Mitochondrial Genome of Fagopyrum esculentum and the Genetic Diversity of Extranuclear Genomes in Buckwheat
Abstract
1. Introduction
2. Results and Discussion
2.1. Genome Assembly: Standard and Custom Tools
- A user provides a starting sequence, which may be a contig from another assembly or just a random sequencing read.
- Elloreas maps reads to the 3′ end of this sequence.
- It finds reads that overlap the ends, such that a part of a read is mapped to the 3′ end while the other part overhangs the contig.
- Elloreas calculates sequence consensus for these “overhanging parts”.
- It extends the contig using this consensus.
- It repeats all steps from “2.” to “6.” for this extended contig.
2.2. Genome Structure and Gene Content
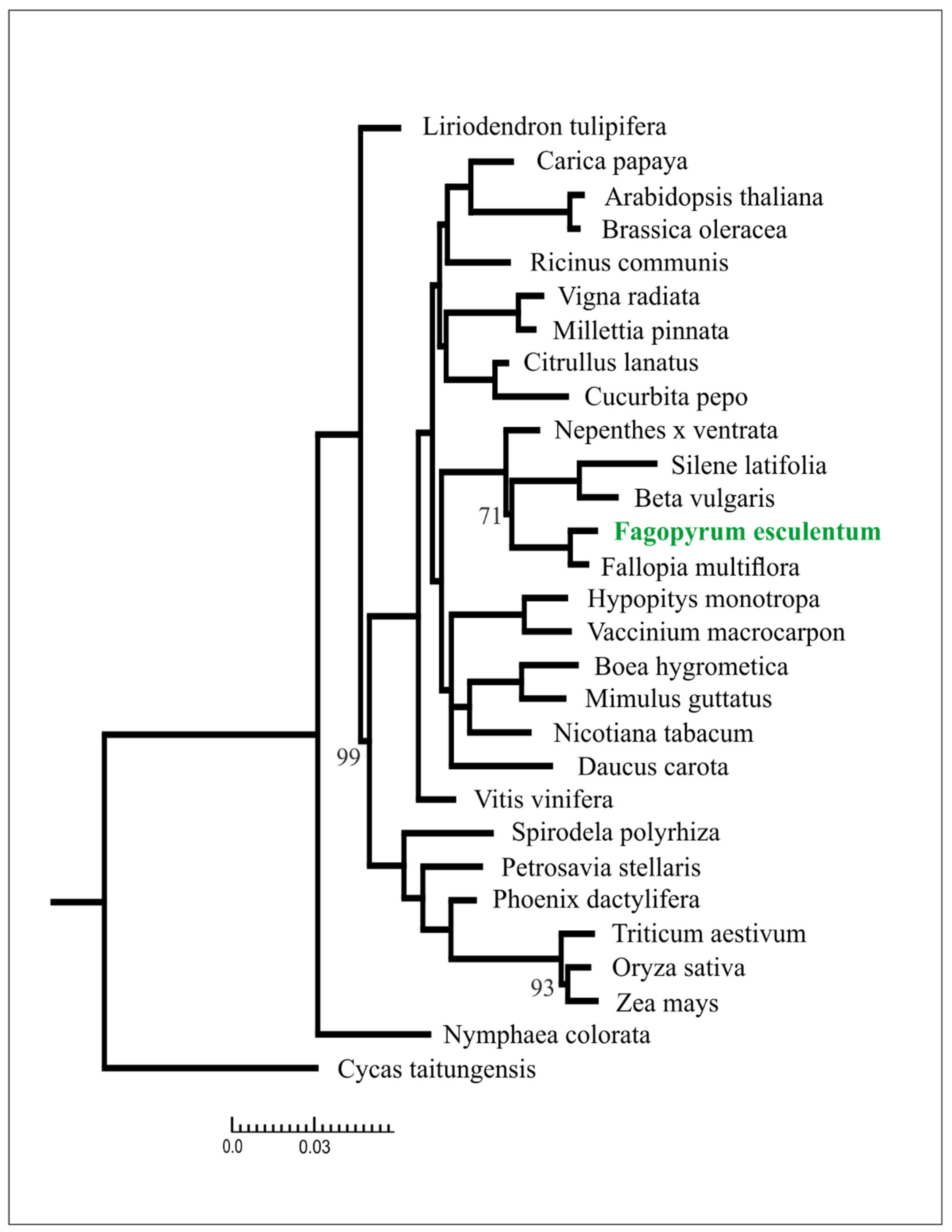
2.3. Phylogenetic Analysis
2.4. Genetic Diversity of the Mitochondrial and Plastid Genome in Common Buckwheat
3. Materials and Methods
3.1. Data Source, DNA Extraction, and Sequencing
3.2. Read Preprocessing for De Novo Assembly
3.3. Mitochondrial Genome Assembly and Assembly Check
3.4. Detection of Structural Variants
3.5. Plastid Genome Assembly
3.6. Annotation
3.7. Alignment and Phylogenetic Analysis
3.8. Mapping and SNP Calling
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Sanchez-Puerta, M.V.; Edera, A.; Gandini, C.L.; Williams, A.V.; Howell, K.A.; Nevill, P.G.; Small, I. Genome-scale transfer of mitochondrial DNA from legume hosts to the holoparasite Lophophytum mirabile (Balanophoraceae). Mol. Phylogenetics Evol. 2019, 132, 243–250. [Google Scholar] [CrossRef]
- Bergthorsson, U.; Richardson, A.O.; Young, G.J.; Goertzen, L.R.; Palmer, J.D. Massive horizontal transfer of mitochondrial genes from diverse land plant donors to the basal angiosperm Amborella. Proc. Natl. Acad. Sci. USA 2004, 101, 17747–17752. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Li, X.; Ma, B.; Gao, Q.; Du, H.; Han, Y.; Li, Y.; Cao, Y.; Qi, M.; Zhu, Y.; et al. The Tartary Buckwheat Genome Provides Insights into Rutin Biosynthesis and Abiotic Stress Tolerance. Mol. Plant 2017, 10, 1224–1237. [Google Scholar] [CrossRef]
- Yasui, Y.; Hirakawa, H.; Ueno, M.; Matsui, K.; Katsube-Tanaka, T.; Yang, S.J.; Aii, J.; Sato, S.; Mori, M. Assembly of the draft genome of buckwheat and its applications in identifying agronomically useful genes. DNA Res. 2016, 23, 215–224. [Google Scholar] [CrossRef] [PubMed]
- Logacheva, M.D.; Samigullin, T.H.; Dhingra, A.; Penin, A.A. Comparative chloroplast genomics and phylogenetics of Fagopyrum esculentum ssp. ancestrale–A wild ancestor of cultivated buckwheat. BMC Plant Biol. 2008, 8, 59. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Zhou, T.; Bai, G.; Zhao, Y. Complete chloroplast genome sequence of Fagopyrum dibotrys: Genome features, comparative analysis and phylogenetic relationships. Sci. Rep. 2018, 8. [Google Scholar] [CrossRef]
- Kim, C.-K.; Kim, Y.-K. The multipartite mitochondrial genome of Fallopia multiflora (Caryophyllales: Polygonaceae). Mitochondrial DNA Part B 2018, 3, 155–156. [Google Scholar] [CrossRef]
- Horn, R.; Gupta, K.J.; Colombo, N. Mitochondrion role in molecular basis of cytoplasmic male sterility. Mitochondrion 2014, 19, 198–205. [Google Scholar] [CrossRef]
- Tuteja, R.; Saxena, R.K.; Davila, J.; Shah, T.; Chen, W.; Xiao, Y.-L.; Fan, G.; Saxena, K.B.; Alverson, A.J.; Spillane, C.; et al. Cytoplasmic Male Sterility-Associated Chimeric Open Reading Frames Identified by Mitochondrial Genome Sequencing of Four Cajanus Genotypes. DNA Res. 2013, 20, 485–495. [Google Scholar] [CrossRef]
- Makarenko, M.S.; Usatov, A.V.; Tatarinova, T.V.; Azarin, K.V.; Logacheva, M.D.; Gavrilova, V.A.; Horn, R. Characterization of the mitochondrial genome of the MAX1 type of cytoplasmic male-sterile sunflower. BMC Plant Biol. 2019, 19. [Google Scholar] [CrossRef]
- Zheleznov, A. Kompleksnyj Podhod k Sozdaniju Ishodnogo Materiala Dlja Selekcionno-Geneticheskih Issledovanij: Na Primere Rjada Vidov Rastenij (A Complex Approach for the Development of Material for Genetics and Breeding Studies, on the Example of Several Plant Species). Ph.D. Thesis, Institute of Cytology and Genetics, Novosibirsk, Russia, 2000. [Google Scholar]
- Mendler-Drienyovszki, N.; Cal, A.J.; Dobránszki, J. Progress and prospects for interspecific hybridization in buckwheat and the genus Fagopyrum. Biotechnol. Adv. 2013, 31, 1768–1775. [Google Scholar] [CrossRef] [PubMed]
- Wick, R.R.; Judd, L.M.; Gorrie, C.L.; Holt, K.E. Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 2017, 13, e1005595. [Google Scholar] [CrossRef] [PubMed]
- Kozik, A.; Rowan, B.A.; Lavelle, D.; Berke, L.; Schranz, M.E.; Michelmore, R.W.; Christensen, A.C. The alternative reality of plant mitochondrial DNA: One ring does not rule them all. PLoS Genet. 2019, 15, e1008373. [Google Scholar] [CrossRef]
- Shearman, J.R.; Sonthirod, C.; Naktang, C.; Pootakham, W.; Yoocha, T.; Sangsrakru, D.; Jomchai, N.; Tragoonrung, S.; Tangphatsornruang, S. The two chromosomes of the mitochondrial genome of a sugarcane cultivar: Assembly and recombination analysis using long PacBio reads. Sci. Rep. 2016, 6. [Google Scholar] [CrossRef] [PubMed]
- Shtratnikova, V.Y.; Schelkunov, M.I.; Penin, A.A.; Logacheva, M.D. Mitochondrial genome of non-photosynthetic mycoheterotrophic plant Hypopitys monotropa, its structure, gene expression and RNA editing. Biorxiv Evol. Biol. 2019. [Google Scholar] [CrossRef]
- Sloan, D.B.; Alverson, A.J.; Chuckalovcak, J.P.; Wu, M.; McCauley, D.E.; Palmer, J.D.; Taylor, D.R. Rapid Evolution of Enormous, Multichromosomal Genomes in Flowering Plant Mitochondria with Exceptionally High Mutation Rates. PLoS Biol. 2012, 10, e1001241. [Google Scholar] [CrossRef]
- Bellot, S.; Cusimano, N.; Luo, S.; Sun, G.; Zarre, S.; Gröger, A.; Temsch, E.; Renner, S.S. Assembled Plastid and Mitochondrial Genomes, as well as Nuclear Genes, Place the Parasite Family Cynomoriaceae in the Saxifragales. Genome Boil. Evol. 2016, 8, 2214–2230. [Google Scholar] [CrossRef]
- Dierckxsens, N.; Mardulyn, P.; Smits, G. NOVOPlasty: De novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 2016, gkw955. [Google Scholar] [CrossRef]
- Kozik, A.; Rowan, B.; Lavelle, D.; Berke, L.; Schranz, M.E.; Michelmore, R.W.; Christensen, A.C. The alternative reality of plant mitochondrial DNA. BioRxiv 2019. [Google Scholar] [CrossRef]
- Palmer, J.D.; Shields, C.R. Tripartite structure of the Brassica campestris mitochondrial genome. Nature 1984, 307, 437–440. [Google Scholar] [CrossRef]
- Alverson, A.J.; Zhuo, S.; Rice, D.W.; Sloan, D.B.; Palmer, J.D. The Mitochondrial Genome of the Legume Vigna radiata and the Analysis of Recombination across Short Mitochondrial Repeats. PLoS ONE 2011, 6, e16404. [Google Scholar] [CrossRef] [PubMed]
- Adams, K.L.; Qiu, Y.-L.; Stoutemyer, M.; Palmer, J.D. Punctuated evolution of mitochondrial gene content: High and variable rates of mitochondrial gene loss and transfer to the nucleus during angiosperm evolution. Proc. Natl. Acad. Sci. USA 2002, 99, 9905–9912. [Google Scholar] [CrossRef] [PubMed]
- Gruzdev, E.V.; Mardanov, A.V.; Beletsky, A.V.; Ravin, N.V.; Skryabin, K.G. The complete mitochondrial genome of the carnivorous flowering plant Nepenthes X Ventrata. Mitochondrial DNA Part B 2018, 3, 1259–1260. [Google Scholar] [CrossRef]
- Kubo, T.; Nishizawa, S.; Sugawara, A.; Itchoda, N.; Estiati, A.; Mikami, T. The complete nucleotide sequence of the mitochondrial genome of sugar beet (Beta vulgaris L.) reveals a novel gene for tRNACys(GCA). Nucleic Acids Res. 2000, 28, 2571–2576. [Google Scholar] [CrossRef] [PubMed]
- Kubo, N.; Arimura, S.-I. Discovery of the rpl10 Gene in Diverse Plant Mitochondrial Genomes and Its Probable Replacement by the Nuclear Gene for Chloroplast RPL10 in Two Lineages of Angiosperms. DNA Res. 2010, 17, 1–9. [Google Scholar] [CrossRef]
- Iorizzo, M.; Senalik, D.; Szklarczyk, M.; Grzebelus, D.; Spooner, D.; Simon, P. De novo assembly of the carrot mitochondrial genome using next generation sequencing of whole genomic DNA provides first evidence of DNA transfer into an angiosperm plastid genome. BMC Plant Biol. 2012, 12, 61. [Google Scholar] [CrossRef]
- Cuenca, A.; Ross, T.G.; Graham, S.W.; Barrett, C.F.; Davis, J.I.; Seberg, O.; Petersen, G. Localized Retroprocessing as a Model of Intron Loss in the Plant Mitochondrial Genome. Genome Boil. Evol. 2016, 8, 2176–2189. [Google Scholar] [CrossRef]
- Sanchez-Puerta, M.V.; Cho, Y.; Mower, J.P.; Alverson, A.J.; Palmer, J.D. Frequent, Phylogenetically Local Horizontal Transfer of the cox1 Group I Intron in Flowering Plant Mitochondria. Mol. Boil. Evol. 2008, 25, 1762–1777. [Google Scholar] [CrossRef]
- Cho, Y.; Qiu, Y.L.; Kuhlman, P.; Palmer, J.D. Explosive invasion of plant mitochondria by a group I intron. Proc. Natl. Acad. Sci. USA 1998, 95, 14244–14249. [Google Scholar] [CrossRef]
- Onodera, Y.; Yamamoto, M.P.; Kubo, T.; Mikami, T. Heterogeneity of the atp6 Presequences in Normal and Different Sources of Male-Sterile Cytoplasms of Sugar Beet. J. Plant Physiol. 1999, 155, 656–660. [Google Scholar] [CrossRef]
- Tsujimura, M.; Kaneko, T.; Sakamoto, T.; Kimura, S.; Shigyo, M.; Yamagishi, H.; Terachi, T. Multichromosomal structure of the onion mitochondrial genome and a transcript analysis. Mitochondrion 2019, 46, 179–186. [Google Scholar] [CrossRef] [PubMed]
- Hiesel, R.; von Haeseler, A.; Brennicke, A. Plant mitochondrial nucleic acid sequences as a tool for phylogenetic analysis. Proce. Natl. Acad. Sci. USA 1994, 91, 634–638. [Google Scholar] [CrossRef] [PubMed]
- Rydin, C.; Wikström, N.; Bremer, B. Conflicting results from mitochondrial genomic data challenge current views of Rubiaceae phylogeny. Am. J. Bot. 2017, 104, 1522–1532. [Google Scholar] [CrossRef] [PubMed]
- Bergthorsson, U.; Adams, K.L.; Thomason, B.; Palmer, J.D. Widespread horizontal transfer of mitochondrial genes in flowering plants. Nature 2003, 424, 197–201. [Google Scholar] [CrossRef] [PubMed]
- Hao, W.; Richardson, A.O.; Zheng, Y.; Palmer, J.D. Gorgeous mosaic of mitochondrial genes created by horizontal transfer and gene conversion. Proce. Natl. Acad. Sci. USA 2010, 107, 21576–21581. [Google Scholar] [CrossRef] [PubMed]
- Ikeda, K. Buckwheat composition, chemistry, and processing. In Advances in Food and Nutrition Research; Elsevier: Amsterdam, The Netherlands, 2002; pp. 395–434. ISBN 978-0-12-016444-8. [Google Scholar]
- Fiebig, A. Rapid evolution of RNA editing sites in a small non-essential plastid gene. Nucleic Acids Res. 2004, 32, 3615–3622. [Google Scholar] [CrossRef]
- Giardi, M.T.; Rea, G.; Lambreva, M.D.; Antonacci, A.; Pastorelli, S.; Bertalan, I.; Johanningmeier, U.; Mattoo, A.K. Mutations of Photosystem II D1 Protein That Empower Efficient Phenotypes of Chlamydomonas reinhardtii under Extreme Environment in Space. PLoS ONE 2013, 8, e64352. [Google Scholar] [CrossRef]
- Amelin, A.V.; Chekalin, E.I.; Zaikin, V.V.; Fesenko, A.N. Photosynthesis response to the changes in light intensity and CO2 concentration in leaves of buckwheat cultivars produced at different periods [Reakcija fotosinteza list”ev sortov grechihi raznyh periodov selekcii na izmenenie intensivnosti sveta i koncentracii so2 v vozduhe]. Vestnik BelGAU 2017, 4, 133–136. [Google Scholar]
- Amelin, A.V.; Fesenko, A.N.; Chekalin, E.I.; Zaikin, V.V. Adaptiveness of productivity and photosynthesis in buckwheat (Fagopyrum esculentum Moench) landraces and varieties produced at different periods [Adaptivnij potencial fotosinteza i produkcionnogo processa u mestnyh form i sortoobrazcov grechihi (Fagopyrum esculentum Moench) raznyh periodov selekcii]. Sel’skokhozyaistvennaya Biol. 2016, 51, 79–88. [Google Scholar] [CrossRef]
- Rieseberg, L.H.; Seiler, G.J. Molecular Evidence and the Origin and Development of the Domesticated Sunflower (Helianthus annum, Asteraceae). Econ. Bot. 1990, 44, 79–91. [Google Scholar] [CrossRef]
- Filippi, C.V.; Merino, G.A.; Montecchia, J.F.; Aguirre, N.C.; Rivarola, M.; Naamati, G.; Fass, M.I.; Álvarez, D.; Di Rienzo, J.; Heinz, R.A.; et al. Genetic Diversity, Population Structure and Linkage Disequilibrium Assessment among International Sunflower Breeding Collections. Genes 2020, 11, 283. [Google Scholar] [CrossRef] [PubMed]
- Doyle, J.; Doyle, J. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bull. 1987, 33886. [Google Scholar]
- Catchen, J.; Hohenlohe, P.A.; Bassham, S.; Amores, A.; Cresko, W.A. Stacks: An analysis tool set for population genomics. Mol. Ecol. 2013, 22, 3124–3140. [Google Scholar] [CrossRef] [PubMed]
- Li, H. Minimap2: Pairwise alignment for nucleotide sequences. Bioinformatics 2018, 34, 3094–3100. [Google Scholar] [CrossRef]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M.; et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef] [PubMed]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and applications. BMC Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef] [PubMed]
- Koren, S.; Walenz, B.P.; Berlin, K.; Miller, J.R.; Bergman, N.H.; Phillippy, A.M. Canu: Scalable and accurate long-read assembly via adaptive k -mer weighting and repeat separation. Genome Res. 2017, 27, 722–736. [Google Scholar] [CrossRef]
- Chin, C.-S.; Peluso, P.; Sedlazeck, F.J.; Nattestad, M.; Concepcion, G.T.; Clum, A.; Dunn, C.; O’Malley, R.; Figueroa-Balderas, R.; Morales-Cruz, A.; et al. Phased diploid genome assembly with single-molecule real-time sequencing. Nat. Methods 2016, 13, 1050–1054. [Google Scholar] [CrossRef]
- Nagano, M.; Aii, J.; Campbell, C.; Kawasaki, S.; Adachi, T. Genome size analysis of the genus Fagopyrum. Fagopyrum 2000, 17, 35–39. [Google Scholar]
- Sedlazeck, F.J.; Rescheneder, P.; Smolka, M.; Fang, H.; Nattestad, M.; von Haeseler, A.; Schatz, M.C. Accurate detection of complex structural variations using single-molecule sequencing. Nat. Methods 2018, 15, 461–468. [Google Scholar] [CrossRef]
- Tcherepanov, V.; Ehlers, A.; Upton, C. Genome Annotation Transfer Utility (GATU): Rapid annotation of viral genomes using a closely related reference genome. BMC Genom. 2006, 7. [Google Scholar] [CrossRef] [PubMed]
- Krzywinski, M.; Schein, J.; Birol, I.; Connors, J.; Gascoyne, R.; Horsman, D.; Jones, S.J.; Marra, M.A. Circos: An information aesthetic for comparative genomics. Genome Res. 2009, 19, 1639–1645. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [PubMed]
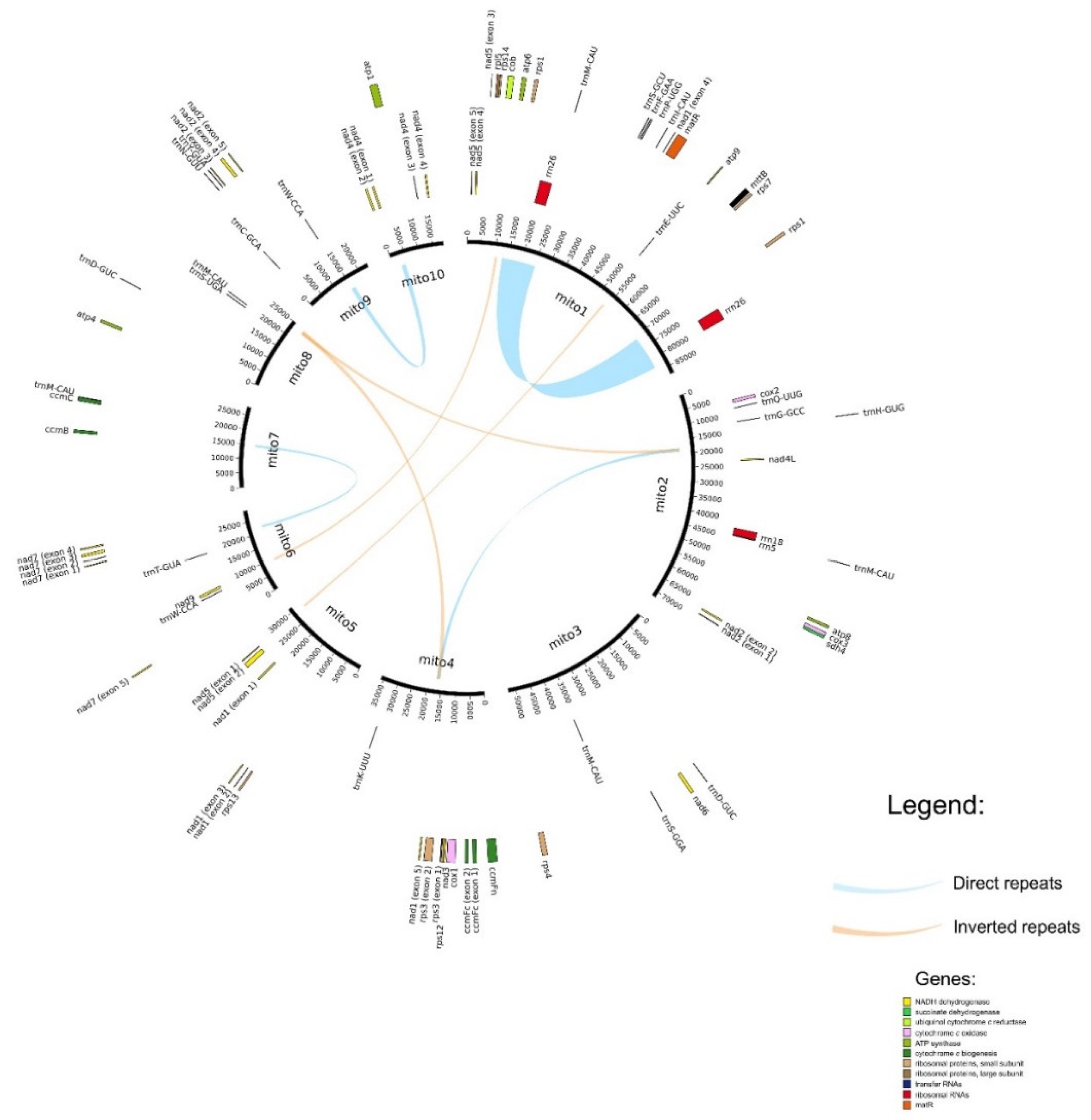
| Number of Chromosomes | Length | Coverage | Accession Number (NCBI) |
|---|---|---|---|
| 1 | 87,722 | 861 | MT318702 |
| 2 | 71,837 | 839 | MT318703 |
| 3 | 52,654 | 872 | MT318705 |
| 4 | 35,904 | 763 | MT318701 |
| 5 | 31,499 | 910 | MT318704 |
| 6 | 28,895 | 864 | MT318706 |
| 7 | 27,675 | 713 | MT318708 |
| 8 | 25,181 | 769 | MT318709 |
| 9 | 23,738 | 988 | MT318710 |
| 10 | 18,958 | 727 | MT318707 |
| Reference Position | Type | Reference (Dasha) | Alternative (F. esculentum ssp. ancestrale) | Coverage | Frequency | Amino Acid Change |
|---|---|---|---|---|---|---|
| 1354 | SNV | G | T | 164 | 100 | psbA:p.Leu49Ile |
| 2105 | SNV | A | C | 144 | 100 | matK:p.Phe465Val |
| 2809 | SNV | C | T | 163 | 99,4 | matK:p.Arg230Gln |
| 3467 | SNV | C | T | 155 | 98,7 | matK:p.Asp11Asn |
| 11370 | SNV | C | A | 143 | 100 | atpF:p.Lys157Asn |
| 19716 | SNV | A | T | 184 | 100 | rpoC2:p.Phe85Ile |
| 34532 | SNV | C | A | 178 | 100 | psbC:p.Leu209Ile |
| 46072 | SNV | G | T | 161 | 100 | rps4:p.Ser148Tyr |
| 58194 | SNV | T | G | 188 | 98,9 | accD:p.Phe61Leu |
| 58536 | SNV | A | C | 149 | 98,7 | accD:p.Glu175Asp |
| 69342 | SNV | G | T | 129 | 100 | rpl20:p.Ser114Tyr |
| 75628 | SNV | T | G | 203 | 100 | psbH:p.Ser48Ala |
| 81203 | SNV | C | T | 152 | 100 | rps8:p.Ser78Asn |
| 83622 | SNV | A | G | 173 | 100 | rps3:p.Met199Thr |
| 90367 | SNV | C | A | 146 | 93,8 | ycf2:p.Gln974Lys |
| 92722 | SNV | A | G | 215 | 83,7 | ycf2:p.Ser1759Gly |
| 110512 | SNV | T | G | 180 | 99,4 | ycf1:p.Phe278Leu |
| 111003 | SNV | C | A | 139 | 100 | ycf1:p.Ser442Tyr |
| 112085 | SNV | T | C | 138 | 100 | ycf1:p.Phe803Leu |
| 113218 | SNV | T | A | 131 | 97,7 | ycf1:p.Phe1180Leu |
| 115685 | SNV | G | A | 144 | 93,1 | ndhF:p.Leu687Phe |
| 115832 | SNV | A | G | 118 | 96,6 | ndhF:p.Phe638Leu |
| 116149 | SNV | G | A | 142 | 96,5 | ndhF:p.Ala532Val |
| 116840 | SNV | C | G | 214 | 98,6 | ndhF:p.Val302Leu |
| 118832 | SNV | C | G | 79 | 100 | rpl32:p.Arg49Gly |
| 127083 | SNV | G | T | 225 | 100 | ndhA:p.Ser92Arg |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Logacheva, M.D.; Schelkunov, M.I.; Fesenko, A.N.; Kasianov, A.S.; Penin, A.A. Mitochondrial Genome of Fagopyrum esculentum and the Genetic Diversity of Extranuclear Genomes in Buckwheat. Plants 2020, 9, 618. https://doi.org/10.3390/plants9050618
Logacheva MD, Schelkunov MI, Fesenko AN, Kasianov AS, Penin AA. Mitochondrial Genome of Fagopyrum esculentum and the Genetic Diversity of Extranuclear Genomes in Buckwheat. Plants. 2020; 9(5):618. https://doi.org/10.3390/plants9050618
Chicago/Turabian StyleLogacheva, Maria D., Mikhail I. Schelkunov, Aleksey N. Fesenko, Artem S. Kasianov, and Aleksey A. Penin. 2020. "Mitochondrial Genome of Fagopyrum esculentum and the Genetic Diversity of Extranuclear Genomes in Buckwheat" Plants 9, no. 5: 618. https://doi.org/10.3390/plants9050618
APA StyleLogacheva, M. D., Schelkunov, M. I., Fesenko, A. N., Kasianov, A. S., & Penin, A. A. (2020). Mitochondrial Genome of Fagopyrum esculentum and the Genetic Diversity of Extranuclear Genomes in Buckwheat. Plants, 9(5), 618. https://doi.org/10.3390/plants9050618




