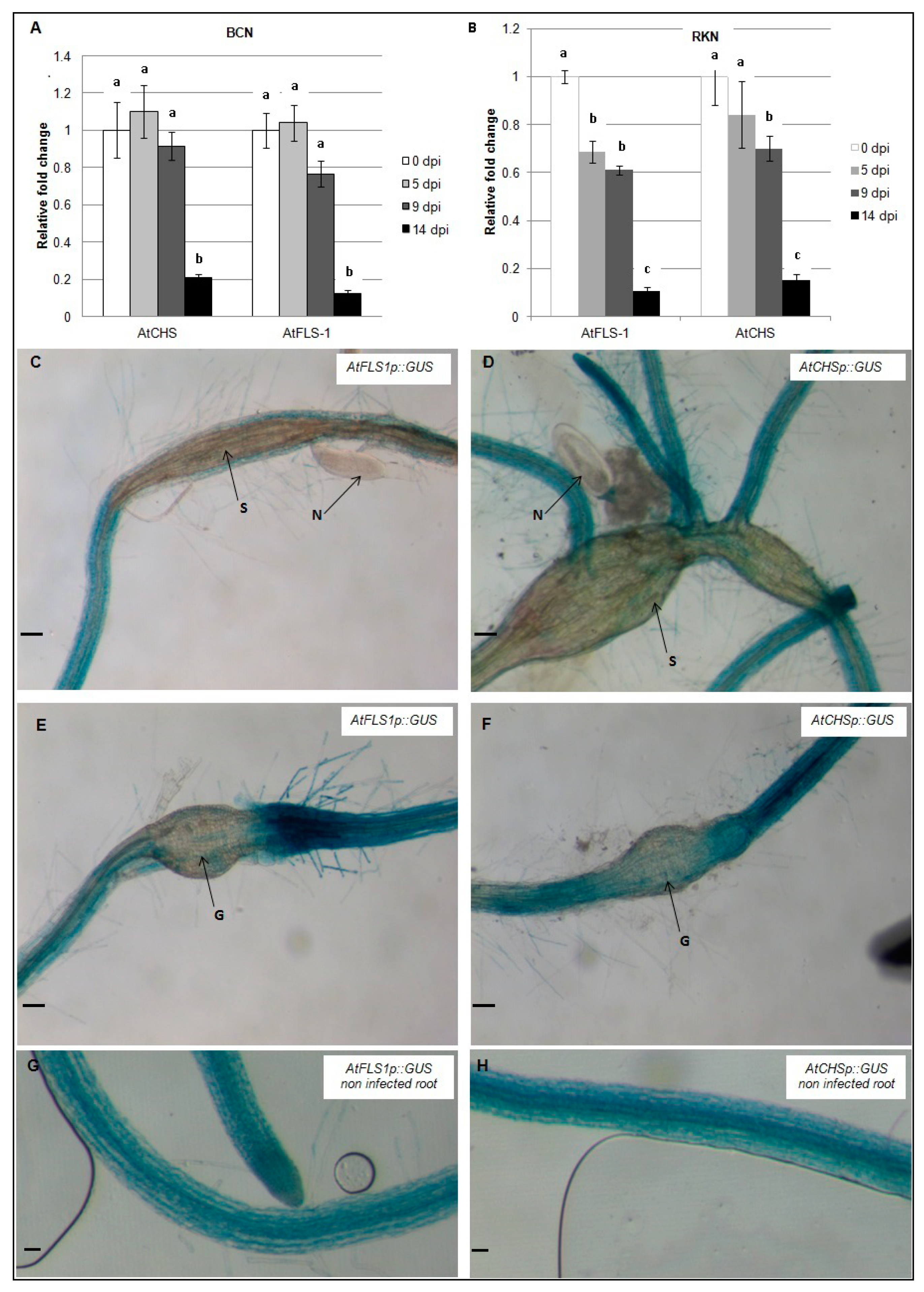
To examine the expression of AtCHS and AtFLS1 during nematode infection, qRT-PCR was used to compare transcript abundance in H. schachtii and M. incognita-infected Arabidopsis roots at 0, 5, 9, and 14 dpi. Our results show that expression of AtCHS and AtFLS1 was strongly reduced in Arabidopsis roots between 9–14 days following infection with H. schachtii. During infection with M. incognita, expression level of AtFLS1 was observed by 5 dpi and 9 dpi, and at 14 dpi, it was strongly downregulated, while expression of AtCHS was reduced between 9–14 dpi (Figure 3A,B).
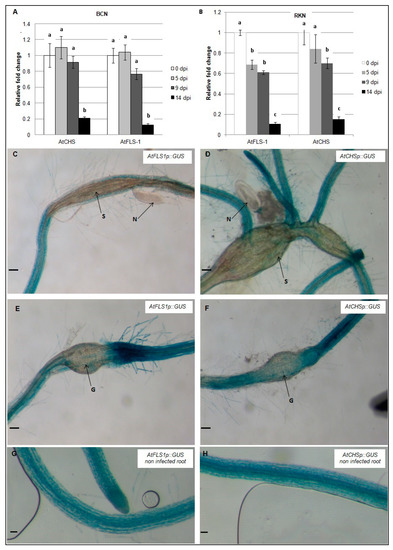
Figure 3.
Temporal patterns of AtCHS and AtFLS1 gene expression during nematode infection. AtCHS and AtFLS1 gene expression was quantified by qRT-PCR in roots of wild-type Arabidopsis plants at 0, 5, 9 and 14 days post-infection (dpi) with the sugar beet cyst nematode, H. schachtii (A) or the southern root-knot nematode M. incognita (B). Histochemical localization of GUS activity directed by AtCHSp::GUS and AtFLS1p::GUS fusion in H. schachtii-induced syncytia (C,D) and in M. incognita-induced gall (E,F) at 9 dpi and in non-infected controls (G,H). G, gall; N, nematode; S, syncytium; R, Arabidopsis root. Expression was normalized to the Arabidopsis Actin8 gene and presented relative to expression at 0 dpi. Values are means ± SE (3 technical replicates) with different letters indicating significant differences between time points p < 0.05 (One-way ANOVA test). Similar results were obtained from a second, independent biological replicate. Scale bar = 200 μm (C,D,G,H), and 100 μm (E,F).
In addition, AtCHSp::GUS and AtFLS1p::GUS reporter genes were used to examine the expression level of AtCHS and AtFLS1 at the nematodes feeding structures 9dpi. Histochemical analysis of GUS gene expression showed that expression of AtCHS and AtFLS1 was strongly downregulated in syncytia and nematode-induced galls suggesting that the downregulation of these genes is important in nematode infection (Figure 3C–F).
2.4. Expression of AtFLS2 in Upregulated in H. schachtii-Induced Syncytia, but Downregulated in M. incognita-Induced Galls, While Expression of AtFLS5 Is Downregulated in Both.
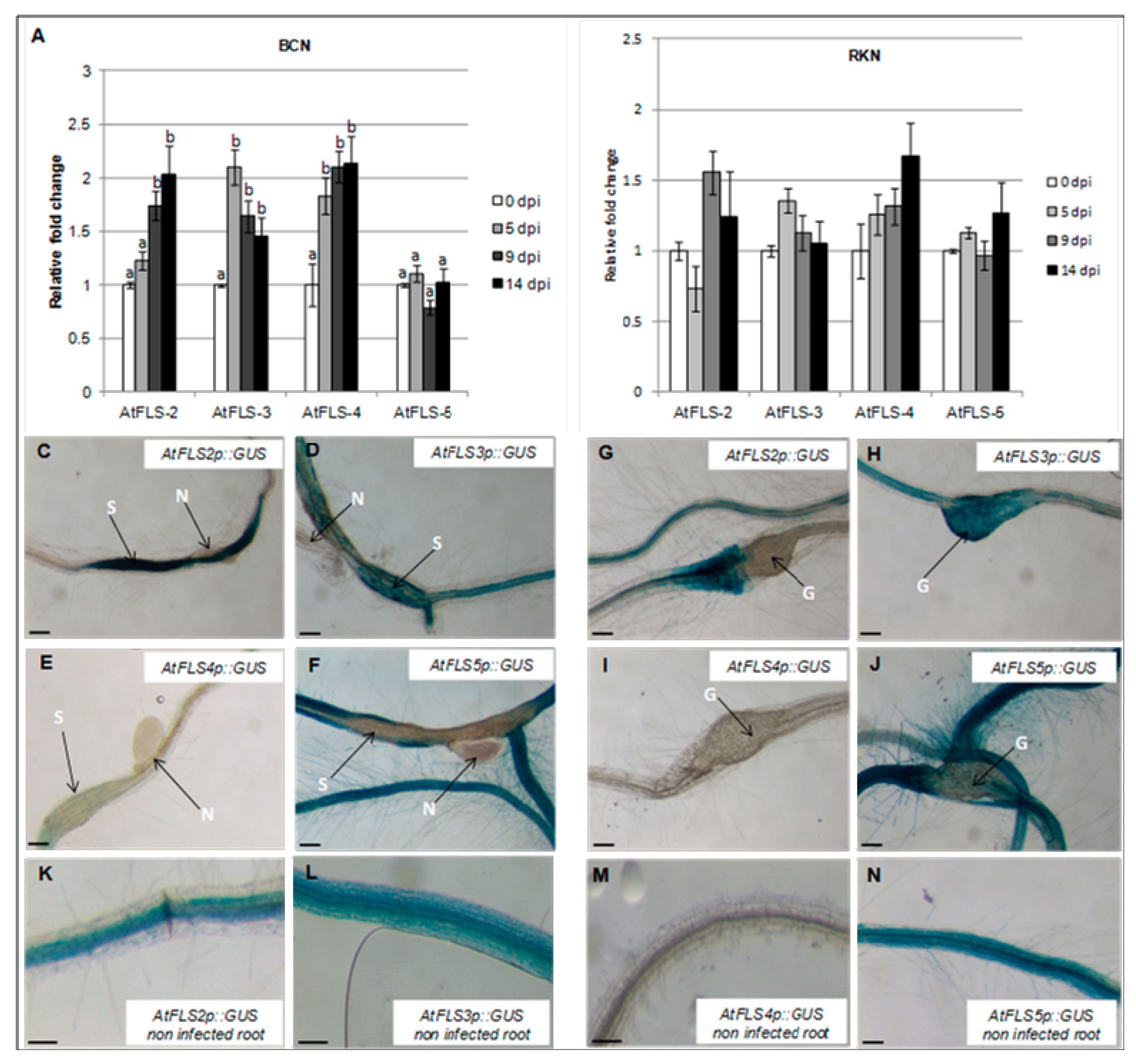
The observed downregulation of AtFLS1 in both nematode-induced syncytia and galls prompted us to examine the expression of four additional FLS genes to determine whether these might have a role in nematode infection. To this end, the abundance of AtFLS2, AtFLS3, AtFLS4, and AtFLS5 transcripts in Arabidopsis whole roots at 0, 5, 9, and 14 dpi by H. schachtii and M. incognita was also measured using qRT-PCR. Our results showed that expression of AtFLS2, AtFLS3, and AtFLS4 increased during H. schachtii infection, while expression of AtFLS5 did not change (Figure 4A). In contrast, during infection with M. incognita, the expression of AtFLS2, AtFLS3, AtFLS4, and AtFLS5 remained relatively unchanged or increased only slightly compared to non-infected roots (Figure 4B).
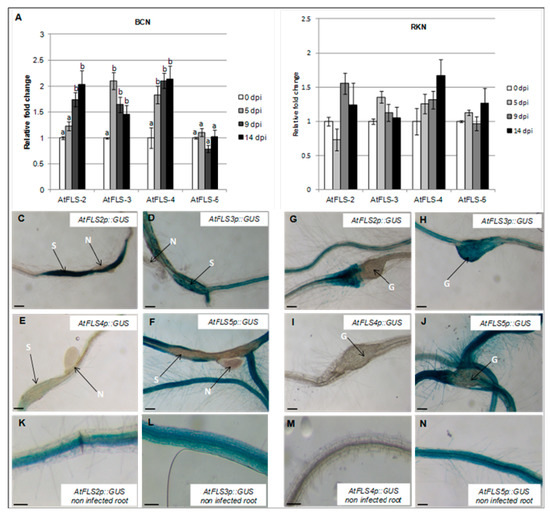
Figure 4.
Temporal expression of Arabidopsis FLS gene family members during nematode infection. Expression of AtFLS2, AtFLS3, AtFLS4, and AtFLS5 was quantified by qRT-PCR in roots of wild-type Arabidopsis plants at 0, 5, 9 and 14 dpi with the sugar beet cyst nematode, H. schachtii (A) of the southern root-knot nematode, M. incognita (B). Histochemical localization of GUS activity directed by AtFLSp::GUS in H. schachtii-induced syncytia (C–F) and in M. incognita-formed galls (G–J) at 9 dpi. (K–N) are non-infected Arabidopsis roots; G, galls; N, nematode; S, syncytium; R, Arabidopsis root. Expression was normalized to the Arabidopsis Actin8 gene and presented relative to expression at 0 dpi. Values are means ± SE (n = 3 technical replicates) with different letters indicating significant differences between time points p < 0.05 (One-way ANOVA test). Similar results were obtained from a second, independent biological replicate. Scale bar = 200 μm (C,J,N), and 150 μm (K–M).
To visualize the expression AtFLS2, AtFLS3, AtFLS4, and AtFLS5 at the nematode feeding sites, Arabidopsis plants expressing the GUS gene under the control of AtFLS2, AtFLS3, AtFLS4, and AtFLS5 promoters were infected with H. schachtii or M. incognita, and GUS expression was visualized at the nematode-induced feeding sites. In H. schachtii-formed syncytia, AtFLS2 was upregulated, suggesting that AtFLS2 may also play a role in syncytia formation and/or maintenance. The expression of AtFLS5 was downregulated at 9 dpi (Figure 4C,F), while the expression of AtFLS3 and AtFLS4 did not change (Figure 4D,E). In M. incognita-formed galls, the expression of AtFLS2 and AtFLS5 was strongly downregulated (Figure 4G,J), while the expression of AtFLS3 and AtFLS4 did not change.
The potential roles of AtFLS1, AtFLS2, AtFLS3, AtFLS4, and AtFLS5 in H. schachtii and M. incognita infection of Arabidopsis were explored further using Atfls T-DNA knockout lines and wild type plants. The number of cyst females in Atfls mutant plants was not significantly different from those developed on the corresponding wild type ecotypes, with the exception of a significant increase in developed cyst nematodes on the Atfls1 mutant (Figure 5A). These results suggest that AtFLS1 plays a role in H. schachtii infection of Arabidopsis roots, while the other four AtFLS genes have little or no effect. A similar significant increase was observed for the Atfls1 and Atfls5 mutants in the number of root-knot nematode galls, with the remainder of the Atfls mutants showing no difference in the number of galls compared to wild-type plants (Figure 5B). This suggests that both AtFLS1 and AtFLS5 may play a role in M. incognita infection.
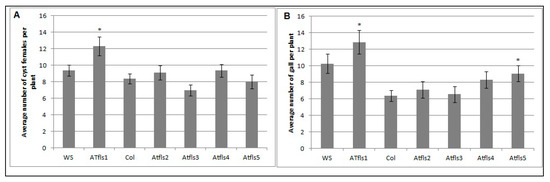
Figure 5.
The effects of Atfls T-DNA mutations on H. schachtii (A) and M. incoinita (B) infection. Atfls1, Atfls2, Atfls3, Atfls4, and Atfls5 knock-outs, two wild-type Arabidopsis plants; Col-0 and WS, were planted on modified Knop’s medium and 10-day-old seedlings were inoculated with approximately 60 surface sterilized J2 of either H. schachtii or M. incognita. Three weeks after inoculation, the number of cyst female nematodes (for sugar beet cyst nematode) per root system or galls (for southern root-knot nematode) were counted. Data are presented as means ± standard errors. Mean values significantly different from the wild type ecotype as determined by t test (p < 0.05). Similar results were obtained from a second, independent biological replicate.
2.5. Expression of AtFLS2, 3, 4, and 5 Is Dependent on AtMYB12
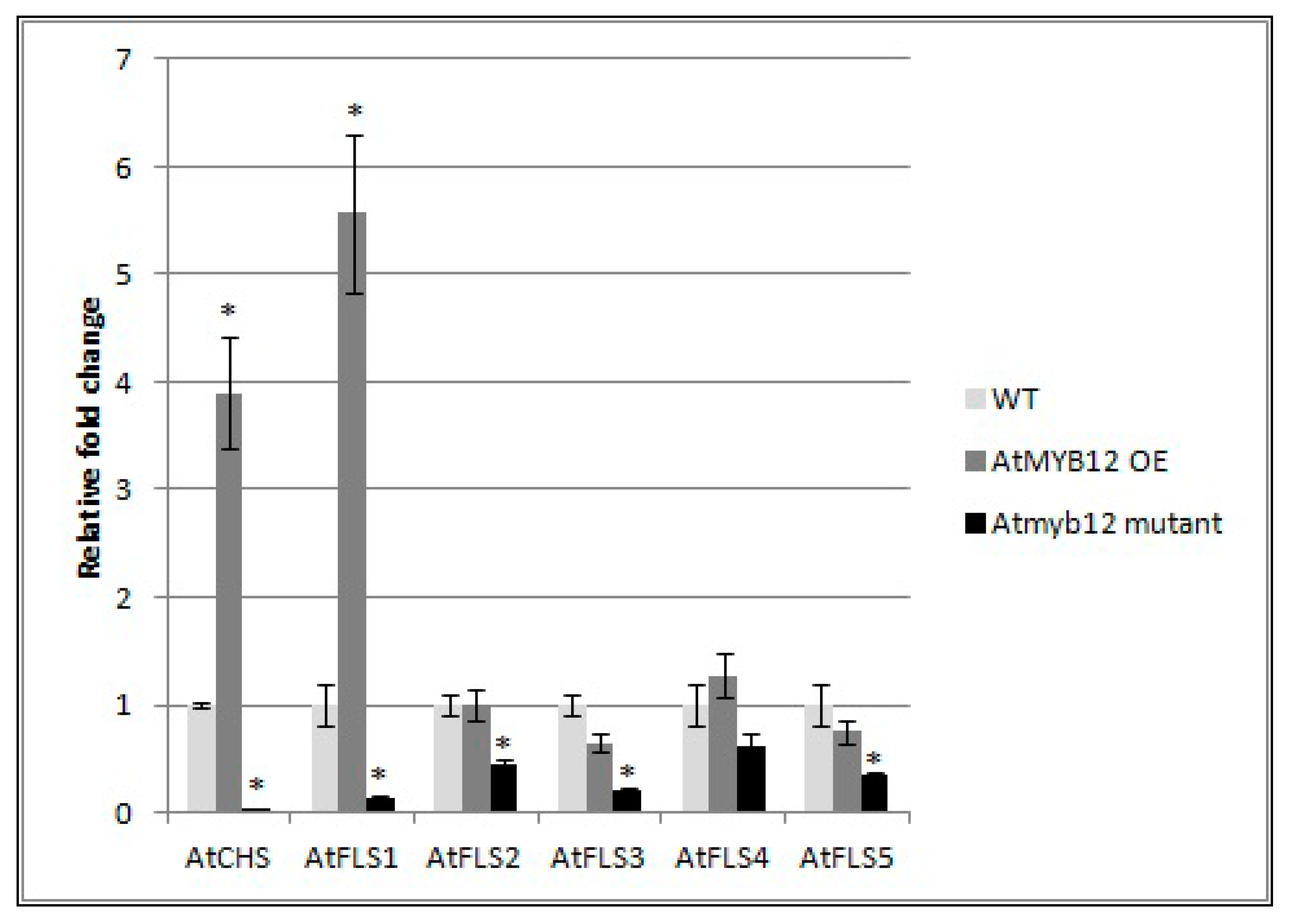
To determine whether AtMYB also affects the expression of the other four AtFLS genes, qRT-PCR was used to compare the mRNA transcript levels of AtFLS2, AtFLS3, AtFLS4, and AtFLS5 in Arabidopsis plants that over-express AtMYB12 as well as in Atmyb12 mutant plants. AtFLS6, which does not appear to be expressed, was not included in this analysis. We also measured the expression levels of AtCHS and AtFLS1 transcripts as controls. Our results did confirm the finding of Mehrtens et al. [48], that the over-expression of AtMYB12 induced expression of AtCHS and AtFLS1, while Atmyb12 knockout reduced AtCHS and AtFLS1 transcript levels to almost undetectable levels (Figure 6). In addition, while over-expression of AtMYB12 had little or no effect on the transcript levels of the other four FLS genes, the Atmyb12 knockout significantly reduced the expression of AtFLS2, AtFLS3, and AtFLS5, with a slight decrease also observed in expression of AtFLS4 (Figure 6), indicating that AtFLS2, 3, 4, and 5 expression is dependent on AtMYB12.
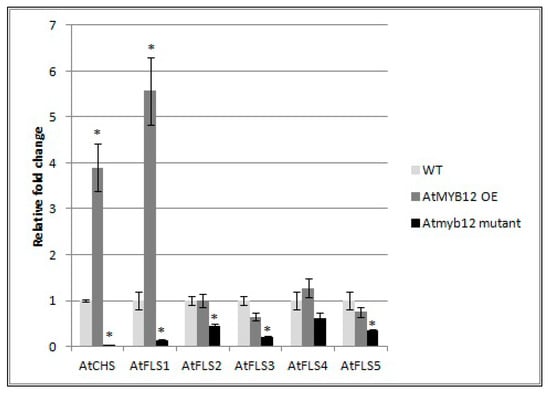
Figure 6.
Effect of AtMYB12 on the expression of AtFLS genes. Expression of AtCHS and AtFLS1, AtFLS2, AtFLS3, AtFLS4, and AtFLS5 in plants overexpressing AtMYB12, Atmyb122 knockout, and wild-type Col-0 (WT) seedlings using quantitative real-time PCR (qRT-PCR). The presented data are the mean fold changes ± standard errors in gene transcript levels relative to uninfected control tissue (baseline set at 1.0). Asterisks (*) indicate that the mean fold change is significantly different from 1.0 as determined by t test (p < 0.05). Similar results were obtained from a second, independent biological replicate.
3. Discussion
Flavonoids are natural plant products involved in diverse plant functions. A previous study using a flavonoid-specific stain showed that these compounds are produced in and around the developing syncytia of H. schachtii and galls of the European dagger nematode, Xiphinema diversicaudatum, respectively [22]. However, a detailed analysis of the expression of flavonoids biosynthesis genes during cyst and root-knot nematode parasitism has not previously been conducted. In this study, we examined the expression patterns of R2R3-MYB12, a flavonol-specific transcription factor; AtCHS, which encodes the first enzyme in the flavonoid pathway; and five flavonol synthase genes (AtFLS1, AtFLS2, AtFLS3, AtFLS4, and AtFLS5) during parasitism by H. schachtii and M. incognita and at the nematode-induced feeding sites. The role of these genes in nematode parasitism was investigated using T-DNA knockouts and over-expression lines.
Quantitative RT-PCR was first used to quantify AtMYB12 transcripts in H. schachtii and M. incognita-infected Arabidopsis roots. It must be noted that only one reference gene (Actin 8) was used in qRT-PCR [51] and that small differences in expression observed among the data are subject to potential variations in reference expression [52,53]. H. schachtii appears to induce AtMYB12 expression in Arabidopsis roots between 5–9 dpi. Similar to this finding, the genome-wide expression analysis of soybean roots infected with soybean cyst nematode showed that a soybean homolog of AtMYB12 (BE0240360) is also upregulated in infected roots [11].
A histochemical analysis of AtMYB12p::GUS lines revealed that expression of AtMYB12 increases in H. schachtii-formed syncytia, suggesting a role of this gene in syncytia formation. Previous studies on the expression of AtMYB12 in syncytia have generated inconsistent results. Szakasits et al. [12] examined gene expression in H. schachtii-formed syncytia using Affymetrix GeneChips and showed that AtMYB12 is downregulated in syncytia. In contrast, transcript profiling of cyst nematode feeding cells in soybean roots showed the soybean homolog of AtMYB12 to be upregulated in syncytia [11]. Using the AtMYB12p:GUS lines offered the advantage of visualizing promoter activity within nematode-induced syncytia without the need for mRNA extraction, suggesting a role of this transcription factor in syncytia formation and/or maintenance.
The role of the transcription factor AtMYB12 in syncytia formation and development is still unclear, but AtMYB12 may induce the expression of downstream genes necessary for the formation and/or maintenance syncytia, such as genes involved in auxin transport. In fact, several auxin-responsive genes are modulated in tobacco plants over-expressing AtMYB12, including downregulation of an auxin efflux carrier [54,55]. Studies on the role of auxin in host-nematode interaction have suggested that changes in auxin levels, possibly achieved by a disruption of local auxin transport, may be important in feeding site formation of cyst nematodes [31,32,56]. A role for auxin in nematode feeding site development is supported by the finding that the cyst nematode effector protein 19C07 interacts specifically with the auxin influx transporter LAX3 [55], possibly to enhance auxin influx into host roots for syncytium development. The expression of AtMYB12 in syncytia may be necessary to modulate host cell metabolism in order to increase the demands of the flux of amino-acids and macromolecules needed for nematode feeding. This latter hypothesis is supported by the increased expression of genes related to carbohydrates and lipid metabolism and the accumulation of amino-acids that have been reported to occur in tobacco plants over-expressing AtMYB12 gene [54]. The reduction in the number of cyst females developing in Arabidopsis Atmyb12 mutant plants compared to wild-type plants further supports the role of AtMYB12 in cyst nematode infection. However, overexpression of AtMYB12 did not increase the susceptibility of Arabidopsis to H. schachtii.
On the other hand, infection with M. incognita decreased the expression of AtMYB12 in the nematode-induced galls, suggesting that downregulation of AtMYB12 may be necessary in establishing giant cells needed for nematode’s survival. The exact role of AtMYB12 downregulation in nematode-induced galls in still unclear, but M. incognita may downregulate the expression of AtMYB12 gene to limit expression of target genes that may interfere with giant cell formation and maintenance. However, the identity of these genes is still unknown and there is little information available on the genes, other than those involved in flavonoid biosynthesis, affected in AtMYB12 knockout lines. The hypothesis that the downregulation of AtMYB12 gene promotes giant cell formation is further supported by the finding that plants impaired in AtMYB12 gene expression showed hyper-susceptibility to infection by M. incognita compared to wild-type plants. The observation that a single gene (AtMYB12) appears to play distinct roles in H. schachtii and M. incognita feeding site development is interesting but not surprising, as these two different nematodes secrete different effector proteins into host root cells [6,57,58], and regulate the expression of distinct genes for the formation and establishment of the feeding sites [7,8,12].
The expression of AtMYB12 in H. schachtii-induced syncytia was not accompanied by an increase in AtFLS1 and AtCHS gene expression, suggesting that, in syncytia, the expression of these two genes may not be under the control of AtMYB12. The observed downregulation of AtFLS1, which has substantial enzymatic activity among the AtFLS genes, in the feeding sites of both H. schachtii and M. incognita is surprising since flavonols are known to have potent free radical-scavenging activity [25] and it has been reported that ROS are produced in both syncytia and giant cells [59,60]. Downregulation of AtFLS1 and AtCHS gene expression in syncytia has been previously reported in studies of syncytia transcriptome [12]. However, these two genes have not been identified to be differentially regulated in M. incognita-formed giant cell in Arabidopsis at least at very early infection stages [7].
Downregulation of key genes in flavonol biosynthesis (CHS and FLS1) at the nematode feeding structures may be necessary for feeding site formation and/or maintenance, perhaps related to the role of flavonols in modulating auxin transport [61]. A study on the effect of selected flavonoid compounds on the behavior of M. incognita showed that flavonols and chalcone, which are the products of FLS1 and CHS, respectively, have inhibitory effects on motility and the hatching of nematodes [33]. The importance of AtFLS1 downregulation in syncytia and giant cells formation was further supported by the hyper-susceptibility of an AtFLS1-deficient line. Atfls1 T-DNA knockout lines appear also to be more susceptible to both H. schachtii and M. incognita compared to wild type plants. The downregulation of AtFLS1 in nematode feeding sites, either directly or indirectly, may reduce potential defense responses from the flavonol pathway that could inhibit successful parasitism by nematodes.
4. Materials and Methods
4.1. Nematode Culture
Cyst nematodes of H. schachtii (BCN) and M. incognita (RKN) were cultured on roots of cabbage plants (Brassica oleracea var. capitata) and tomato plants (Solanum lycopersicon cv. Rutgers) grown in soil, respectively. Eggs of H. schachtii were collected from crushed cysts as previously described for cyst species [62], while eggs of M. incognita were extracted from tomato roots with sodium hypochlorite as previously described [63]. Eggs were hatched over water in Baermann pans at 28 °C for 48 h, and the hatched preparasitic J2 (pre-J2) were collected, surface-sterilized by incubation for 10 min in sterilization solution (0.004% mercuric chloride, 0.004% sodium azide and 0.002% Triton X-100), and rinsed three times with sterile distilled water.
4.2. Nematode Infection Assay and Data Collection
Arabidopsis thaliana (Columbia and Wassilewskija ecotypes) seeds were surface sterilized and transferred (one seed per well) into six-well culture plates (Falcon) containing 6 mL of sterile modified Knops medium [64] solidified with 0.8% Daishin agar (Brunschwig Chemie) as previously described [8]. Seeds on plates were placed in a 24 °C growth chamber under a 16 h light/8 h dark cycle for 2 weeks. Surface sterilized pre-J2 nematodes were suspended in 1.5% low-melting-point agarose to allow even distribution and to facilitate their movement into solid Knops medium. Plants were inoculated with approximately 60 J2 per plant and developed cysts (for sugar beet cyst nematodes) and galls (for root-knot nematodes) were counted 3–4 weeks post-infection, using a dissecting microscope. Nematode infection assays were conducted on two independent biological replicates, and similar results were obtained (the data of one experiment is presented). The means and standard errors of 18 replicates per treatment were calculated. Statistical differences were determined by the paired t-test with an alpha level of 0.05 using SAS software (Cary, NC, USA).
4.3. Plant Material
Arabidopsis lines containing AtCHSp::GUS and AtMYB12P::GUS were previously described [48] and were kindly provided by Dr. Bernd Weisshaar of the Max Planck Institute, Cologne, Germany. The AtMYB12 T-DNA insertion line (Salk_046675C, insertion in third exon) and AtMYB12 ectopic over-expression line (35S-MYB12, CS9603; [48] were obtained from the Arabidopsis Biological Resource Center (ABRC, The Ohio State University, Columbus, OH, USA). AtCHSp::GUS, AtFLSp::GUS, and T-DNA insertion lines for AtFLS1, 2, 3, 4, and 5 were previously described [34,65]. The T-DNA lines were AJ588535 (insertion in the 5′ untranslated region of AtFLS1), SALK_076420 (AtFLS1 promoter), GABI 429B10 (second intron of AtFLS2), SALK_050041 (third exon of AtFLS3), SALK_002309 (third exon of AtFLS4), and GABI 317E12 (first intron of AtFLS5).
4.4. Histochemical Localization of GUS
Arabidopsis roots infected with H. schachtii and M. incognita were excised 9 dpi. Histochemical staining for GUS expression was performed at 37 °C for 4 h using X-Gluc solution [(0.1 M NaH2PO4, 10 mM EDTA, 0.5 mM each of K3Fe(CN)6 and K4F2(CN)6, 3H2O, 0.1% Triton X-100 and 1 mg/mL 5-bromo-4-chloro-3-indolyl-beta-D-glucuronic acid (cyclohexylammonium salt) (Gold Biotechnology, St. Louis, MO, USA)], and then mounted onto glass slides. Samples were examined using a Nikon SMZ 800 stereo microscope, and images were captured with a SPOT 2 digital camera (Diagnostic Instruments, Inc., Sterling Heights, MI, USA).
4.5. RNA Isolation and Quantitative RT-PCR
Total RNA was isolated from whole roots of six Arabidopsis plants using the RNeasy Plant Mini Kit (QIAgen, Valencia, CA, USA) following the manufacturer’s instructions. Prior to quantitative RT-PCR, total RNA was treated with RNase-free DNase I (Ambion, Austin, TX, USA) to eliminate any contaminating genomic DNA. First-strand cDNA was synthesized from 2–3 µg of total RNA using SuperScript-II RT (Invitrogen, Carlsbad, CA, USA) and oligo-dT18 primers following the manufacturer’s instructions.
All reactions were performed in a DNA Engine Mx3000P (Agilent Technologies, Santa Clara, CA, USA). Each 20 μL qRT-PCR reaction contained 1X Brilliant II SYBR Green qPCR Master Mix (Agilent Technologies, Santa Clara, CA, USA), 5 μL cDNA template and 5 μM each forward and reverse primers (Table 1). The PCR cycling parameters were set at 95 °C for 10 min followed by 40 cycles of 95 °C for 15 s, 60 °C for 1 min, and 72 °C for 1 min. At the completion of each reaction, dissociation melt curve analyses (60–90 °C every 0.5 °C for 1 s) were conducted to discount the effects of primer-dimer formation and contamination. The qRT-PCR reactions were performed in three technical triplicates. Using the 2−ΔΔCT method [66], the data are presented as the fold change in gene expression normalized against the endogenous Arabidopsis actin 8 gene (At1g49240), and presented relative to expression at 0 dpi. qRT-PCR experiments were conducted on two independent biological replicates and similar results were obtained (the data of one experiment was presented). Values are means ± SE (n = 3 technical replicates). One-way ANOVA with a Student–Newman–Keuls post hoc test was used to determine significant differences between means.

Table 1.
Primers used in Real-time PCR.
Author Contributions
N.H., B.S.J.W., and E.L.D. were responsible for project conception and design. N.H., B.S.J.W., C.L., contributed to technical design of experiments and E.L.D. provided project support. N.H., B.S.J.W., C.L., and E.L.D. contributed to data analyses and preparation of the report for publication. All authors have read and agreed to the published version of the manuscript.
Funding
This work was funded by the US Department of Agriculture National Research Initiative (grant no. 2006-35607-16601, and grant no. 2008-35302-18824), and by the United Soybean Board (project no. 7214).
Conflicts of Interest
The authors declare no conflict of interest.
References
- Chitwood, D.J. Research on plant-parasitic nematode biology conducted by the United States Department of Agriculture-Agricultural Research Service. Pest Manag. Sci. 2003, 59, 748–753. [Google Scholar] [CrossRef] [PubMed]
- Davis, E.L.; Mitchum, M.G. Nematodes: Sophisticated parasites of legumes. Plant Physiol. 2005, 137, 1182–1188. [Google Scholar] [CrossRef] [PubMed]
- Jones, M.G.K. The development and function of plant cells modified by endoparasitic nematodes. In Plant Parasitic Nematodes; Zuckerman, B.M., Rohde, R.A., Eds.; Academic Press: New York, NY, USA, 1981; Volume 3, pp. 265–279. [Google Scholar]
- Wyss, U.; Grundler, F.M.W.; Munch, A. The parasitic behaviour of second-stage juveniles of Meloidogyne incognita in roots of Arabidopsis thaliana. Nematologica 1992, 38, 98–111. [Google Scholar]
- Golinowski, W.; Grundler, F.; Sobczak, M. Changes in the structure of Arabidopsis thaliana during female development of the pant-parasitic nematode Heterodera schachtii. Protoplasma 1996, 194, 103–116. [Google Scholar] [CrossRef]
- Mitchum, M.G.; Hussey, R.S.; Baum, T.J.; Wang, X.; Elling, A.A.; Wubben, M.; Davis, E.L. Nematode effector proteins: An emerging paradigm of parasitism. New Phytol. 2013, 199, 879–894. [Google Scholar] [CrossRef] [PubMed]
- Barcala, M.; Garcia, A.; Cabrera, J.; Casson, S.; Lindsey, K.; Favery, B.; Garcia-Casado, G.; Solano, R.; Fenoll, C.; Escobar, C. Early transcriptomic events in microdissected Arabidopsis nematode-induced giant cells. Plant J. 2010, 61, 698–712. [Google Scholar] [CrossRef]
- Jammes, F.; Lecomte, P.; Almeida-Engler, J.; Bitton, F.; Martin-Magniette, M.L.; Renou, J.P.; Abad, P.; Favery, B. Genome-Wide expression profiling of the host response to root-knot nematode infection in Arabidopsis. Plant J. 2005, 44, 447–458. [Google Scholar] [CrossRef]
- Puthoff, D.P.; Nettleton, D.; Rodermel, S.R.; Baum, T.J. Arabidopsis gene expression changes during cyst nematode parasitism revealed by statistical analyses of microarray expression profiles. Plant J. 2003, 33, 911–921. [Google Scholar] [CrossRef]
- Alkharouf, N.W.; Klink, V.P.; Choukha, I.B.; Beard, H.S.; McDonald, M.H.; Meyer, S.; Knap, H.T.; Khan, R.; Mathews, B.F. Timecourse microarray analyses reveal global changes in gene expression of susceptible Glycine max (soybean) roots during infecton y Heterodera glycines (soybean cyst nematode). Planta 2006, 224, 838–852. [Google Scholar] [CrossRef]
- Ithal, N.; Recknor, J.; Nettleton, D.; Maier, T.; Baum, T.J.; Mitchum, M.G. Developmental transcript profiling of cyst nematode feeding cells in soybean roots. Mol. Plant Microbe Interact. 2007, 20, 510–525. [Google Scholar] [CrossRef]
- Szakasits, D.; Heinen, P.; Wieczorek, K.; Hofmann, J.; Wagner, F.; Kreil, D.P.; Sykacek, P.; Grundler, F.M.W.; Bohlmann, H. The transcriptome of syncytia induced by the cyst nematode Heterodera schachtii in Arabidopsis roots. Plant J. 2009, 57, 771–784. [Google Scholar] [CrossRef] [PubMed]
- Ververidis, F.; Trantas, E.; Douglas, C.; Vollmer, G.; Kretzschmar, G.; Panopoulos, N. Biotechnology of flavonoids and other phenylpropanoid-derived natural products. Part I: Chemical diversity, impacts on plant biology and human health. Biotech. J. 2007, 2, 1214–1234. [Google Scholar] [CrossRef]
- Izaguirre, M.M.; Mazza, C.A.; Svatos, A.; Baldwin, I.T.; Ballare, C.L. Solar ultraviolet-B radiation and insect herbivory trigger partially overlapping phenolic responses in Nicotiana attenuata and Nicotiana longiflora. Ann. Bot. 2007, 99, 103–109. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, Y.; Sasaki, N.; Ohmiya, A. Biosynthesis of plant pigments: Anthocyanins, betalains and carotenoids. Plant J. 2008, 54, 733–749. [Google Scholar] [CrossRef]
- Christensen, A.B.; Gregersen, P.L.; Schroder, J.; Collinge, D.B. A chalcone synthase with an unusual substrate preference is expressed in barley leaves in response to UV light and pathogen attack. Plant Mol. Biol. 1998, 37, 849–857. [Google Scholar] [CrossRef]
- Schmid, J.; Doerner, P.W.; Clouse, S.D.; Dixon, R.A.; Lamb, C.J. Developmental and environmental-regulation of a bean chalcone synthase promoter in transgenic tobacco. Plant Cell 1990, 2, 619–631. [Google Scholar]
- Shirley, B.W. Flavonoid biosynthesis: ‘New’ functions for an ‘old’ pathway. Trends Plant Sci. 1996, 1, 377–382. [Google Scholar]
- Treutter, D. Significance of flavonoids in plant resistance and enhancement of their biosynthesis. Plant Biol. 2005, 7, 581–591. [Google Scholar] [CrossRef]
- Lu, Y.; Chen, Q.; Bu, Y.; Luo, R.; Hao, S.; Zhang, J.; Tian, J.; Yao, Y. Flavonoid accumulation plays an important role in the rust resistance of Malus plant leaves. Front. Plant Sci. 2017, 8, 1286. [Google Scholar] [CrossRef]
- Chin, S.; Behm, C.A.; Methesius, U. Functions of flavonoids in plant-nematode interactions. Plants 2018, 7, 85. [Google Scholar] [CrossRef]
- Jones, J.T.; Furlanetto, C.; Phillips, M.S. The role of flavonoids produced in response to cyst nematode infection of Arabidopsis thaliana. Nematology 2007, 9, 671–677. [Google Scholar]
- Sugiyama, A.; Yazaki, K. Favonoids in plant rhizospheres: Secretion, fate and their effects on biological communication. Plant Biotech. 2014, 31, 431–443. [Google Scholar] [CrossRef]
- Stafford, H.A. Flavonoid evolution—An enzymatic approach. Plant Physiol. 1991, 96, 680–685. [Google Scholar] [CrossRef] [PubMed]
- Braca, A.; Fico, G.; Morelli, I.; De Simone, F.; Tome, F.; De Tommasi, N. Antioxidant and free radical scavenging activity of flavonol glycosides from different Aconitum species. J. Ethnopharmacol. 2003, 86, 63–67. [Google Scholar] [CrossRef]
- Simmonds, M.S.J. Flavonoid-Insect interactions: Recent advances in our knowledge. Phytochemistry 2003, 64, 21–30. [Google Scholar] [CrossRef]
- Hoffmann-Campo, C.B.; Ramos, J.A.; de Oliveira, M.C.N.; Oliveira, L.J. Detrimental effect of rutin on Anticarsia gemmatalis. Pesqui. Agropecu. Bras. 2006, 41, 1453–1459. [Google Scholar] [CrossRef]
- Buer, C.S.; Muday, G.K. The transparent testa4 mutation prevents flavonoid synthesis and alters auxin transport and the response of Arabidopsis roots to gravity and light. Plant Cell 2004, 16, 1191–1205. [Google Scholar] [CrossRef]
- Maloney, G.; DiNapoli, K.; Muday, G. The anthocyanin reduced tomato mutant demonstrates the role of flavonols in tomato lateral root and root hair development. Plant Physiol. 2014, 166, 614–631. [Google Scholar] [CrossRef]
- Gheysen, G.; Mitchum, M.G. How nematodes manipulate plant development pathways for infection. Curr. Opin. Plant Biol. 2011, 14, 415–421. [Google Scholar] [CrossRef]
- Goverse, A.; Overmars, H.; Engelbertink, J.; Schots, A.; Bakker, J.; Helder, J. Both induction and morphogenesis of cyst nematode feeding cells are mediated by auxin. Mol. Plant Microbe Interact. 2000, 13, 1121–1129. [Google Scholar] [CrossRef]
- Karczmarek, A.; Overmars, H.; Helder, J.; Goverse, A. Feeding cell development by cyst and root-knot nematodes involves a similar early, local and transient activation of a specific auxin-inducible promoter element. Mol. Plant Pathol. 2004, 5, 343–346. [Google Scholar] [CrossRef] [PubMed]
- Wuyts, N.; Swennen, R.; De Waele, D. Effects of plant phenylpropanoid pathway products and selected terpenoids and alkaloids on the behaviour of the plant-parasitic nematodes Radopholus similis, Pratylenchus penetrans and Meloidogyne incognita. Nematology 2006, 8, 89–101. [Google Scholar] [CrossRef]
- Owens, D.K.; Alerding, A.B.; Crosby, K.C.; Bandara, A.B.; Westwood, J.H.; Winkel, B.S.J. Functional analysis of a predicted flavonol synthase gene family in Arabidopsis. Plant Physiol. 2008, 147, 1046–1061. [Google Scholar] [CrossRef]
- Wisman, E.; Hartmann, U.; Sagasser, M.; Baumann, E.; Palme, K.; Hahlbrock, K.; Saedler, H.; Weisshaar, B. Knock-Out mutants from an En-1 mutagenized Arabidopsis thaliana population generate phenylpropanoid biosynthesis phenotypes. Proc. Natl. Acad. Sci. USA 1998, 95, 12432–12437. [Google Scholar] [CrossRef] [PubMed]
- Preuss, A.; Stracke, R.; Weisshaar, B.; Hillebrecht, A.; Matern, U.; Martens, S. Arabidopsis thaliana expresses a second functional flavonol synthase. FEBS Lett. 2009, 583, 1981–1986. [Google Scholar] [CrossRef] [PubMed]
- Higginson, T.; Li, S.F.; Parish, R.W. AtMYB103 regulates tapetum and trichome development in Arabidopsis thaliana. Plant J. 2003, 35, 177–192. [Google Scholar] [CrossRef]
- Lee, M.M.; Schiefelbein, J. WEREWOLF, a MYB-related protein in Arabidopsis, is a position-dependent regulator of epidermal cell patterning. Cell 1999, 99, 473–483. [Google Scholar] [CrossRef]
- Shikazono, N.; Tanaka, A.; Yokota, Y.; Watanabe, H.; Tano, S. Nucleotide sequence of the GLABROUS1 gene of Arabidopsis thaliana ecotype Columbia. DNA Seq. 1998, 9, 177–181. [Google Scholar] [CrossRef]
- Bender, J.; Fink, G.R. A Myb homologue, ATR1, activates tryptophan gene expression in Arabidopsis. Proc. Natl. Acad. Sci. USA 1998, 95, 5655–5660. [Google Scholar] [CrossRef]
- Urao, T.; Yamaguchishinozaki, K.; Urao, S.; Shinozaki, K. An Arabidopsis myb homolog is induced by dehydration stress and its gene-product binds to the conserved MYB recognition sequence. Plant Cell 1993, 5, 1529–1539. [Google Scholar]
- Hirayama, T.; Shinozaki, K. A cdc5+ homolog of a higher plant, Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 1996, 93, 13371–13376. [Google Scholar] [CrossRef] [PubMed]
- Baudry, A.; Heim, M.A.; Dubreucq, B.; Caboche, M.; Weisshaar, B.; Lepiniec, L. TT2, TT8, and TTG1 synergistically specify the expression of BANYULS and proanthocyanidin biosynthesis in Arabidopsis thaliana. Plant J. 2004, 39, 366–380. [Google Scholar] [CrossRef] [PubMed]
- Borevitz, J.O.; Xia, Y.J.; Blount, J.; Dixon, R.A.; Lamb, C. Activation tagging identifies a conserved MYB regulator of phenylpropanoid biosynthesis. Plant Cell 2000, 12, 2383–2393. [Google Scholar] [CrossRef] [PubMed]
- Penfield, S.; Meissner, R.C.; Shoue, D.A.; Carpita, N.C.; Bevan, M.W. MYB61 is required for mucilage deposition and extrusion in the Arabidopsis seed coat. Plant Cell 2001, 13, 2777–2791. [Google Scholar] [CrossRef] [PubMed]
- Daniel, X.; Lacomme, C.; Morel, J.B.; Roby, D. A novel myb oncogene homologue in Arabidopsis thaliana related to hypersensitive cell death. Plant J. 1999, 20, 57–66. [Google Scholar] [CrossRef]
- Luo, J.; Butelli, E.; Hill, L.; Parr, A.; Niggeweg, R.; Bailey, P.; Weisshaar, B.; Martin, C. AtMYB12 regulates caffeoyl quinic acid and flavonol synthesis in tomato: Expression in fruit results in very high levels of both types of polyphenol. Plant J. 2008, 56, 316–326. [Google Scholar] [CrossRef]
- Mehrtens, F.; Kranz, H.; Bednarek, P.; Weisshaar, B. The Arabidopsis transcription factor MYB12 is a flavonol-specific regulator of phenylpropanoid biosynthesis. Plant Physiol. 2005, 138, 1083–1096. [Google Scholar] [CrossRef]
- Stracke, R.; Ishihara, H.; Barsch, G.H.A.; Mehrtens, F.; Niehaus, K.; Weisshaar, B. Differential regulation of closely related R2R3-MYB transcription factors controls flavonol accumulation in different parts of the Arabidopsis thaliana seedling. Plant J. 2007, 50, 660–677. [Google Scholar] [CrossRef]
- Feinbaum, R.L.; Ausubel, F.M. Transcriptional regulation of the Arabidopsis thaliana chalcone synthase gene. Mol. Cell. Biol. 1988, 8, 1985–1992. [Google Scholar] [CrossRef]
- Bustin, A.S.; Benes, V.; Garson, A.J.; Hellemans, J.; Hugett, J.; Kubsta, M.; Mueller, R.; Nolan, T.; Pfaffl, W.M.; Shipley, L.G.; et al. The MIQE guidelines: Minimum information for publication of quantitative Real-Time PCR experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef]
- Verma, A.; Lee, C.; Morriss, S.; Odu, F.; Kenning, C.; Rizzo, N.; Spollen, W.G.; Lin, M.; McRae, A.G.; Givan, S.A.; et al. The novel cyst nematode effector protein 30D08 targets host nuclear functions to alter gene expression in feeding sites. New Phytol. 2018, 219, 697–713. [Google Scholar] [CrossRef] [PubMed]
- Zhao, D.; You, Y.; Fan, H.; Zhu, X.; Wang, Y.; Duan, Y.; Xuan, Y.; Chen, L. The role of sugar transporter genes during early infection of root-knot nematodes. Int. J. Mol. Sci. 2018, 19, 302. [Google Scholar] [CrossRef]
- Misra, P.; Pandey, A.; Tiwari, M.; Chandrashekar, K.; Sidhu, O.P.; Asif, M.H.; Chakrabarty, D.; Singh, P.K.; Trivedi, P.K.; Nath, P.; et al. Modulation of transcriptome and metabolome of tobacco by Arabidopsis transcription factor, AtMYB12, leads to insect resistance. Plant Physiol. 2010, 152, 2258–2268. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.; Chronis, D.; Kenning, C.; Peret, B.; Hewezi, T.; Davis, E.L.; Baum, T.J.; Hussey, R.; Bennett, M.; Mitchum, M.G. The novel cyst nematode effector protein 19C07 interacts with the Arabidopsis auxin influx transporter LAX3 to control feeding site development. Plant Physiol. 2011, 155, 866–880. [Google Scholar] [CrossRef] [PubMed]
- Grunewald, W.; Cannoot, B.; Friml, J.; Gheysen, G. Parasitic nematodes modulate PIN-mediated auxin transport to facilitate infection. PLoS Pathog. 2009, 5, e1000266. [Google Scholar] [CrossRef]
- Gao, B.L.; Allen, R.; Maier, T.; Davis, E.L.; Baum, T.J.; Hussey, R.S. The parasitome of the phytonematode Heterodera glycines. Mol. Plant Microbe Interact. 2003, 16, 720–726. [Google Scholar] [CrossRef]
- Huang, G.Z.; Gao, B.L.; Maier, T.; Allen, R.; Davis, E.L.; Baum, T.J.; Hussey, R.S. A profile of putative parasitism genes expressed in the esophageal gland cells of the root-knot nematode Meloidogyne incognita. Mol. Plant Microbe Interact. 2003, 16, 376–381. [Google Scholar] [CrossRef]
- Siddique, S.; Matera, C.; Radakovic, Z.S.; Shamim Hasan, M.; Gutbrod, P.; Rozanska, E.; Sobczak, M.; Angel Torres, M.; Grundler, F.M. Parasitic worms stimulate host NADPH oxidases to produce reactive oxygen species that limit plant cell death and promote infection. Sci. Signal. 2014, 7, ra33. [Google Scholar] [CrossRef]
- Melillo, M.T.; Leonetti, P.; Bongiovanni, M.; Castagnone-Sereno, P.; Bleve-Zacheo, T. Modulation of reactive oxygen species activities and H2O2 accumulation during compatible and incompatible tomato-root-knot nematode interactions. New Phytol. 2006, 170, 501–512. [Google Scholar] [CrossRef]
- Brown, D.E.; Rashotte, A.M.; Murphy, A.S.; Tague, B.W.; Peer, W.A.; Taiz, L.; Muday, G.K. Flavonoids act as negative regulators of auxin transport in vivo in Arabidopsis. Plant Physiol. 2001, 126, 524–535. [Google Scholar] [CrossRef]
- Goellner, M.; Wang, X.H.; Davis, E.L. Endo-beta-1,4-glucanase expression in compatible plant-nematode interactions. Plant Cell 2001, 13, 2241–2255. [Google Scholar] [CrossRef] [PubMed]
- Hussey, R.S.; Barker, K.R. A comparison of methods of collecting inocula of Meloidogyne spp., including a new technique. Plant Dis. Report. 1973, 57, 1025–1028. [Google Scholar]
- Sijmons, P.C.; Grundler, F.M.W.; Vonmende, N.; Burrows, P.R.; Wyss, U. Arabidopsis thaliana as a new model host for plant-parasitic nematodes. Plant J. 1991, 1, 245–254. [Google Scholar] [CrossRef]
- Feinbaum, R.L.; Storz, G.; Ausubel, F. High intensity and blue light regulated expression of chimeric chalcone synthase genes in transgenic Arabidopsis thaliana plants. Mol. Gen. Genet. 1991, 226, 449–456. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).