Germination Data Analysis by Time-to-Event Approaches
Abstract
1. Introduction
2. Results and Discussion
2.1. Kaplan-Meier Estimator
2.2. Cox’s Proportional Hazard
2.3. Accelerated Failure Time Model
2.4. General Considerations
3. Material and Methods
3.1. Germination Experiment
3.2. Data Collection and Statistical Analysis
3.3. Non-Parametric Approach
3.4. Semi-Parametric Approach
3.5. Parametric Approach
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| AFT | Accelerated Failure Time |
| F-H | Fleming- Harrington |
| KM | Kaplan Meier |
| M-H | Mantel-Haenszel |
| PH | Proportional Hazard |
References
- Baskin, C.C.; Baskin, J.M. Seeds: Ecology, biogeography, and evolution of dormancy and germination; Academic Press: New York, NY, USA, 2001; Chapter 2; ISBN 9780124166776. [Google Scholar]
- Hay, F.; Mead, A.; Bloomberg, M. Modelling seed germination in response to continuous variables: Use and limitations of probit analysis and alternative approaches. Seed Sci. Res. 2014, 24, 165–186. [Google Scholar] [CrossRef]
- Ranal, M.A.; de Santana, D.G. How and why to measure the germination process? Braz. J. Bot. 2000, 29, 1–11. [Google Scholar] [CrossRef]
- Shafii, B.; Price, W.J.; Swensen, J.B.; Murray, G.A. Nonlinear Estimation of Growth Curve Models for Germination Data Analysis. In Proceedings of the 1991 Kansas State University Conference on Applied Statistics in Agriculture; Manhattan, K.S., Milliken, G.A., Schwenke, J.R., Eds.; Kansas State University: Manhattan, KS, USA, 1991; pp. 19–42. [Google Scholar]
- Romano, A.; Stevanato, P.; Sorgonà, A.; Cacco, G.; Abenavoli, M.R. Dynamic Response of Key Germination Traits to NaCl Stress in Sugar Beet Seeds. Sugar Tech. 2018. [Google Scholar] [CrossRef]
- Cox, D.R.; Oakes, D. Analysis of survival data; Chapman and Hall: London, UK, 1984; ISBN 9780412244902. [Google Scholar]
- Wu, L.; Teräväinen, T.; Kaiser, G.; Anderson, R.; Boulanger, A.; Rudin, C. Estimation of System Reliability Using a Semiparametric Model. In Proceedings of the IEEE EnergyTech, Cleveland, OH, USA, 25–26 May 2011; Case Western Reserve University: Cleveland, OH, USA. [Google Scholar] [CrossRef]
- Burhanuddin, M.A.; Ghani, M.K.A.; Ahmad, A.; Abal Abas, Z.; Izzah, Z. Reliability analysis of the failure data in industrial repairable systems due to equipment risk factors. Appl. Math. Sci. 2014, 8, 1543–1555. [Google Scholar] [CrossRef]
- Abadi, A.; Amanpour, F.; Bajdik, C.; Yavari, P. Breast cancer survival analysis: Applying the generalized gamma distribution under different conditions of the proportional hazards and accelerated failure time assumptions. Int J. Prev. Med. 2012, 3, 644–651. [Google Scholar]
- Austin, P.C. The use of propensity score methods with survival or time-to-event outcomes: Reporting measures of effect similar to those used in randomized experiments. Stat. Med. 2013, 33, 1242–1258. [Google Scholar] [CrossRef]
- Keiley, M.K.; Martin, N.C. Survival Analysis in Family Research. J. Fam. Psychol. 2005, 19, 142–156. [Google Scholar] [CrossRef]
- Scherm, H.; Ojiambo, P.S. Applications of survival analysis in botanical epidemiology. Phytopathology 2004, 94, 1022–1026. [Google Scholar] [CrossRef]
- He, F.; Alfaro, R.I. White pine weevil attack on white spruce: A survival time analysis. Ecol. Appl. 2000, 10, 225–232. [Google Scholar] [CrossRef]
- Magnussen, S.; Alfaro, R.I.; Boudewyn, P. Survival-time analysis of white spruce during spruce budworm defoliation. Silva Fenn. 2005, 39, 177–189. [Google Scholar] [CrossRef]
- Peto, R.; Pike, M.C.; Armitage, P.; Breslow, N.E.; Cox, D.R.; Howard, S.V.; Mantel, N.; McPherson, K.; Peto, J.; Smith, P.G. Design and analysis of randomized clinical trials requiring prolonged observation of each patient. Part II: Analysis and Examples. Brit. J. Cancer. 1977, 35, 1–39. [Google Scholar] [CrossRef]
- Lewicki, P.; Hill, T. Statistics: Methods and Applications, 1st ed.; StatSoft, Inc.: Tulsa, OK, USA, 2006; p. 578. ISBN 1884233597. [Google Scholar]
- McNair, J.N.; Sunkara, A.; Frobish, D. How to analyse seed germination data using statistical time to event analysis: Nonparametric and semiparametric methods. Seed Sci. Res. 2012, 22, 77–95. [Google Scholar] [CrossRef]
- Onofri, A.; Gresta, F.; Tei, F. A new method for the analysis of germination and emergence data of weed species. Weed Res. 2010, 50, 187–198. [Google Scholar] [CrossRef]
- Scott, S.J.; Jones, R.A. Low temperature seed germination of Lycopersicon species evaluated by survival analysis. Euphytica 1982, 31, 869–883. [Google Scholar] [CrossRef]
- Gunjača, J.; Šarčević, H. Survival analysis of the wheat germination data. In Proceedings of the 22nd International Conference on Information Technology Interfaces (ITI 2000), Pula, Croatia, 13–16 June 2000; pp. 307–310. [Google Scholar]
- Manso, R.; Fortin, M.; Calama, R.; Pardos, M. Modelling seed germination in forest tree species through survival analysis. The Pinus pinea L. case study. Forest. Ecol. Manag. 2013, 289, 515–524. [Google Scholar] [CrossRef]
- Kauth, P.; Biber, P. Testa imposed dormancy in Vallisneria americana seeds from the Mississippi Gulf Coast. J. Torrey Bot. Soc. 2014, 141, 80–90. [Google Scholar] [CrossRef]
- Andersen, G.L.; Krzywinski, K.; Gjessing, H.K.; Pierce, R.H. Seed viability and germination success of Acacia tortilis along land-use and aridity gradients in the Eastern Sahara. Ecol. Evol. 2015, 6, 256–266. [Google Scholar] [CrossRef]
- Chhetri, S.B.; Rawal, D.S. Germination Phenological Response Identifies Flora Risk to Climate Change. Climate 2017, 5, 73. [Google Scholar] [CrossRef]
- Cumming, E.; Jarvis, J.C.; Sherman, C.D.H.; York, P.H.; Smith, T.M. Seed germination in a southern Australian temperate seagrass. Peer J. 2017, 5, e3114. [Google Scholar] [CrossRef] [PubMed]
- Mamani, G.; Chuquillanqui Soto, H.; Chumbiauca Mateo, S.L.; Sahley, C.T.; Alonso, A.; Linares-Palomino, R. Substrate, moisture, temperature and seed germination of the threatened endemic tree Eriotheca vargasii (Malvaceae). Rev. Biol. Trop. 2018, 66, 1162–1170. [Google Scholar] [CrossRef]
- Barak, R.S.; Lichtenberger, T.; Wellman-Houde, A.; Kramer, A.T.; Larkin, D.J. Cracking the case: Seed traits and phylogeny predict time to germination in prairie restoration species. Ecol. Evol. 2018, 8, 5551–5562. [Google Scholar] [CrossRef] [PubMed]
- Winkler, M.; Hülber, K.; Hietz, P. Effect of canopy position on germination and seedling survival of epiphytic bromeliads in a Mexican humid montane forest. Ann Bot. 2005, 95, 1039–1047. [Google Scholar] [CrossRef]
- Hirsch, H.; Wypior, C.; von Wehrden, H.; Wesche, K.; Renison, D.; Hensen, I. Germination performance of native and non-native Ulmus pumila populations. NeoBiota 2012, 15, 53–68. [Google Scholar] [CrossRef]
- Lawless, J.F. Statistical models and methods for lifetime data; John Wiley and Sons: New York, NY, USA, 2000; pp. 14–16. ISBN 978-0-471-37215-8. [Google Scholar]
- Kleinbaum, D.; Klein, M. Survival analysis: A self-learning text; Springer: New York, NY, USA, 2005; pp. 292–300. ISBN 978-1-4419-6645-2. [Google Scholar]
- Onofri, A.; Mesgaran, M.B.; Tei, F.; Cousens, R.D. The cure model: An improved way to describe seed germination? Weed Res. 2011, 51, 516–524. [Google Scholar] [CrossRef]
- Michel, B.E. Evaluation of the water potentials of solutions of Polyethylene Glycol 8000 both in the absence and presence of other solutes. Plant Physiol. 1983, 72, 66–70. [Google Scholar] [CrossRef]
- Klein, J.P.; Moeschberger, M.L. Survival analysis: Techniques for censored and truncated data, 2nd ed.; Springer-Verlag: New York, NY, USA, 2003; ISBN 978-0-387-95399-1. [Google Scholar]
- Blagoev, K.B.; Wilkerson, J.; Fojo, T. Hazard ratios in cancer clinical trials-a primer. Nat. Rev. Clin. Oncol. 2012, 9, 178–183. [Google Scholar] [CrossRef]
- George, B.; Seals, S.; Aban, I. Survival analysis and regression models. J. Nucl. Cardiol. 2014, 21, 686–694. [Google Scholar] [CrossRef]
- Kaplan, E.L.; Meier, P. Nonparametric estimation from incomplete observations. J. Am. Stat. Assoc. 1958, 53, 457–481. [Google Scholar] [CrossRef]
- Mantel, N. Evaluation of survival data and two new rank order statistics arising in its consideration. Cancer Chemoth. Rep. 1966, 50, 163–170. [Google Scholar]
- Tarone, R.E.; Ware, J. On distribution-free tests for equality of survival distributions. Biometrika 1977, 64, 156–160. [Google Scholar] [CrossRef]
- Kalbfleisch, J.D.; Prentice, R.L. Statistical Analysis of Failure Time Data, 2nd ed.; Wiley: New York, NY, USA, 2002; pp. 20–26. ISBN 9780471363576. [Google Scholar]
- Le, C.T. Applied survival analysis; John Wiley and Sons: New York, NY, USA, 1997; ISBN 0471170852. [Google Scholar]
- Peto, R.; Peto, J. Asymptotically efficient rank invariant procedures. J. R. Stat. Soc. 1972, 135, 185–207. [Google Scholar] [CrossRef]
- Fleming, T.R.; Harrington, D.P. A class of hypothesis tests for one and two samples censored survival data. Commun. Stat. 1981, 10, 763–794. [Google Scholar] [CrossRef]
- Klein, J.P.; Rizzo, J.D.; Zhang, M.J.; Keiding, N. Statistical methods for the analysis and presentation of the results of bone marrow transplants. Part I: Unadjusted analysis. Bone Marrow Transpl. 2001, 28, 909–915. [Google Scholar] [CrossRef]
- Gomez, G.; Calle, M.L.; Oller, R.; Langohr, K. Tutorial on methods for interval-censored data and their implementation in R. Stat. Model. 2009, 9, 259–297. [Google Scholar] [CrossRef]
- Cox, D.R. Regression models and life tables (with discussion). J.R. Statist. Soc. B. 1972, 34, 187–220. [Google Scholar]
- Hess, K.R. Graphical methods for assessing violations of the proportional hazards assumption in Cox regression. Stat. Med. 1995, 14, 1707–1723. [Google Scholar] [CrossRef]
- Therneau, T.M.; Grambsch, P.M. Modeling survival data: Extending the Cox model; Springer-Verlag: New York, NY, USA, 2000; pp. 48–53. ISBN 978-0-387-98784-2. [Google Scholar]
- Therneau, T.M.; Grambsch, P.M.; Pankratz, V.S. Penalized Survival Models and Frailty. J. Comput. Graph. Stat. 2003, 12, 156–175. [Google Scholar] [CrossRef]
- Akaike, H. A new look at the statistical model identification. IEEE Trans. Autom. Control 1974, 19, 716–723. [Google Scholar] [CrossRef]

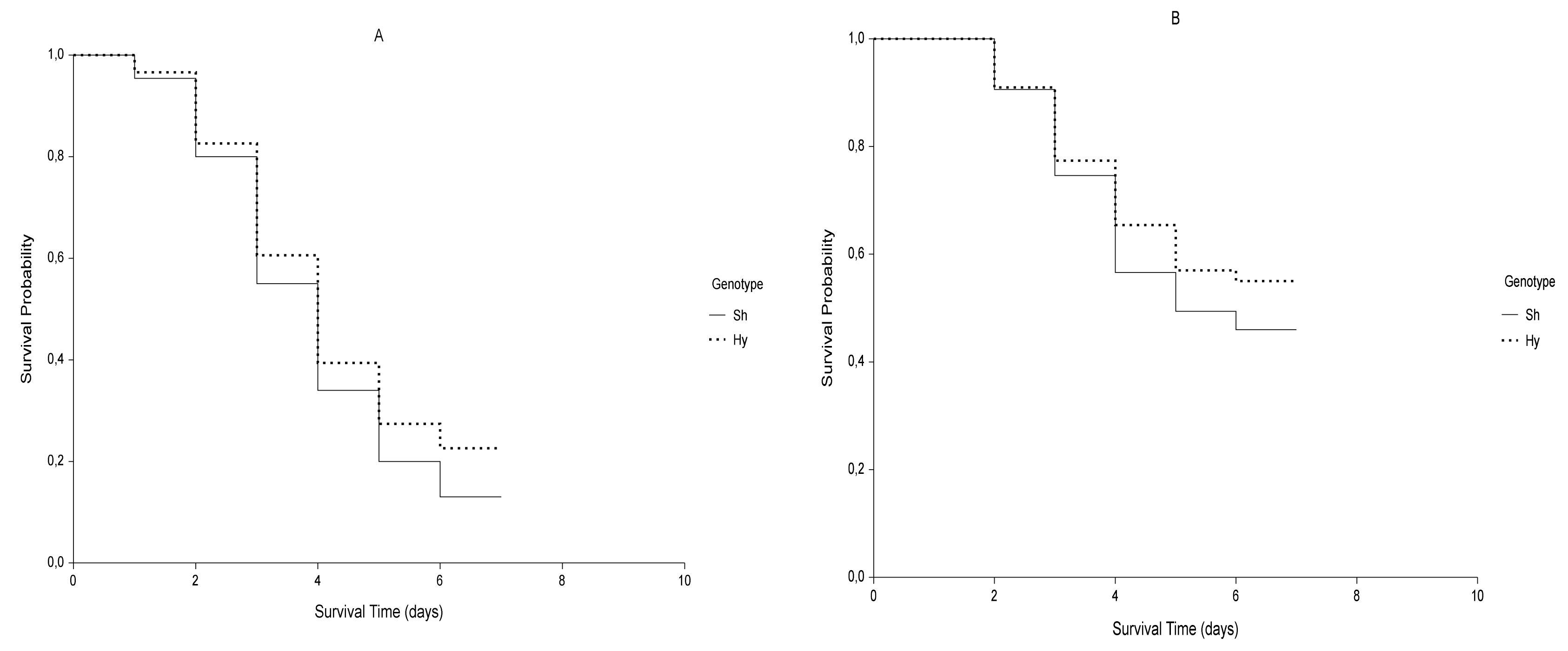

| G | 0 MPa vs. −0.6 MPa | T | Sh vs. Hy | |||||||
| Sh | Test | χ2 | df | p-value | z-value | No-Stress | χ2 | df | p-value | z-value |
| M-H Log-rank | 50.012 | 1 | <0.0001 | ±7.072 | 4.432 | 1 | 0.0353 * | ±2.105 | ||
| Gehan-Wilcoxon | 38.361 | 1 | <0.0001 | ±6.194 | 2.588 | 1 | 0.1077 | ±1.609 | ||
| Tarone-Ware | 43.982 | 1 | <0.0001 | ±6.632 | 3.359 | 1 | 0.0668 | ±1.833 | ||
| Peto-Peto | 38.428 | 1 | <0.0001 | ±6.199 | 2.354 | 1 | 0.1249 | ±1.534 | ||
| Modified Peto-Peto | 38.380 | 1 | <0.0001 | ±6.195 | 2.348 | 1 | 0.1254 | ±1.532 | ||
| Fleming-Harrington | 48.561 | 1 | <0.0001 | ±6.969 | 5.602 | 1 | 0.0179 * | ±2.367 | ||
| Hy | M-H Log-rank | 43.580 | 1 | <0.0001 | ±6.602 | Stress | 2.972 | 1 | 0.0847 | ±1.724 |
| Gehan-Wilcoxon | 36.546 | 1 | <0.0001 | ±6.045 | 2.588 | 1 | 0.1077 | ±1.609 | ||
| Tarone-Ware | 40.225 | 1 | <0.0001 | ±6.342 | 2.799 | 1 | 0.0943 | ±1.673 | ||
| Peto-Peto | 36.321 | 1 | <0.0001 | ±6.027 | 2.452 | 1 | 0.1174 | ±1.566 | ||
| Modified Peto-Peto | 36.292 | 1 | <0.0001 | ±6.024 | 2.451 | 1 | 0.1175 | ±1.565 | ||
| Fleming-Harrington | 39.032 | 1 | <0.0001 | ±6.248 | 2.741 | 1 | 0.0978 | ±1.656 | ||
| Explanatory Variable | βi | SE of βi | Exp (βi) | Wald z-Value | p-Value | Exp (βi) 95% CI |
|---|---|---|---|---|---|---|
| Genotype (Hy) | −0.2344 | 0.087 | 0.791 | −2.6875 | 0.0072 * | 0.6667–0.9385 |
| Treatment (stress) | −0.8685 | 0.090 | 0.419 | −9.6221 | <0.00001 | 0.3515–0.5008 |
| Log-Normal Distribution | ||||||
| Genotype | Treatment (ψ in MPa) | Shape (s) | SE | Scale (m) | SE | −2 Log-Likelihood |
| Sh | 0 | 1.311 | 0.038 | 0.538 | 0.029 | 747.08 |
| −0.6 | 1.550 | 0.092 | 1.109 | 0.084 | 618.34 | |
| Hy | 0 | 1.425 | 0.044 | 0.601 | 0.036 | 740.10 |
| −0.6 | 1.835 | 0.114 | 1.242 | 0.105 | 565.32 | |
| Log-Logistic Distribution | ||||||
| Sh | 0 | 1.315 | 0.037 | 0.304 | 0.019 | 742.94 |
| −0.6 | 1.497 | 0.095 | 0.709 | 0.058 | 620.46 | |
| Hy | 0 | 1.394 | 0.043 | 0.355 | 0.024 | 738.74 |
| −0.6 | 1.792 | 0.117 | 0.800 | 0.074 | 567.14 | |
| Log-Normal Model | |||||||
| Parameter | Parameter Estimate | SE | z-Value | p-Value | 95% CI | T50 | Time Ratio (γ) |
| Intercept | 1.3185 (α0) | 0.0403 | 32.68 | <0.0001 | 1.24–1.40 | 3.7381 | 1 |
| Treatment | 0.4520 (α1) | 0.0482 | 9.36 | <0.0001 | 0.35–0.54 | 5.8747 | 1.5715 |
| Genotype | 0.1151 (α2) | 0.0481 | 2.39 | 0.0167 * | 0.02–0.20 | 4.1944 | 1.1220 |
| Log-Logistic Model | |||||||
| Parameter | Parameter Estimate | SE | z-value | p-value | 95% CI | T50 | Time Ratio (γ) |
| Intercept | 1.3086 (α0) | 0.0397 | 32.92 | <0.0001 | 1.23–1.38 | 3.7013 | 1 |
| Treatment | 0.4554 (α1) | 0.0487 | 9.34 | <0.0001 | 0.36–0.55 | 5.8364 | 1.5768 |
| Genotype | 0.1184 (α2) | 0.0493 | 2.40 | 0.0163* | 0.02–0.21 | 4.1666 | 1.1257 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Romano, A.; Stevanato, P. Germination Data Analysis by Time-to-Event Approaches. Plants 2020, 9, 617. https://doi.org/10.3390/plants9050617
Romano A, Stevanato P. Germination Data Analysis by Time-to-Event Approaches. Plants. 2020; 9(5):617. https://doi.org/10.3390/plants9050617
Chicago/Turabian StyleRomano, Alessandro, and Piergiorgio Stevanato. 2020. "Germination Data Analysis by Time-to-Event Approaches" Plants 9, no. 5: 617. https://doi.org/10.3390/plants9050617
APA StyleRomano, A., & Stevanato, P. (2020). Germination Data Analysis by Time-to-Event Approaches. Plants, 9(5), 617. https://doi.org/10.3390/plants9050617






