Insight into the Chromosome Structure of the Cultivated Tetraploid Alfalfa (Medicago sativa subsp. sativa L.) by a Combined Use of GISH and FISH Techniques
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. DNA Extraction and Preparation of Probes
2.3. Chromosome Preparations and C-Banding
2.4. Fluorescence In Situ Hybridization
3. Results
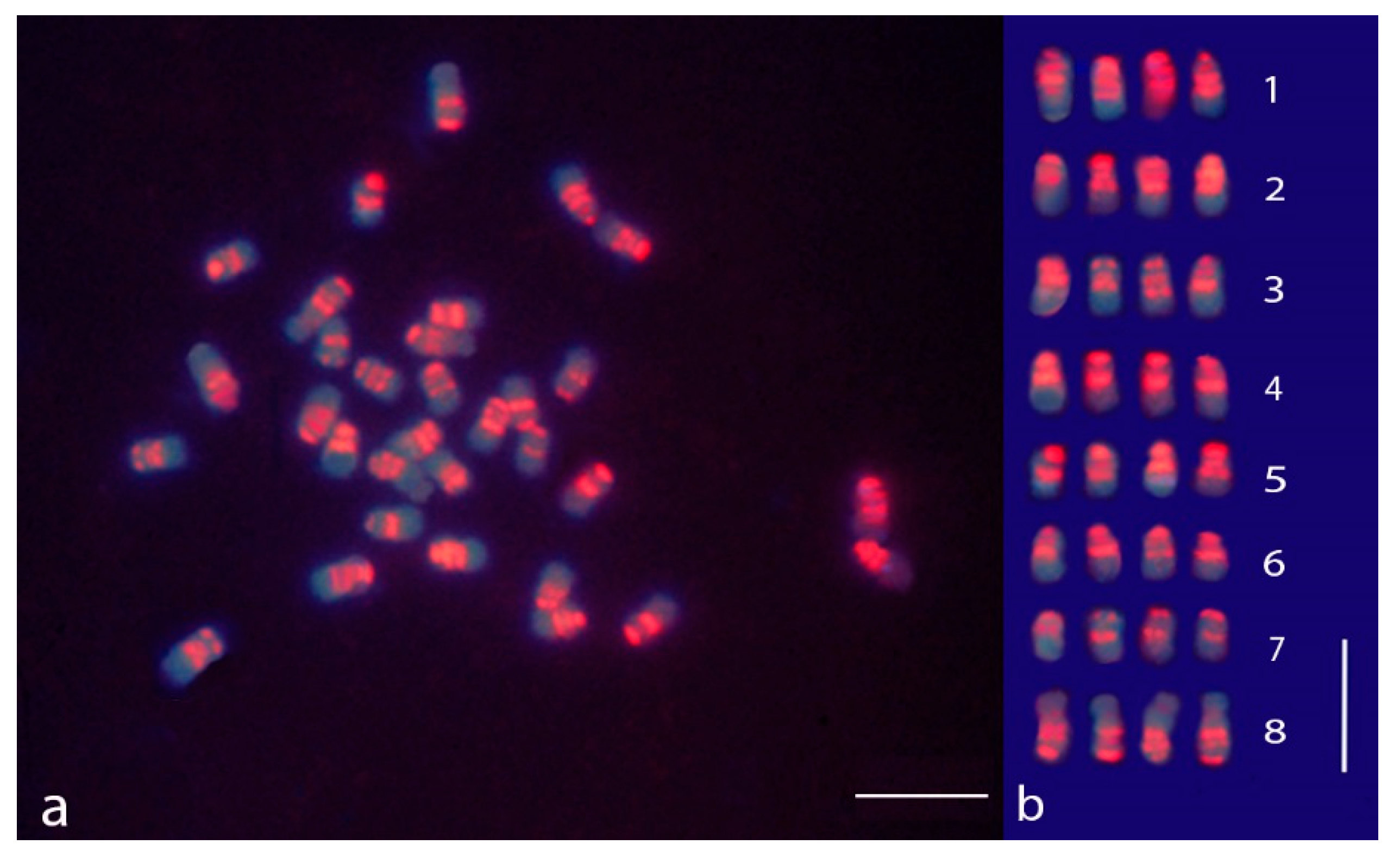
3.1. sGISH and C-Banding in M. sativa subsp. sativa
3.2. sGISH Signal Pattern
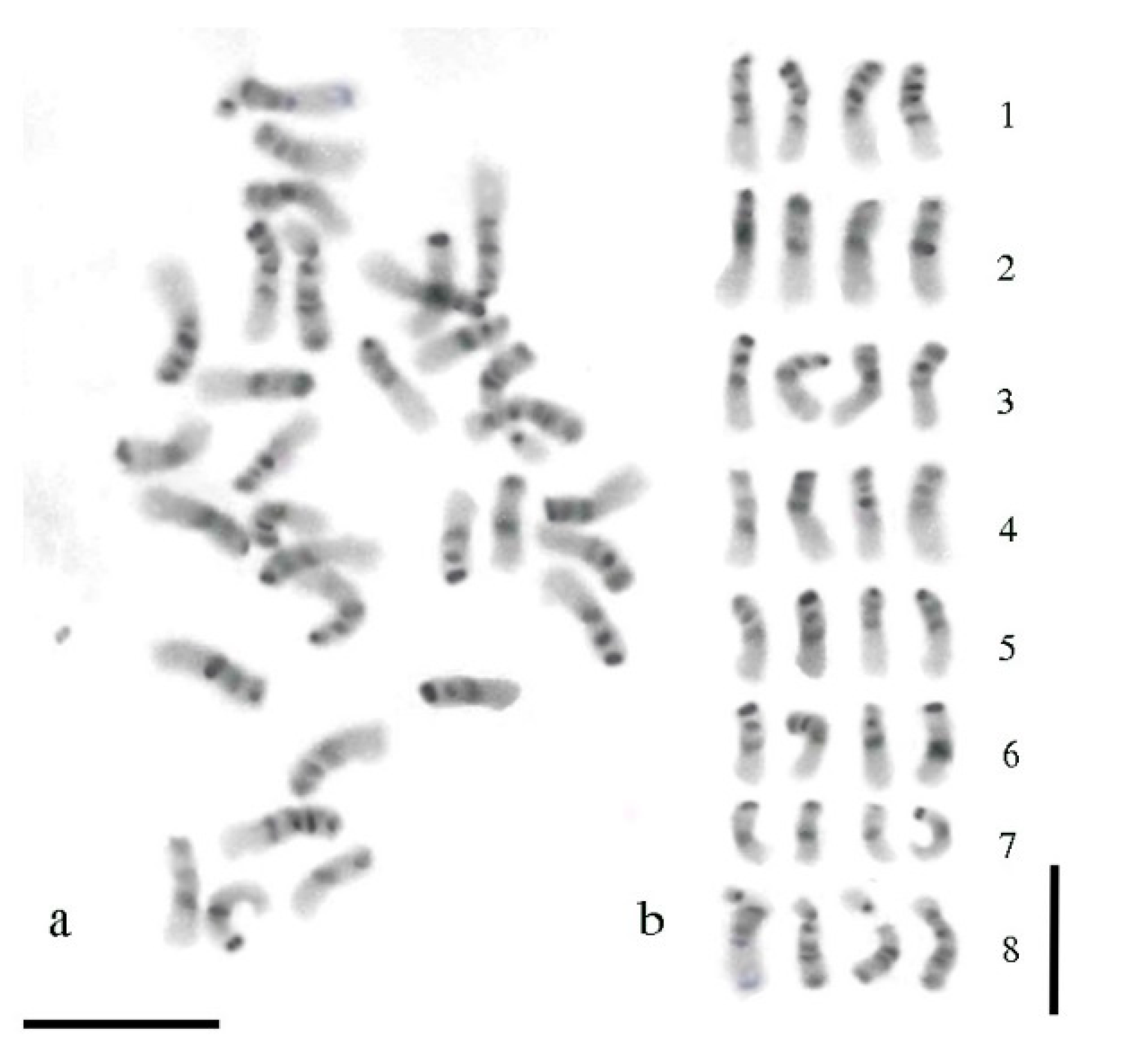
3.3. Giemsa C-Banding Pattern
3.4. Comparison between sGISH and C-Banding Patterns
3.5. FISH Mapping of Telomeric (TTTAGGG) Repeats
4. Discussion
Funding
Conflicts of Interest
References
- Bennet, M.D. The development and use of genomic in situ hybridization (GISH) as a new tool in plant cytogenetics. In Kew Chromos Conference IV; Royal Botanical Garden: Kew, UK, 1995; pp. 167–184. [Google Scholar]
- Falistocco, E. Physical mapping of rRNA genes in Medicago sativa and M. glomerata by fluorescent in situ hybridization. J. Hered. 2000, 91, 256–260. [Google Scholar] [CrossRef] [PubMed]
- Yu, F.; Lei, Y.; Li, Y.; Dou, Q.; Wang, H.; Chen, Z. Cloning and characterization of chromosomal markers in alfalfa (Medicago sativa L.). Theor. Appl. Genet. 2013, 126, 1885–1896. [Google Scholar] [CrossRef] [PubMed]
- Charlesworth, B.; Sniegowski, P.; Stephan, W. The evolutionary dynamics of repetitive DNA in eukaryotes. Nature 1994, 371, 215–220. [Google Scholar] [CrossRef] [PubMed]
- Mehrotra, S.; Goyal, V. Repetitive sequences in plant nuclear DNA: Types, distribution. Evolution and function. Genom. Proteom. Bioinform. 2014, 12, 164–171. [Google Scholar] [CrossRef]
- Mukai, Y. Multicolor Fluorescence In Situ Hybridization: A New Tool for Genome Analysis. In Methods of Genome Analysis in Plants; CRC Press: New York, NY, USA, 1996; pp. 181–192. [Google Scholar]
- She, C.; Liu, J.; Diau, Y.; Hu, Z.; Song, Y. The distribution of repetitive DNAs along chromosomes in plants revealed by self-genomic in situ hybridization. J. Genet. Genom. 2007, 34, 437–448. [Google Scholar] [CrossRef]
- She, C.; Wei, L.; Jiang, X. Molecular cytogenetic characterization and comparison of the two cultivated Canavalia species (Fabaceae). Comp. Cytogenet. 2017, 11, 579–600. [Google Scholar] [CrossRef]
- Maluszynska, J.; Hasterok, R. Identification of individual chromosomes and parental genomes in Brassica juncea using GISH and FISH. Cytogenet. Genome. Res. 2005, 109, 310–314. [Google Scholar] [CrossRef]
- Ansari, H.A.; Ellison, N.W.; Williams, W.M. Molecular and cytogenetic evidence for an allotetraploid origin of Trifolium dubium (Leguminosae). Chromosoma 2008, 117, 159–167. [Google Scholar] [CrossRef]
- Wolny, E.; Hasterok, R. Comparative cytogenetic analysis of the genomes of the model grass Brachypodium distachyon and its close relatives. Ann. Bot. 2009, 104, 873–881. [Google Scholar] [CrossRef]
- Falistocco, E. Cytogenetics and chromosome number evolution of annual Medicago species (Fabaceae) with emphasis on the origin of the polyploid Medicago rugosa and M. scutellata. Plant Biosyst. 2018, 153, 1–7. [Google Scholar]
- Fuchs, J.; Brandes, A.; Schubert, I. Telomere sequence localization and karyotype evolution in higher plants. Plant. Syst. Evol. 1995, 196, 227–241. [Google Scholar] [CrossRef]
- Sykorovà, E.; Yoong, L.K.; Fajkus, J.; Leitch, A.R. The signature of the Cestrum genome suggests an evolutionaty response to the loss of (TTTAGGG)n telomeres. Chromosoma 2003, 112, 164–172. [Google Scholar] [CrossRef] [PubMed]
- He, L.; Liu, J.; Torres, G.A.; Zhang, H.; Jiang, J.; Xie, C. Interstitial telomeric repeats are enriched in the centromeres of chromosomes in Solanum species. Chromosome Res. 2013, 21, 5–13. [Google Scholar] [CrossRef] [PubMed]
- Sousa, A.; Renner, S.S. Interstitial telomere-like repeats in the monocot family Araceae. Bot. J. Linn. Soc. 2015, 177, 15–26. [Google Scholar] [CrossRef]
- Bolzàn, A.D. Interstitial telomeric sequences in vertebrate chromosomes: Origin, function, instability and evolution. Mutat. Res. 2017, 773, 51–65. [Google Scholar] [CrossRef]
- Gerlach, W.L.; Bedbrook, J.R. Cloning and characterization of ribosomal RNA genes from wheat and barley. Nucleic Acid. Res. 1979, 7, 1869–1885. [Google Scholar] [CrossRef]
- Falistocco, E.; Falcinelli, M.; Veronesi, F. Karyotype and C-banding pattern of mitotic chromosomes in alfalfa, Medicago sativa L. Plant Breed. 1995, 114, 451–453. [Google Scholar] [CrossRef]
- Irifune, K.; Hirai, K.; Zheng, J.; Tanaka, R. Nucleotide sequence of a highly repeated DNA sequence and its chromosomal localization in Allium fistolosum. Theor. Appl. Genet. 1995, 90, 312–316. [Google Scholar] [CrossRef]
- Kuipers, A.G.J.; van Os, D.P.M.; de Jong, J.H.; Ramanna, M.S. Molecular cytogenetics of Alstroemeria: Identification of parental genomes in interspecific hybrids and characterization of repetitive DNA families in constitutive heterochromatin. Chromosome Res. 1997, 5, 31–39. [Google Scholar] [CrossRef]
- Markova, M.; Lengerova, M.; Zluvova, J.; Janousek, B.; Vyscot, B. Karyological analysis of an interspecific hybrid between the dioecious Silene latifolia and the hermaproditic Silene viscosa. Genome 2006, 49, 373–379. [Google Scholar] [CrossRef]
- Zhou, J.; Yang, Z.; Li, G.; Liu, C.; Ren, Z. Discrimination of repetitive sequences polymorphism in Secale cereale by Genomic In Situ Hybridization-Banding. J. Integr. Plant Biol. 2008, 50, 452–456. [Google Scholar] [CrossRef] [PubMed]
- Pontvianne, F.; Carpentier, M.C.; Durut, N.; Pavlištová, V.; Jaške, K.; Schořová, Š.; Parrinello, H.; Rohmer, M.; Pikaard, C.S.; Fojtová, M.; et al. Identification of nucleolus-associated chromatin domains reveals a role for the nucleolus in 3D organization of the A. thaliana genome. Cell Rep. 2016, 16, 1574–1587. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Chunyan Cheng, C.; Li, J.; Yang, S.; Wang, Y.; Li, Z.; Chen, J.; Lou, Q. Chromosomal structures and repetitive sequences divergence in Cucumis species revealed by comparative cytogenetic mapping. BMC Genomics 2015, 16, 730. [Google Scholar] [CrossRef] [PubMed]
- Souza, G.; Vanzela, A.L.L.; Crosa, O.; Guerra, M. Interstitial telomeric sites and Robertsonian translocations in species of Ipheion and Nothoscordum (Amaryllidaceae). Genetica 2016, 144, 157–166. [Google Scholar] [CrossRef] [PubMed]
- Aksenova, A.Y.; Sergei, M.; Mirkin, S.M. At the beginning of the end and in the middle of the beginning: Structure and maintenance of telomeric DNA repeats and interstitial telomeric sequences. Genes 2019, 10, 118. [Google Scholar] [CrossRef]

© 2020 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Falistocco, E. Insight into the Chromosome Structure of the Cultivated Tetraploid Alfalfa (Medicago sativa subsp. sativa L.) by a Combined Use of GISH and FISH Techniques. Plants 2020, 9, 542. https://doi.org/10.3390/plants9040542
Falistocco E. Insight into the Chromosome Structure of the Cultivated Tetraploid Alfalfa (Medicago sativa subsp. sativa L.) by a Combined Use of GISH and FISH Techniques. Plants. 2020; 9(4):542. https://doi.org/10.3390/plants9040542
Chicago/Turabian StyleFalistocco, Egizia. 2020. "Insight into the Chromosome Structure of the Cultivated Tetraploid Alfalfa (Medicago sativa subsp. sativa L.) by a Combined Use of GISH and FISH Techniques" Plants 9, no. 4: 542. https://doi.org/10.3390/plants9040542
APA StyleFalistocco, E. (2020). Insight into the Chromosome Structure of the Cultivated Tetraploid Alfalfa (Medicago sativa subsp. sativa L.) by a Combined Use of GISH and FISH Techniques. Plants, 9(4), 542. https://doi.org/10.3390/plants9040542



