Characterization of Trehalose-6-phosphate Synthase and Trehalose-6-phosphate Phosphatase Genes and Analysis of its Differential Expression in Maize (Zea mays) Seedlings under Drought Stress
Abstract
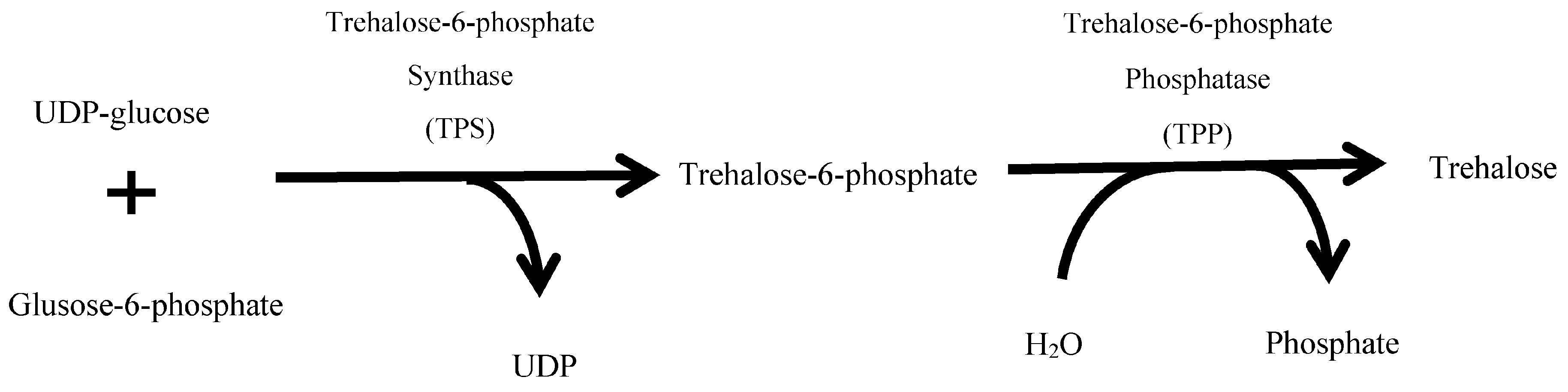
:1. Introduction
2. Results
2.1. Cloning and Characterization of TPS and TPP cDNA
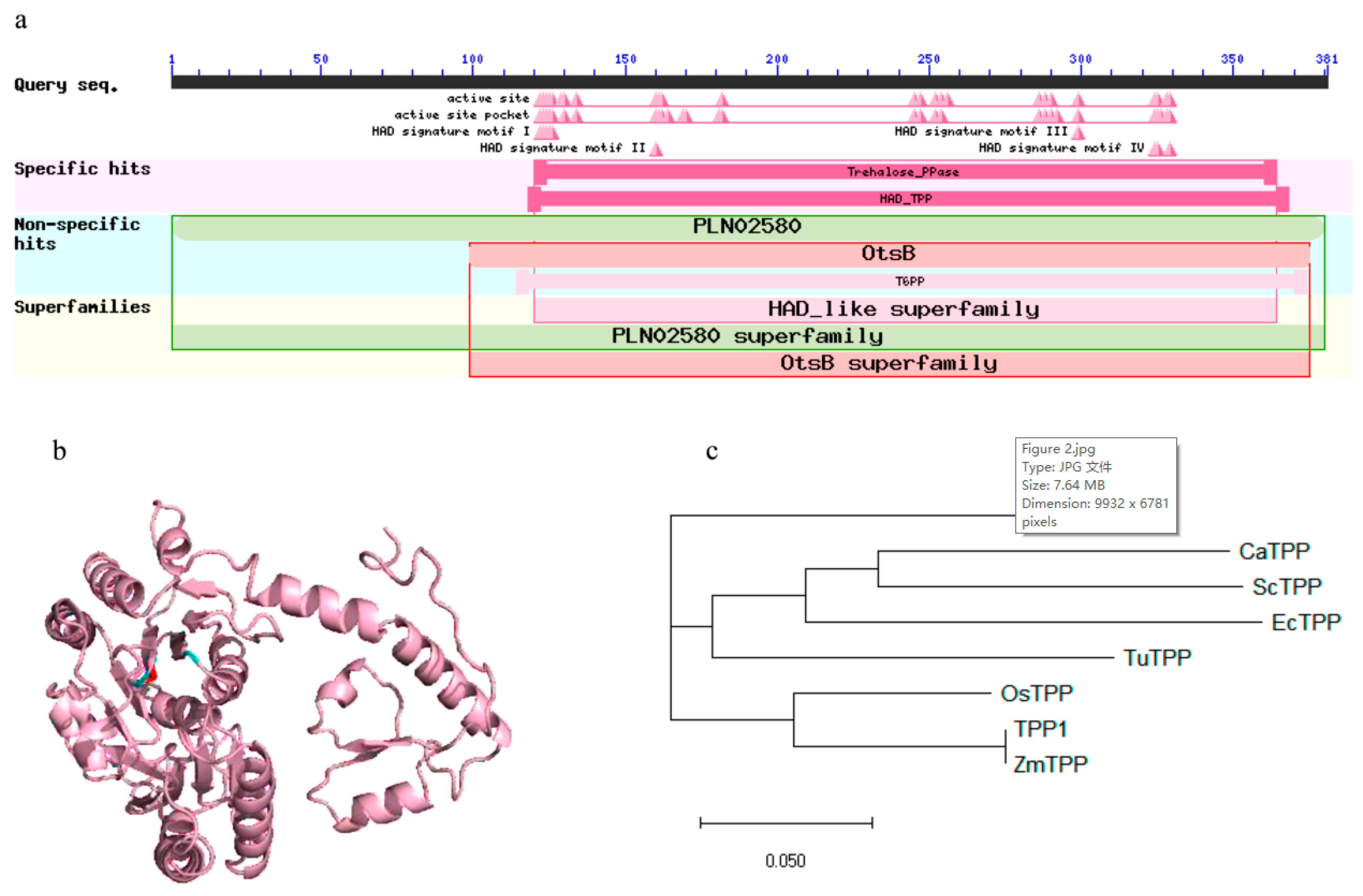
2.2. Amino Acid Sequence Analysis and Phylogenetic Relationship
2.3. In Silico Analysis of Catalytic Sites
2.4. Real-Time Quantitative PCR
3. Discussion
4. Materials and Methods
4.1. Plant Material, Growth Conditions and Stress Treatment
4.2. Watering Treatments
4.3. RNA Extraction and cDNA Synthesis
4.4. Cloning TPS/TPP cDNA and Sequence Analysis
4.5. Real-Time Quantitative PCR
4.6. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Wingler, A. The Function of Trehalose Biosynthesis in Plants. Phytochemistry 2002, 60, 437–440. [Google Scholar] [CrossRef]
- Mostafa, M.R.; Mervat, S.S.; Safaa, R.E.-L.; Ebtihal, M.A.E.; Magdi, T.A. Exogenous α-Tocopherol Has a Beneficial Effect on Glycine Max (L.) Plants Irrigated with Diluted Sea Water. J. Hortic. Sci. Biotechnol. 2015, 90, 195–202. [Google Scholar] [CrossRef]
- Patist, A.; Zoerb, H. Preservation Mechanisms of Trehalose in Food and Biosystems. Colloids Surf. B Biointerfaces 2005, 40, 107–113. [Google Scholar] [CrossRef]
- Iturriaga, G.; Suárez, R.; Nova-Franco, B. Trehalose Metabolism: From Osmoprotection to Signaling. Int. J. Mol. Sci. 2009, 10, 3793–3810. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lunn, J.E.; Delorge, I.; Figueroa, C.M.; Van Dijck, P.; Stitt, M. Trehalose Metabolism in Plants. Plant J. 2014, 79, 544–567. [Google Scholar] [CrossRef] [PubMed]
- Klähn, S.; Hagemann, M. Compatible Solute Biosynthesis in Cyanobacteria: Cyanobacterial Solutes. Environ. Microbiol. 2011, 13, 551–562. [Google Scholar] [CrossRef]
- Chang, S.-W.; Chang, W.-H.; Lee, M.-R.; Yang, T.-J.; Yu, N.-Y.; Chen, C.-S.; Shaw, J.-F. Simultaneous Production of Trehalose, Bioethanol, and High-Protein Product from Rice by an Enzymatic Process. J. Agric. Food Chem. 2010, 58, 2908–2914. [Google Scholar] [CrossRef]
- Ma, C.; Wang, Z.; Kong, B.; Lin, T. Exogenous Trehalose Differentially Modulate Antioxidant Defense System in Wheat Callus during Water Deficit and Subsequent Recovery. Plant Growth Regul. 2013, 70, 275–285. [Google Scholar] [CrossRef]
- Mostofa, M.G.; Hossain, M.A.; Fujita, M. Trehalose Pretreatment Induces Salt Tolerance in Rice (Oryza Sativa L.) Seedlings: Oxidative Damage and Co-Induction of Antioxidant Defense and Glyoxalase Systems. Protoplasma 2015, 252, 461–475. [Google Scholar] [CrossRef]
- Abdelgawad, Z.; Hathout, T.; El-Khallal, S.; Said, E.; Al-Mokadem, A. Accumulation of Trehalose Mediates Salt Adaptation in Rice Seedlings. Am. Eurasian J. Agric. Env. Sci. 2014, 14, 1450–1463. [Google Scholar]
- Bae, H.; Herman, E.; Bailey, B.; Bae, H.-J.; Sicher, R. Exogenous Trehalose Alters Arabidopsis Transcripts Involved in Cell Wall Modification, Abiotic Stress, Nitrogen Metabolism, and Plant Defense. Physiol. Plant. 2005, 125, 114–126. [Google Scholar] [CrossRef]
- Avonce, N.; Mendoza-Vargas, A.; Morett, E.; Iturriaga, G. Insights on the Evolution of Trehalose Biosynthesis. BMC Evol. Biol. 2006, 6, 109. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Svanström, A.; van Leeuwen, M.R.; Dijksterhuis, J.; Melin, P. Trehalose Synthesis in Aspergillus Niger: Characterization of Six Homologous Genes, All with Conserved Orthologs in Related Species. BMC Microbiol. 2014, 14, 90. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, S.; Ouyang, K.; Wang, K. Genome-Wide Identification, Evolution, and Expression Analysis of TPS and TPP Gene Families in Brachypodium Distachyon. Plants 2019, 8, 362. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Satoh-Nagasawa, N.; Nagasawa, N.; Malcomber, S.; Sakai, H.; Jackson, D. A Trehalose Metabolic Enzyme Controls Inflorescence Architecture in Maize. Nature 2006, 441, 227–230. [Google Scholar] [CrossRef] [PubMed]
- Miranda, J.A.; Avonce, N.; Suárez, R.; Thevelein, J.M.; Van Dijck, P.; Iturriaga, G. A Bifunctional TPS–TPP Enzyme from Yeast Confers Tolerance to Multiple and Extreme Abiotic-Stress Conditions in Transgenic Arabidopsis. Planta 2007, 226, 1411–1421. [Google Scholar] [CrossRef] [Green Version]
- Lin, Q.; Yang, J.; Wang, Q.; Zhu, H.; Chen, Z.; Dao, Y.; Wang, K. Overexpression of the Trehalose-6-Phosphate Phosphatase Family Gene AtTPPF Improves the Drought Tolerance of Arabidopsis Thaliana. BMC Plant Biol. 2019, 19, 381. [Google Scholar] [CrossRef] [Green Version]
- Garg, A.K.; Kim, J.-K.; Owens, T.G.; Ranwala, A.P.; Choi, Y.D.; Kochian, L.V.; Wu, R.J. Trehalose Accumulation in Rice Plants Confers High Tolerance Levels to Different Abiotic Stresses. Proc. Natl. Acad. Sci. USA 2002, 99, 15898–15903. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.-J.; Hao, Y.-J.; Zhang, Z.-G.; Chen, T.; Zhang, J.-S.; Chen, S.-Y. Isolation of Trehalose-6-Phosphate Phosphatase Gene from Tobacco and Its Functional Analysis in Yeast Cells. J. Plant Physiol. 2005, 162, 215–223. [Google Scholar] [CrossRef]
- Wu, W.; Pang, Y.; Shen, G.-A.; Lu, J.; Lin, J.; Wang, J.; Sun, X.; Tang, K. Molecular Cloning, Characterization and Expression of a Novel Trehalose-6-Phosphate Synthase Homologue from Ginkgo Biloba. BMB Rep. 2006, 39, 158–166. [Google Scholar] [CrossRef] [Green Version]
- Jiang, W.; Fu, F.-L.; Zhang, S.-Z.; Wu, L.; Li, W.-C. Cloning and Characterization of Functional Trehalose-6-Phosphate Synthase Gene in Maize. J. Plant Biol. 2010, 53, 134–141. [Google Scholar] [CrossRef]
- Wang, G.; Zhao, G.; Feng, Y.; Xuan, J.; Sun, J.; Guo, B.; Jiang, G.; Weng, M.; Yao, J.; Wang, B.; et al. Cloning and Comparative Studies of Seaweed Trehalose-6-Phosphate Synthase Genes. Mar. Drugs 2010, 8, 2065–2079. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, X.; Hou, Z.; Huang, C.; Chen, Q.; Gao, W.; Zhang, J. Cloning, Purification and Characterization of Trehalose-6-Phosphate Synthase from Pleurotus Tuoliensis. PeerJ 2018, 6, e5230. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martinez-Cruz, E. Cultivo in Vitro Mediante Embriogénesis Somática Y Transformación Genética DE Maíz (Zea Mays L.) Con Genes DE Biosíntesis DE Trehalosa. Ph.D. Thesis, Universidad Autonoma Chapingo, Mexico, 2014. [Google Scholar]
- Efeoğlu, B.; Ekmekçi, Y.; Çiçek, N. Physiological Responses of Three Maize Cultivars to Drought Stress and Recovery. South Afr. J. Bot. 2009, 75, 34–42. [Google Scholar] [CrossRef] [Green Version]
- Khan, M.B.; Hussain, M.; Raza, A.; Farooq, S.; Jabran, K. Seed Priming with CaCl 2 and Ridge Planting for Improved Drought Resistance in Maize. Turk. J. Agric. For. 2015, 39, 193–203. [Google Scholar] [CrossRef]
- Nayyar, H.; Gupta, D. Differential Sensitivity of C3 and C4 Plants to Water Deficit Stress: Association with Oxidative Stress and Antioxidants. Environ. Exp. Bot. 2006, 58, 106–113. [Google Scholar] [CrossRef]
- Gibson, R.P.; Lloyd, R.M.; Charnock, S.J.; Davies, G.J. Characterization of Escherichia Coli OtsA, a Trehalose-6-Phosphate Synthase from Glycosyltransferase Family 20. Acta Crystallogr. D Biol. Crystallogr. 2002, 58, 349–351. [Google Scholar] [CrossRef] [Green Version]
- Kosmas, S.A.; Argyrokastritis, A.; Loukas, M.G.; Eliopoulos, E.; Tsakas, S.; Kaltsikes, P.J. Isolation and Characterization of Drought-Related Trehalose 6-Phosphate-Synthase Gene from Cultivated Cotton (Gossypium Hirsutum L.). Planta 2006, 223, 329–339. [Google Scholar] [CrossRef]
- Tang, B.; Chen, J.; Yao, Q.; Pan, Z.; Xu, W.; Wang, S.; Zhang, W. Characterization of a Trehalose-6-Phosphate Synthase Gene from Spodoptera Exigua and Its Function Identification through RNA Interference. J. Insect Physiol. 2010, 56, 813–821. [Google Scholar] [CrossRef]
- Kaasen, I.; McDougall, J.; Strøm, A.R. Analysis of the OtsBA Operon for Osmoregulatory Trehalose Synthesis in Escherichia Coli and Homology of the OtsA and OtsB Proteins to the Yeast Trehalose-6-Phosphate Synthase/Phosphatase Complex. Gene 1994, 145, 9–15. [Google Scholar] [CrossRef]
- Sanders, D.; Brownlee, C.; Harper, J.F. Communicating with Calcium. Plant Cell 1999, 11, 691–706. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Knight, H. Calcium Signaling during Abiotic Stress in Plants. In International Review of Cytology; Elsevier: Amsterdam, The Netherlands, 1999; Volume 195, pp. 269–324. [Google Scholar] [CrossRef]
- Ge, L.-F.; Chao, D.-Y.; Shi, M.; Zhu, M.-Z.; Gao, J.-P.; Lin, H.-X. Overexpression of the Trehalose-6-Phosphate Phosphatase Gene OsTPP1 Confers Stress Tolerance in Rice and Results in the Activation of Stress Responsive Genes. Planta 2008, 228, 191–201. [Google Scholar] [CrossRef] [PubMed]
- Avonce, N.; Leyman, B.; Mascorro-Gallardo, J.O.; Van Dijck, P.; Thevelein, J.M.; Iturriaga, G. The Arabidopsis Trehalose-6-P Synthase AtTPS1 Gene Is a Regulator of Glucose, Abscisic Acid, and Stress Signaling. Plant Physiol. 2004, 136, 3649–3659. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nuccio, M.L.; Wu, J.; Mowers, R.; Zhou, H.-P.; Meghji, M.; Primavesi, L.F.; Paul, M.J.; Chen, X.; Gao, Y.; Haque, E.; et al. Expression of Trehalose-6-Phosphate Phosphatase in Maize Ears Improves Yield in Well-Watered and Drought Conditions. Nat. Biotechnol. 2015, 33, 862–869. [Google Scholar] [CrossRef]
- Stiekema, W.J.; Heidekamp, F.; Dirkse, W.G.; van Beckum, J.; de Haan, P.; ten Bosch, C.; Louwerse, J.D. Molecular Cloning and Analysis of Four Potato Tuber MRNAs. Plant Mol. Biol. 1988, 11, 255–269. [Google Scholar] [CrossRef]




© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Acosta-Pérez, P.; Camacho-Zamora, B.D.; Espinoza-Sánchez, E.A.; Gutiérrez-Soto, G.; Zavala-García, F.; Abraham-Juárez, M.J.; Sinagawa-García, S.R. Characterization of Trehalose-6-phosphate Synthase and Trehalose-6-phosphate Phosphatase Genes and Analysis of its Differential Expression in Maize (Zea mays) Seedlings under Drought Stress. Plants 2020, 9, 315. https://doi.org/10.3390/plants9030315
Acosta-Pérez P, Camacho-Zamora BD, Espinoza-Sánchez EA, Gutiérrez-Soto G, Zavala-García F, Abraham-Juárez MJ, Sinagawa-García SR. Characterization of Trehalose-6-phosphate Synthase and Trehalose-6-phosphate Phosphatase Genes and Analysis of its Differential Expression in Maize (Zea mays) Seedlings under Drought Stress. Plants. 2020; 9(3):315. https://doi.org/10.3390/plants9030315
Chicago/Turabian StyleAcosta-Pérez, Phamela, Bianka Dianey Camacho-Zamora, Edward A. Espinoza-Sánchez, Guadalupe Gutiérrez-Soto, Francisco Zavala-García, María Jazmín Abraham-Juárez, and Sugey Ramona Sinagawa-García. 2020. "Characterization of Trehalose-6-phosphate Synthase and Trehalose-6-phosphate Phosphatase Genes and Analysis of its Differential Expression in Maize (Zea mays) Seedlings under Drought Stress" Plants 9, no. 3: 315. https://doi.org/10.3390/plants9030315




