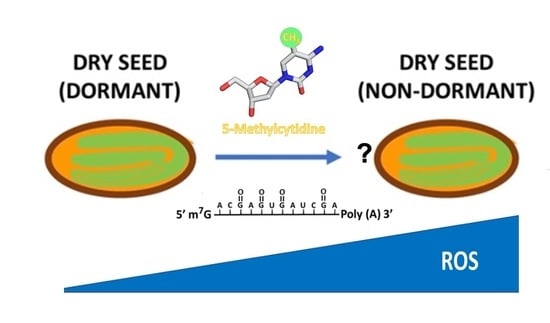
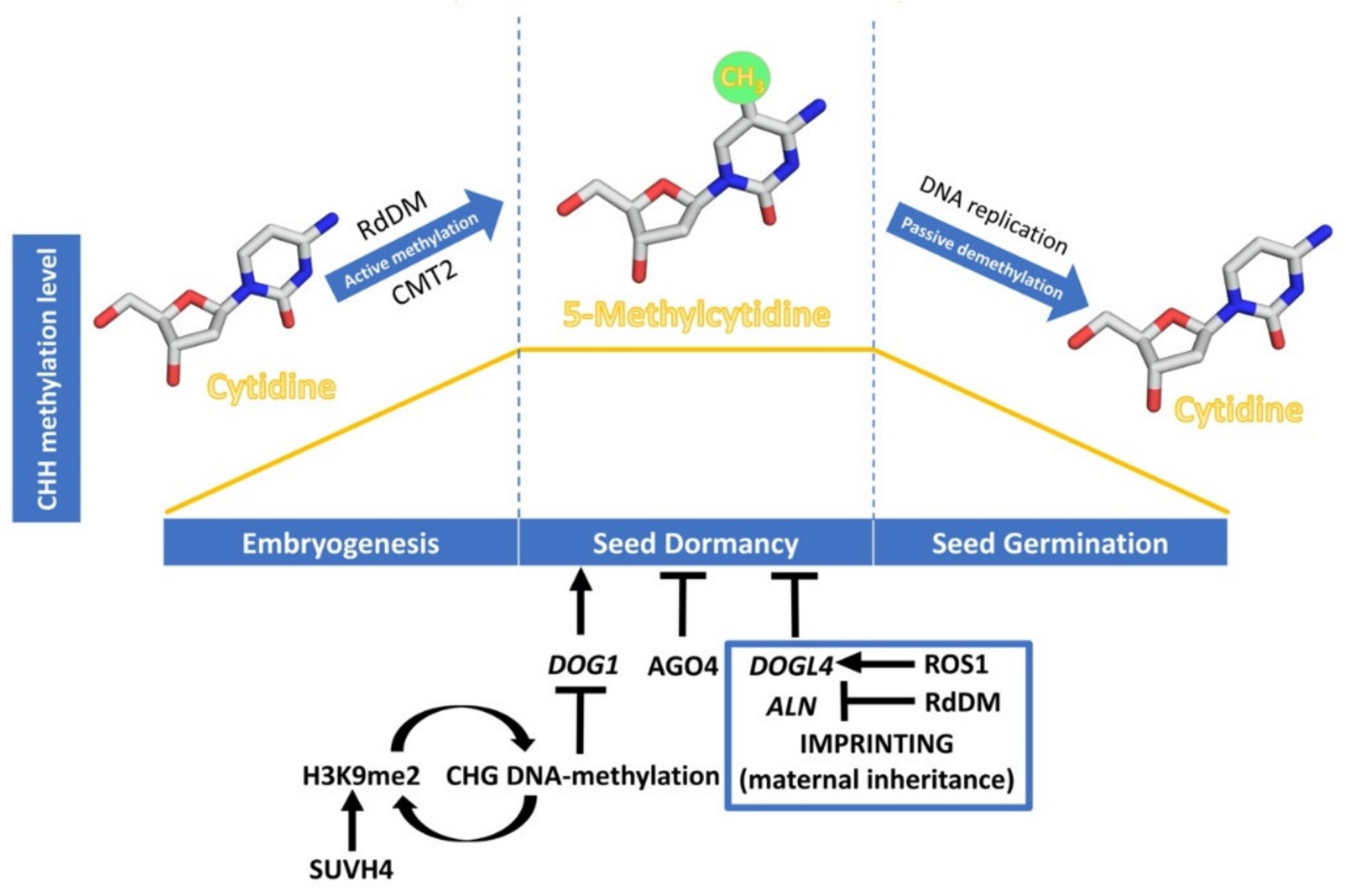
Reactive Oxygen Species (ROS) and Nucleic Acid Modifications during Seed Dormancy
Abstract
1. Introduction: Seed Dormancy
2. Reactive Oxygen Species (ROS) Promote Dormancy Release and Modify Stability of Nucleic Acids
2.1. ROS Affect Dormancy Release
2.2. Oxidation Modifies Nucleic Acid Stability during Seed Dormancy
3. mRNA and DNA Methylation upon Seed Development
3.1. The Seed Epitranscriptome
3.2. Regulation of Seed Development through DNA Methylation
3.2.1. DNA Methylation Basis
3.2.2. DNA-Methylation and Seed Development
4. Concluding Remarks and Future Perspectives
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Vicente-Carbajosa, J.; Carbonero, P. Seed maturation: Developing an intrusive phase to accomplish a quiescent state. Int. J. Dev. Biol. 2005, 49, 645–651. [Google Scholar] [CrossRef]
- Cadman, C.S.C.; Toorop, P.E.; Hilhorst, H.W.M.; Finch-Savage, W.E. Gene expression profiles of Arabidopsis Cvi seeds during dormancy cycling indicate a common underlying dormancy control mechanism. Plant J. 2006, 46, 805–822. [Google Scholar] [CrossRef] [PubMed]
- Holdsworth, M.J.; Bentsink, L.; Soppe, W.J.J. Molecular networks regulating Arabidopsis seed maturation, after-ripening, dormancy and germination. New Phytol. 2008, 179, 33–54. [Google Scholar] [CrossRef] [PubMed]
- Penfield, S. Seed dormancy and germination. Curr. Biol. 2017, 27, R874–R878. [Google Scholar] [CrossRef] [PubMed]
- Gubler, F.; Millar, A.A.; Jacobsen, J.V. Dormancy release, ABA and pre-harvest sprouting. Curr. Opin. Plant Biol. 2005, 8, 183–187. [Google Scholar] [CrossRef] [PubMed]
- Finkelstein, R.; Reeves, W.; Ariizumi, T.; Steber, C. Molecular aspects of seed dormancy. Annu. Rev. Plant Biol. 2008, 59, 387–415. [Google Scholar] [CrossRef]
- Barrero, J.M.; Millar, A.A.; Griffiths, J.; Czechowski, T.; Scheible, W.R.; Udvardi, M.; Reid, J.B.; Ross, J.J.; Jacobsen, J.V.; Gubler, F. Gene expression profiling identifies two regulatory genes controlling dormancy and ABA sensitivity in Arabidopsis seeds. Plant J. 2010, 61, 611–622. [Google Scholar] [CrossRef]
- Abraham, Z.; Iglesias-Fernández, R.; Martínez, M.; Rubio-Somoza, I.; Díaz, I.; Carbonero, P.; Vicente-Carbajosa, J. A developmental switch of gene expression in the barley seed mediated by HvVP1 (Viviparous-1) and HvGAMYB interactions. Plant Physiol. 2016, 170, 2146–2158. [Google Scholar] [CrossRef]
- Shu, K.; Liu, X.; Xie, Q.; He, Z. Two Faces of One Seed: Hormonal regulation of dormancy and germination. Mol. Plant 2016, 9, 34–45. [Google Scholar] [CrossRef]
- Carbonero, P.; Iglesias-Fernández, R.; Vicente-Carbajosa, J. The AFL subfamily of B3 transcription factors: Evolution and function in angiosperm seeds. J. Exp. Bot. 2017, 68, 871–880. [Google Scholar] [CrossRef]
- Nonogaki, H. The long-standing paradox of seed dormancy unfolded? Trends Plant Sci. 2019, 24, 989–998. [Google Scholar] [CrossRef] [PubMed]
- Nakabayashi, K.; Bartsch, M.; Xiang, Y.; Miatton, E.; Pellengahr, S.; Yano, R.; Seo, M.; Soppe, W.J.J. The time required for dormancy release in Arabidopsis is determined by DELAY OF GERMINATION1 protein levels in freshly harvested seeds. Plant Cell 2012, 24, 2826–2838. [Google Scholar] [CrossRef] [PubMed]
- Alonso-Blanco, C.; Bentsink, L.; Hanhart, C.J.; Blankestijn-de Vries, H.; Koornneef, M. Analysis of natural allelic variation at seed dormancy loci of Arabidopsis thaliana. Genetics 2003, 164, 711–729. [Google Scholar] [CrossRef] [PubMed]
- Bentsink, L.; Jowett, J.; Hanhart, C.J.; Koornneef, M. Cloning of DOG1, a quantitative trait locus controlling seed dormancy in Arabidopsis. Proc. Natl. Acad. Sci. USA 2006, 103, 17042–17047. [Google Scholar] [CrossRef] [PubMed]
- Née, G.; Xiang, Y.; Soppe, W.J.J. The release of dormancy, a wake-up call for seeds to germinate. Curr. Opin. Plant Biol. 2017, 35, 8–14. [Google Scholar] [CrossRef] [PubMed]
- Nishimura, N.; Tsuchiya, W.; Moresco, J.J.; Hayashi, Y.; Satoh, K.; Kaiwa, N.; Irisa, T.; Kinoshita, T.; Schroeder, J.I.; Yates, J.R.; et al. Control of seed dormancy and germination by DOG1-AHG1 PP2C phosphatase complex via binding to heme. Nat. Commun. 2018, 9. [Google Scholar] [CrossRef]
- Nonogaki, H. A repressor complex silencing ABA signaling in seeds? J. Exp. Bot. 2020. [Google Scholar] [CrossRef]
- Carrillo-Barral, N.; del Carmen Rodríguez-Gacio, M.; Matilla, A.J. Delay of Germination-1 (DOG1): A key to understanding seed dormancy. Plants 2020, 9, 480. [Google Scholar] [CrossRef]
- Baskin, J.M.; Baskin, C.C. A classification system for seed dormancy. Seed Sci. Res. 2004, 14, 1–16. [Google Scholar] [CrossRef]
- Iglesias-Fernández, R.; Matilla, A. After-ripening alters the gene expression pattern of oxidases involved in the ethylene and gibberellin pathways during early imbibition of Sisymbrium officinale L. seeds. J. Exp. Bot. 2009, 60, 1645–1661. [Google Scholar] [CrossRef]
- Chahtane, H.; Kim, W.; Lopez-Molina, L. Primary seed dormancy: A temporally multilayered riddle waiting to be unlocked. J. Exp. Bot. 2017, 68, 857–869. [Google Scholar] [CrossRef] [PubMed]
- Buijs, G.; Vogelzang, A.; Nijveen, H.; Bentsink, L. Dormancy cycling: Translation-related transcripts are the main difference between dormant and non-dormant seeds in the field. Plant J. 2019. [Google Scholar] [CrossRef] [PubMed]
- Donohue, K.; Dorn, L.; Griffith, C.; Kim, E.S.; Aguilera, A.; Polisetty, C.R.; Schmitt, J. The evolutionary ecology of seed germination of Arabidopsis thaliana: Variable natural selection on germination timing. Evolution 2005, 59, 758–770. [Google Scholar] [CrossRef] [PubMed]
- Schmuths, H.; BACHMANN, K.; WEBER, W.E.; HORRES, R.; HOFFMANN, M.H. Effects of preconditioning and temperature during germination of 73 natural accessions of Arabidopsis thaliana. Ann. Bot. 2006, 97, 623–634. [Google Scholar] [CrossRef]
- Nakabayashi, K.; Okamoto, M.; Koshiba, T.; Kamiya, Y.; Nambara, E. Genome-wide profiling of stored mRNA in Arabidopsis thaliana seed germination: Epigenetic and genetic regulation of transcription in seed. Plant J. 2005, 41, 697–709. [Google Scholar] [CrossRef]
- Carrera, E.; Holman, T.; Medhurst, A.; Dietrich, D.; Footitt, S.; Theodoulou, F.L.; Holdsworth, M.J. Seed after-ripening is a discrete developmental pathway associated with specific gene networks in Arabidopsis. Plant J. 2008, 53, 214–224. [Google Scholar] [CrossRef]
- Carrillo-Barral, N.; Matilla, A.J.; Iglesias-Fernández, R.; del Carmen Rodríguez-Gacio, M. Nitrate-induced early transcriptional changes during imbibition in non-after-ripenedSisymbrium officinaleseeds. Physiol. Plant. 2013, 148, 560–573. [Google Scholar] [CrossRef]
- Carrillo-Barral, N.; Matilla, A.J.; del Carmen Rodríguez-Gacio, M.; Iglesias-Fernández, R. Nitrate affects sensu-stricto germination of after-ripened Sisymbrium officinale seeds by modifying expression of SoNCED5, SoCYP707A2 and SoGA3ox2 genes. Plant Sci. 2014, 217–218, 99–108. [Google Scholar] [CrossRef]
- Dekkers, B.J.W.; Pearce, S.P.; van Bolderen-Veldkamp, R.P.M.; Holdsworth, M.J.; Bentsink, L. Dormant and after-Ripened Arabidopsis thaliana seeds are distinguished by early transcriptional differences in the imbibed state. Front. Plant Sci. 2016, 7. [Google Scholar] [CrossRef]
- Bazin, J.; Langlade, N.; Vincourt, P.; Arribat, S.; Balzergue, S.; El-Maarouf-Bouteau, H.; Bailly, C. Targeted mRNA oxidation regulates sunflower seed dormancy alleviation during dry after-ripening. Plant Cell 2011, 23, 2196–2208. [Google Scholar] [CrossRef]
- Bailly, C. The signalling role of ROS in the regulation of seed germination and dormancy. Biochem. J. 2019, 476, 3019–3032. [Google Scholar] [CrossRef] [PubMed]
- Finch-Savage, W.E.; Cadman, C.S.C.; Toorop, P.E.; Lynn, J.R.; Hilhorst, H.W.M. Seed dormancy release in Arabidopsis Cvi by dry after-ripening, low temperature, nitrate and light shows common quantitative patterns of gene expression directed by environmentally specific sensing. Plant J. 2007, 51, 60–78. [Google Scholar] [CrossRef] [PubMed]
- Sano, N.; Permana, H.; Kumada, R.; Shinozaki, Y.; Tanabata, T.; Yamada, T.; Hirasawa, T.; Kanekatsu, M. Proteomic analysis of embryonic proteins synthesized from long-Lived mRNAs during germination of rice seeds. Plant Cell Physiol. 2012, 53, 687–698. [Google Scholar] [CrossRef] [PubMed]
- Galland, M.; Huguet, R.; Arc, E.; Cueff, G.; Job, D.; Rajjou, L. Dynamic proteomics emphasizes the importance of selective mRNA translation and protein turnover during Arabidopsis seed germination. Mol. Cell. Proteom. 2014, 13, 252–268. [Google Scholar] [CrossRef]
- Sano, N.; Rajjou, L.; North, H.M. Lost in translation: Physiological roles of stored mRNAs in seed germination. Plants 2020, 9, 347. [Google Scholar] [CrossRef]
- Xiao, W.; Custard, R.D.; Brown, R.C.; Lemmon, B.E.; Harada, J.J.; Goldberg, R.B.; Fischer, R.L. DNA methylation is critical for Arabidopsis embroyogenesis and seed viability. Plant Cell 2006, 18, 805–814. [Google Scholar] [CrossRef]
- Jeevan Kumar, S.P.; Rajendra Prasad, S.; Banerjee, R.; Thammineni, C. Seed birth to death: Dual functions of reactive oxygen species in seed physiology. Ann. Bot. 2015, 116, 663–668. [Google Scholar] [CrossRef]
- Buijs, G.; Kodde, J.; Groot, S.P.C.; Bentsink, L. Seed dormancy release accelerated by elevated partial pressure of oxygen is associated with DOG loci. J. Exp. Bot. 2018, 69, 3601–3608. [Google Scholar] [CrossRef]
- Kurek, K.; Plitta-Michalak, B.; Ratajczak, E. Reactive Oxygen species as potential drivers of the seed aging process. Plants 2019, 8, 174. [Google Scholar] [CrossRef]
- Oracz, K.; Bouteau, H.E.-M.; Farrant, J.M.; Cooper, K.; Belghazi, M.; Job, C.; Job, D.; Corbineau, F.; Bailly, C. ROS production and protein oxidation as a novel mechanism for seed dormancy alleviation. Plant J. 2007, 50, 452–465. [Google Scholar] [CrossRef]
- Müller, K.; Carstens, A.C.; Linkies, A.; Torres, M.A.; Leubner-Metzger, G. The NADPH-oxidase AtrbohB plays a role in Arabidopsis seed after-ripening. New Phytol. 2009, 184, 885–897. [Google Scholar] [CrossRef] [PubMed]
- Ebone, L.A.; Caverzan, A.; Chavarria, G. Physiologic alterations in orthodox seeds due to deterioration processes. Plant Physiol. Biochem. 2019, 145, 34–42. [Google Scholar] [CrossRef] [PubMed]
- De Giorgi, J.; Piskurewicz, U.; Loubery, S.; Utz-Pugin, A.; Bailly, C.; Mène-Saffrané, L.; Lopez-Molina, L. An endosperm-associated cuticle is required for Arabidopsis seed viability, dormancy and early control of germination. PLOS Genet. 2015, 11, e1005708. [Google Scholar] [CrossRef] [PubMed]
- Nakabayashi, K.; Bartsch, M.; Ding, J.; Soppe, W.J.J. Seed dormancy in Arabidopsis requires self-binding ability of DOG1 Protein and the presence of multiple isoforms generated by alternative splicing. PLOS Genet. 2015, 11, e1005737. [Google Scholar] [CrossRef]
- Liu, Y.; Ye, N.; Liu, R.; Chen, M.; Zhang, J. H2O2 mediates the regulation of ABA catabolism and GA biosynthesis in Arabidopsis seed dormancy and germination. J. Exp. Bot. 2010, 61, 2979–2990. [Google Scholar] [CrossRef]
- Barba-Espín, G.; Díaz-Vivancos, P.; Job, D.; Belghazi, M.; Job, C.; Hernández, J.A. Understanding the role of H2O2 during pea seed germination: A combined proteomic and hormone profiling approach. Plant. Cell Environ. 2011, 34, 1907–1919. [Google Scholar] [CrossRef]
- Yu, Y.; Zhen, S.; Wang, S.; Wang, Y.; Cao, H.; Zhang, Y.; Li, J.; Yan, Y. Comparative transcriptome analysis of wheat embryo and endosperm responses to ABA and H2O2 stresses during seed germination. BMC Genom. 2016, 17. [Google Scholar] [CrossRef]
- Ishibashi, Y.; Aoki, N.; Kasa, S.; Sakamoto, M.; Kai, K.; Tomokiyo, R.; Watabe, G.; Yuasa, T.; Iwaya-Inoue, M. The interrelationship between abscisic acid and reactive oxygen species plays a key role in barley seed dormancy and germination. Front. Plant Sci. 2017, 8. [Google Scholar] [CrossRef]
- Leymarie, J.; Vitkauskaité, G.; Hoang, H.H.; Gendreau, E.; Chazoule, V.; Meimoun, P.; Corbineau, F.; El-Maarouf-Bouteau, H.; Bailly, C. Role of reactive oxygen species in the regulation of Arabidopsis seed dormancy. Plant Cell Physiol. 2012, 53, 96–106. [Google Scholar] [CrossRef]
- Morscher, F.; Kranner, I.; Arc, E.; Bailly, C.; Roach, T. Glutathione redox state, tocochromanols, fatty acids, antioxidant enzymes and protein carbonylation in sunflower seed embryos associated with after-ripening and ageing. Ann. Bot. 2015, 116, 669–678. [Google Scholar] [CrossRef]
- Vigliocco, A.; Del Bel, Z.; Pérez-Chaca, M.V.; Molina, A.; Zirulnik, F.; Andrade, A.M.; Alemano, S. Spatiotemporal variations in salicylic acid and hydrogen peroxide in sunflower seeds during transition from dormancy to germination. Physiol. Plant. 2019. [Google Scholar] [CrossRef] [PubMed]
- Müller, K.; Linkies, A.; Vreeburg, R.A.M.; Fry, S.C.; Krieger-Liszkay, A.; Leubner-Metzger, G. In vivo cell wall loosening by hydroxyl radicals during cress seed germination and elongation growth. Plant Physiol. 2009, 150, 1855–1865. [Google Scholar] [CrossRef] [PubMed]
- Oracz, K.; Voegele, A.; Tarkowská, D.; Jacquemoud, D.; Turečková, V.; Urbanová, T.; Strnad, M.; Sliwinska, E.; Leubner-Metzger, G. Myrigalone A inhibits Lepidium sativum seed germination by interference with gibberellin metabolism and apoplastic superoxide production required for embryo extension growth and endosperm rupture. Plant Cell Physiol. 2012, 53, 81–95. [Google Scholar] [CrossRef] [PubMed]
- Waszczak, C.; Carmody, M.; Kangasjärvi, J. Reactive Oxygen Species in plant signaling. Annu. Rev. Plant Biol. 2018, 69, 209–236. [Google Scholar] [CrossRef]
- Oracz, K.; El-Maarouf-Bouteau, H.; Kranner, I.; Bogatek, R.; Corbineau, F.; Bailly, C. The mechanisms involved in seed dormancy alleviation by hydrogen cyanide unravel the role of eactive Oxygen Species as key factors of cellular signaling during germination. Plant Physiol. 2009, 150, 494–505. [Google Scholar] [CrossRef]
- Bahin, E.; Bailly, C.; Sotta, B.; Kranner, I.; Corbineau, F.; Leymarie, J. Crosstalk between reactive oxygen species and hormonal signalling pathways regulates grain dormancy in barley. Plant Cell Environ. 2011, 34, 980–993. [Google Scholar] [CrossRef]
- Kaur, K.; Zhawar, V.K. Antioxidant potential of fresh and after-ripened dry embryos of two wheat cultivars contrasting in drought tolerance. Indian J. Plant Physiol. 2016, 21, 350–354. [Google Scholar] [CrossRef]
- Farooq, M.A.; Niazi, A.K.; Akhtar, J.; Saifullah Farooq, M.; Souri, Z.; Karimi, N.; Rengel, Z. Acquiring control: The evolution of ROS-Induced oxidative stress and redox signaling pathways in plant stress responses. Plant Physiol. Biochem. 2019, 141, 353–369. [Google Scholar] [CrossRef]
- Serrato, A.J.; Crespo, J.L.; Florencio, F.J.; Cejudo, F.J. Characterization of two thioredoxins h with predominant localization in the nucleus of aleurone and scutellum cells of germinating wheat seeds. Plant Mol. Biol. 2001, 46, 361–371. [Google Scholar] [CrossRef]
- Ortiz-Espín, A.; Iglesias-Fernández, R.; Calderón, A.; Carbonero, P.; Sevilla, F.; Jiménez, A. Mitochondrial AtTrxo1 is transcriptionally regulated by AtbZIP9 and AtAZF2 and affects seed germination under saline conditions. J. Exp. Bot. 2017, 68, 1025–1038. [Google Scholar] [CrossRef]
- Xu, F.; Tang, J.; Gao, S.; Cheng, X.; Du, L.; Chu, C. Control of rice pre-harvest sprouting by glutaredoxin-mediated abscisic acid signaling. Plant J. 2019, 100, 1036–1051. [Google Scholar] [CrossRef] [PubMed]
- Chmielowska-Bąk, J.; Arasimowicz-Jelonek, M.; Deckert, J. In search of the mRNA modification landscape in plants. BMC Plant Biol. 2019, 19. [Google Scholar] [CrossRef] [PubMed]
- Simms, C.L.; Hudson, B.H.; Mosior, J.W.; Rangwala, A.S.; Zaher, H.S. An active role for the ribosome in determining the fate of oxidized mRNA. Cell Rep. 2014, 9, 1256–1264. [Google Scholar] [CrossRef] [PubMed]
- Taddei, F.; Hayakawa, H.; Bouton, M.-F.; Cirinesi, A.-M.; Matic, I.; Sekiguchi, M.; Radman, M. Counteraction by MutT protein of transcriptional errors caused by oxidative damage. Science 1997, 278, 128–130. [Google Scholar] [CrossRef] [PubMed]
- Gao, F.; Rampitsch, C.; Chitnis, V.R.; Humphreys, G.D.; Jordan, M.C.; Ayele, B.T. Integrated analysis of seed proteome and mRNA oxidation reveals distinct post-transcriptional features regulating dormancy in wheat (Triticum aestivum L.). Plant Biotechnol. J. 2013, 11, 921–932. [Google Scholar] [CrossRef]
- Gao, F.; Jordan, M.C.; Ayele, B.T. Microarray dataset of after-ripening induced mRNA oxidation in wheat seeds. Data Br. 2018, 21, 852–855. [Google Scholar] [CrossRef]
- Nelson, S.K.; Ariizumi, T.; Steber, C.M. Biology in the dry seed: Transcriptome changes associated with dry seed dormancy and dormancy loss in the arabidopsis GA-insensitive sleepy1-2 mutant. Front. Plant Sci. 2017, 8. [Google Scholar] [CrossRef]
- Cléry, A.; Blatter, M.; Allain, F.H.-T. RNA recognition motifs: Boring? Not quite. Curr. Opin. Struct. Biol. 2008, 18, 290–298. [Google Scholar] [CrossRef]
- Chai, Q.; Singh, B.; Peisker, K.; Metzendorf, N.; Ge, X.; Dasgupta, S.; Sanyal, S. Organization of ribosomes and nucleoids in Escherichia coli cells during growth and in quiescence. J. Biol. Chem. 2014, 289, 11342–11352. [Google Scholar] [CrossRef]
- Bai, B.; van der Horst, S.; Cordewener, J.H.G.; America, T.A.H.P.; Hanson, J.; Bentsink, L. Seed-stored mRNAs that are specifically associated to monosomes are translationally regulated during germination. Plant Physiol. 2020, 182, 378–392. [Google Scholar] [CrossRef]
- El-Maarouf-Bouteau, H.; Meimoun, P.; Job, C.; Job, D.; Bailly, C. Role of protein and mRNA oxidation in seed dormancy and germination. Front. Plant Sci. 2013, 4. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Chu, P.; Zhou, Y.; Li, Y.; Liu, J.; Ding, Y.; Tsang, E.W.T.; Jiang, L.; Wu, K.; Huang, S. Overexpression of AtOGG1, a DNA glycosylase/AP lyase, enhances seed longevity and abiotic stress tolerance in Arabidopsis. J. Exp. Bot. 2012, 63, 4107–4121. [Google Scholar] [CrossRef] [PubMed]
- Madugundu, G.S.; Cadet, J.; Wagner, J.R. Hydroxyl-radical-induced oxidation of 5-methylcytosine in isolated and cellular DNA. Nucleic Acids Res. 2014, 42, 7450–7460. [Google Scholar] [CrossRef] [PubMed]
- Michalak, M.; Barciszewska, M.Z.; Barciszewski, J.; Plitta, B.P.; Chmielarz, P. Global Changes in DNA methylation in seeds and seedlings of Pyrus communis after Seed desiccation and storage. PLoS ONE 2013, 8, e70693. [Google Scholar] [CrossRef]
- Michalak, M.; Plitta-Michalak, B.P.; Naskręt-Barciszewska, M.; Barciszewski, J.; Bujarska-Borkowska, B.; Chmielarz, P. Global 5-methylcytosine alterations in DNA during ageing of Quercus robur seeds. Ann. Bot. 2015, 116, 369–376. [Google Scholar] [CrossRef]
- Shen, L.; Liang, Z.; Wong, C.E.; Yu, H. Messenger RNA modifications in plants. Trends Plant Sci. 2019, 24, 328–341. [Google Scholar] [CrossRef]
- Cui, X.; Liang, Z.; Shen, L.; Zhang, Q.; Bao, S.; Geng, Y.; Zhang, B.; Leo, V.; Vardy, L.A.; Lu, T.; et al. 5-Methylcytosine RNA methylation in Arabidopsis thaliana. Mol. Plant 2017, 10, 1387–1399. [Google Scholar] [CrossRef]
- Vandivier, L.E.; Campos, R.; Kuksa, P.P.; Silverman, I.M.; Wang, L.-S.; Gregory, B.D. Chemical modifications mark alternatively spliced and uncapped messenger RNAs in Arabidopsis. Plant Cell 2015, 27, 3024–3037. [Google Scholar] [CrossRef]
- Fray, R.G.; Simpson, G.G. The Arabidopsis epitranscriptome. Curr. Opin. Plant Biol. 2015, 27, 17–21. [Google Scholar] [CrossRef]
- Zhong, S.; Li, H.; Bodi, Z.; Button, J.; Vespa, L.; Herzog, M.; Fray, R.G. MTA Iis an Arabidopsis Messenger RNA adenosine methylase and interacts with a homolog of a sex-specific splicing factor. Plant Cell 2008, 20, 1278–1288. [Google Scholar] [CrossRef]
- Bodi, Z.; Zhong, S.; Mehra, S.; Song, J.; Graham, N.; Li, H.; May, S.; Fray, R.G. Adenosine methylation in Arabidopsis mRNA is associated with the 3′ end and reduced levels cause developmental defects. Front. Plant Sci. 2012, 3. [Google Scholar] [CrossRef]
- Diez, C.M.; Roessler, K.; Gaut, B.S. Epigenetics and plant genome evolution. Curr. Opin. Plant Biol. 2014, 18, 1–8. [Google Scholar] [CrossRef]
- Zhang, X.; Yazaki, J.; Sundaresan, A.; Cokus, S.; Chan, S.W.L.; Chen, H.; Henderson, I.R.; Shinn, P.; Pellegrini, M.; Jacobsen, S.E.; et al. Genome-wide high-resolution mapping and functional analysis of DNA methylation in Arabidopsis. Cell 2006, 126, 1189–1201. [Google Scholar] [CrossRef]
- Bewick, A.J.; Schmitz, R.J. Gene body DNA methylation in plants. Curr. Opin. Plant Biol. 2017, 36, 103–110. [Google Scholar] [CrossRef]
- Zilberman, D.; Gehring, M.; Tran, R.K.; Ballinger, T.; Henikoff, S. Genome-wide analysis of Arabidopsis thaliana DNA methylation uncovers an interdependence between methylation and transcription. Nat. Genet. 2007, 39, 61–69. [Google Scholar] [CrossRef]
- Law, J.A.; Jacobsen, S.E. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat. Rev. Genet. 2010, 11, 204–220. [Google Scholar] [CrossRef] [PubMed]
- Matzke, M.A.; Kanno, T.; Matzke, A.J.M. RNA-Directed DNA methylation: The evolution of a complex epigenetic pathway in flowering plants. Annu. Rev. Plant Biol. 2015, 66, 243–267. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Lang, Z.; Zhu, J.-K. Dynamics and function of DNA methylation in plants. Nat. Rev. Mol. Cell Biol. 2018, 19, 489–506. [Google Scholar] [CrossRef] [PubMed]
- Parrilla-Doblas, J.T.; Roldán-Arjona, T.; Ariza, R.R.; Córdoba-Cañero, D. Active DNA Demethylation in plants. Int. J. Mol. Sci. 2019, 20, 4683. [Google Scholar] [CrossRef] [PubMed]
- van Zanten, M.; Koini, M.A.; Geyer, R.; Liu, Y.; Brambilla, V.; Bartels, D.; Koornneef, M.; Fransz, P.; Soppe, W.J.J. Seed maturation in Arabidopsis thaliana is characterized by nuclear size reduction and increased chromatin condensation. Proc. Natl. Acad. Sci. USA 2011, 108, 20219–20224. [Google Scholar] [CrossRef]
- Kawakatsu, T.; Nery, J.R.; Castanon, R.; Ecker, J.R. Dynamic DNA methylation reconfiguration during seed development and germination. Genome Biol. 2017, 18, 171. [Google Scholar] [CrossRef] [PubMed]
- An, Y.C.; Goettel, W.; Han, Q.; Bartels, A.; Liu, Z.; Xiao, W. Dynamic changes of genome-wide DNA methylation during soybean seed development. Sci. Rep. 2017, 7. [Google Scholar] [CrossRef] [PubMed]
- Bouyer, D.; Kramdi, A.; Kassam, M.; Heese, M.; Schnittger, A.; Roudier, F.; Colot, V. DNA methylation dynamics during early plant life. Genome Biol. 2017, 18, 179. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.-Y.; Le, B.H.; Chen, M.; Henry, K.F.; Hur, J.; Hsieh, T.-F.; Chen, P.-Y.; Pelletier, J.M.; Pellegrini, M.; Fischer, R.L.; et al. Similarity between soybean and Arabidopsis seed methylomes and loss of non-CG methylation does not affect seed development. Proc. Natl. Acad. Sci. USA 2017, 114, E9730–E9739. [Google Scholar] [CrossRef]
- Narsai, R.; Gouil, Q.; Secco, D.; Srivastava, A.; Karpievitch, Y.V.; Liew, L.C.; Lister, R.; Lewsey, M.G.; Whelan, J. Extensive transcriptomic and epigenomic remodelling occurs during Arabidopsis thaliana germination. Genome Biol. 2017, 18. [Google Scholar] [CrossRef]
- Choi, Y.; Gehring, M.; Johnson, L.; Hannon, M.; Harada, J.J.; Goldberg, R.B.; Jacobsen, S.E.; Fischer, R.L. DEMETER, a DNA glycosylase domain protein, is required for endosperm gene imprinting and seed viability in Arabidopsis. Cell 2002, 110, 33–42. [Google Scholar] [CrossRef]
- Xiao, W.; Brown, R.C.; Lemmon, B.E.; Harada, J.J.; Goldberg, R.B.; Fischer, R.L. Regulation of seed size by hypomethylation of maternal and paternal genomes. Plant Physiol. 2006, 142, 1160–1168. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef]
- Li, P.; Ni, H.; Ying, S.; Wei, J.; Hu, X. Teaching an old dog a new trick: Multifaceted strategies to control primary seed germination by DELAY OF GERMINATION 1 (DOG1). Phyton (B. Aires). 2020, 89, 1–12. [Google Scholar] [CrossRef]
- Zheng, J.; Chen, F.; Wang, Z.; Cao, H.; Li, X.; Deng, X.; Soppe, W.J.J.; Li, Y.; Liu, Y. A novel role for histone methyltransferase KYP/SUVH4 in the control of Arabidopsis primary seed dormancy. New Phytol. 2012, 193, 605–616. [Google Scholar] [CrossRef]
- Singh, M.; Singh, J. Seed development-related expression of ARGONAUTE4_9 class of genes in barley: Possible role in seed dormancy. Euphytica 2012, 188, 123–129. [Google Scholar] [CrossRef]
- Singh, M.; Singh, S.; Randhawa, H.; Singh, J. Polymorphic homoeolog of key gene of RdDM pathway, ARGONAUTE4_9 class is associated with pre-harvest sprouting in Wheat (Triticum aestivum L.). PLoS ONE 2013, 8. [Google Scholar] [CrossRef] [PubMed]
- Zha, P.; Liu, S.; Li, Y.; Ma, T.; Yang, L.; Jing, Y.; Lin, R. The evening complex and the chromatin-remodeling factor PICKLE coordinately control seed dormancy by directly repressing DOG1 in Arabidopsis. Plant Commun. 2020, 1, 100011. [Google Scholar] [CrossRef]
- Köhler, C.; Wolff, P.; Spillane, C. Epigenetic mechanisms underlying genomic imprinting in plants. Annu. Rev. Plant Biol. 2012, 63, 331–352. [Google Scholar] [CrossRef] [PubMed]
- Piskurewicz, U.; Iwasaki, M.; Susaki, D.; Megies, C.; Kinoshita, T.; Lopez-Molina, L. Dormancy-specific imprinting underlies maternal inheritance of seed dormancy in Arabidopsis thaliana. eLife 2016. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.; Xie, W.; Xu, D.; Miki, D.; Tang, K.; Huang, C.-F.; Zhu, J.-K. DNA demethylase ROS1 negatively regulates the imprinting of DOGL4 and seed dormancy in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2018, 115, E9962–E9970. [Google Scholar] [CrossRef]
- Iwasaki, M.; Hyvärinen, L.; Piskurewicz, U.; Lopez-Molina, L. Non-canonical RNA-directed DNA methylation participates in maternal and environmental control of seed dormancy. eLife 2019, 8. [Google Scholar] [CrossRef]
- Kim, J.-S.; Lim, J.Y.; Shin, H.; Kim, B.-G.; Yoo, S.-D.; Kim, W.T.; Huh, J.H. ROS1-dependent DNA demethylation is required for ABA-inducible NIC3 expression. Plant Physiol. 2019, 179, 1810–1821. [Google Scholar] [CrossRef]
- Iglesias-Fernández, R.; Del Carmen Rodrguez-Gacio, M.; Matilla, A.J. Progress in research on dry afterripening. Seed Sci. Res. 2011, 21, 69–80. [Google Scholar] [CrossRef]
- Baskin, C.C.; Baskin, J.M. Seeds: Ecology, Biogeography, and, Evolution of Dormancy and Germination, 2nd ed.; Academic Press: Cambridge, MA, USA, 2014; pp. 79–117. [Google Scholar]
- Klausmeyer, K.R.; Shaw, M.R. Climate change, habitat loss, protected areas and the climate adaptation potential of species in mediterranean ecosystems worldwide. PLoS ONE 2009, 4. [Google Scholar] [CrossRef]

© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Katsuya-Gaviria, K.; Caro, E.; Carrillo-Barral, N.; Iglesias-Fernández, R. Reactive Oxygen Species (ROS) and Nucleic Acid Modifications during Seed Dormancy. Plants 2020, 9, 679. https://doi.org/10.3390/plants9060679
Katsuya-Gaviria K, Caro E, Carrillo-Barral N, Iglesias-Fernández R. Reactive Oxygen Species (ROS) and Nucleic Acid Modifications during Seed Dormancy. Plants. 2020; 9(6):679. https://doi.org/10.3390/plants9060679
Chicago/Turabian StyleKatsuya-Gaviria, Kai, Elena Caro, Néstor Carrillo-Barral, and Raquel Iglesias-Fernández. 2020. "Reactive Oxygen Species (ROS) and Nucleic Acid Modifications during Seed Dormancy" Plants 9, no. 6: 679. https://doi.org/10.3390/plants9060679
APA StyleKatsuya-Gaviria, K., Caro, E., Carrillo-Barral, N., & Iglesias-Fernández, R. (2020). Reactive Oxygen Species (ROS) and Nucleic Acid Modifications during Seed Dormancy. Plants, 9(6), 679. https://doi.org/10.3390/plants9060679