Melatonin Regulatory Mechanisms and Phylogenetic Analyses of Melatonin Biosynthesis Related Genes Extracted from Peanut under Salinity Stress
Abstract
1. Introduction
2. Results
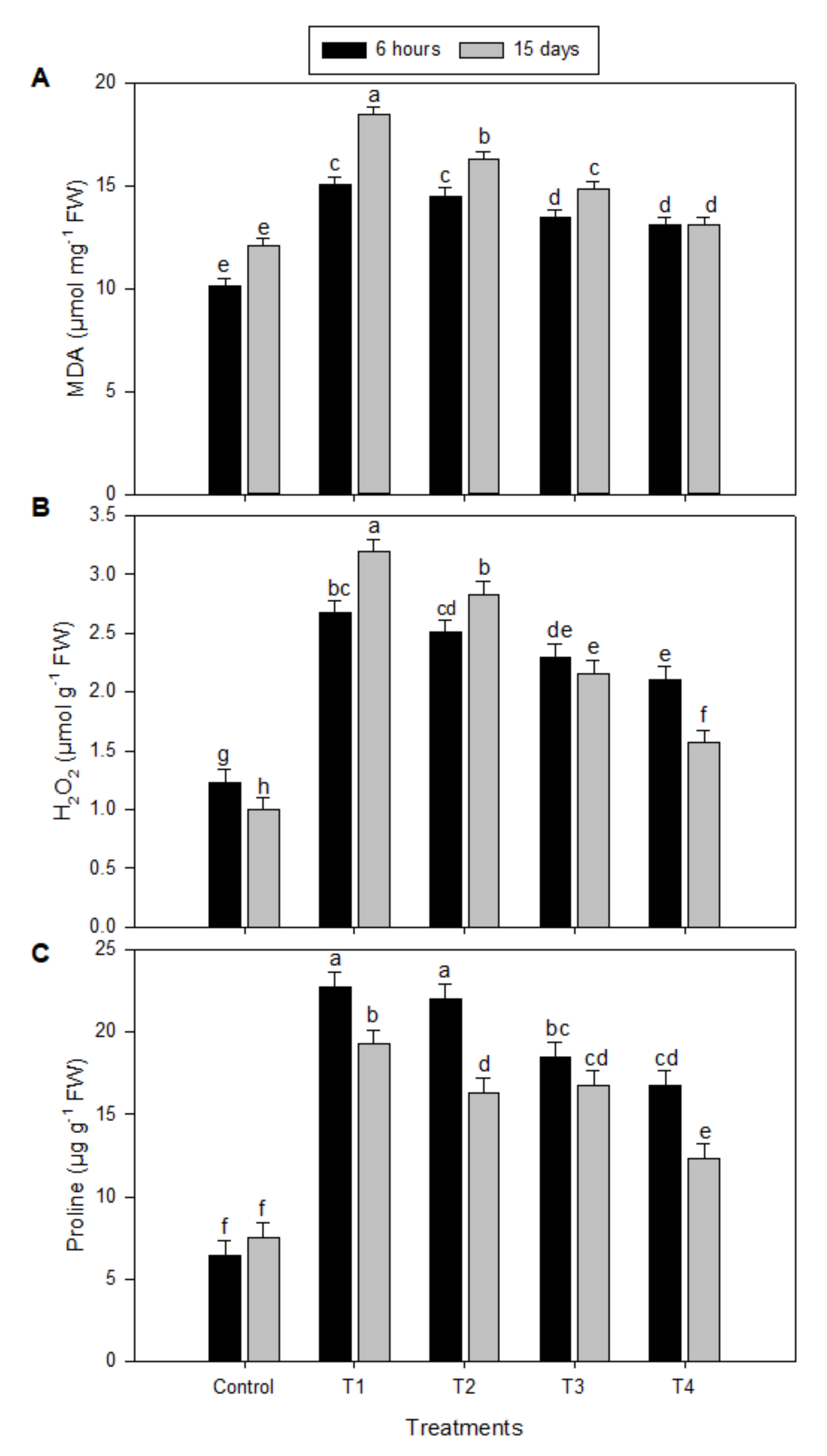
2.1. Levels of Malondialdehyde (MDA), Hydrogen Peroxide (H2O2), and Proline under Salinity Stress and Melatonin (MT) Treatments
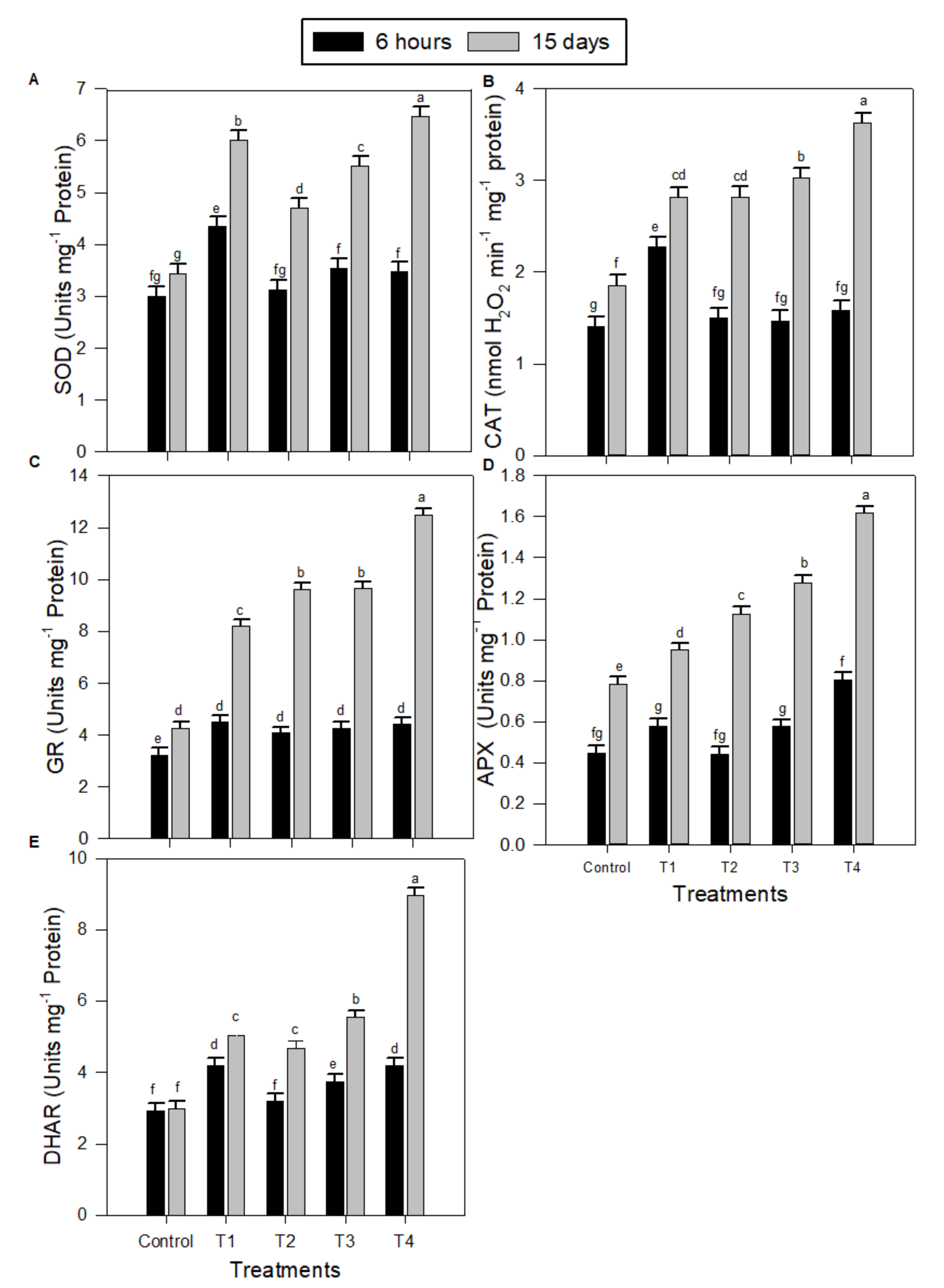
2.2. Levels of Antioxidant System Components in Peanut Seedlings under Salinity Stress and Melatonin (MT) Treatments
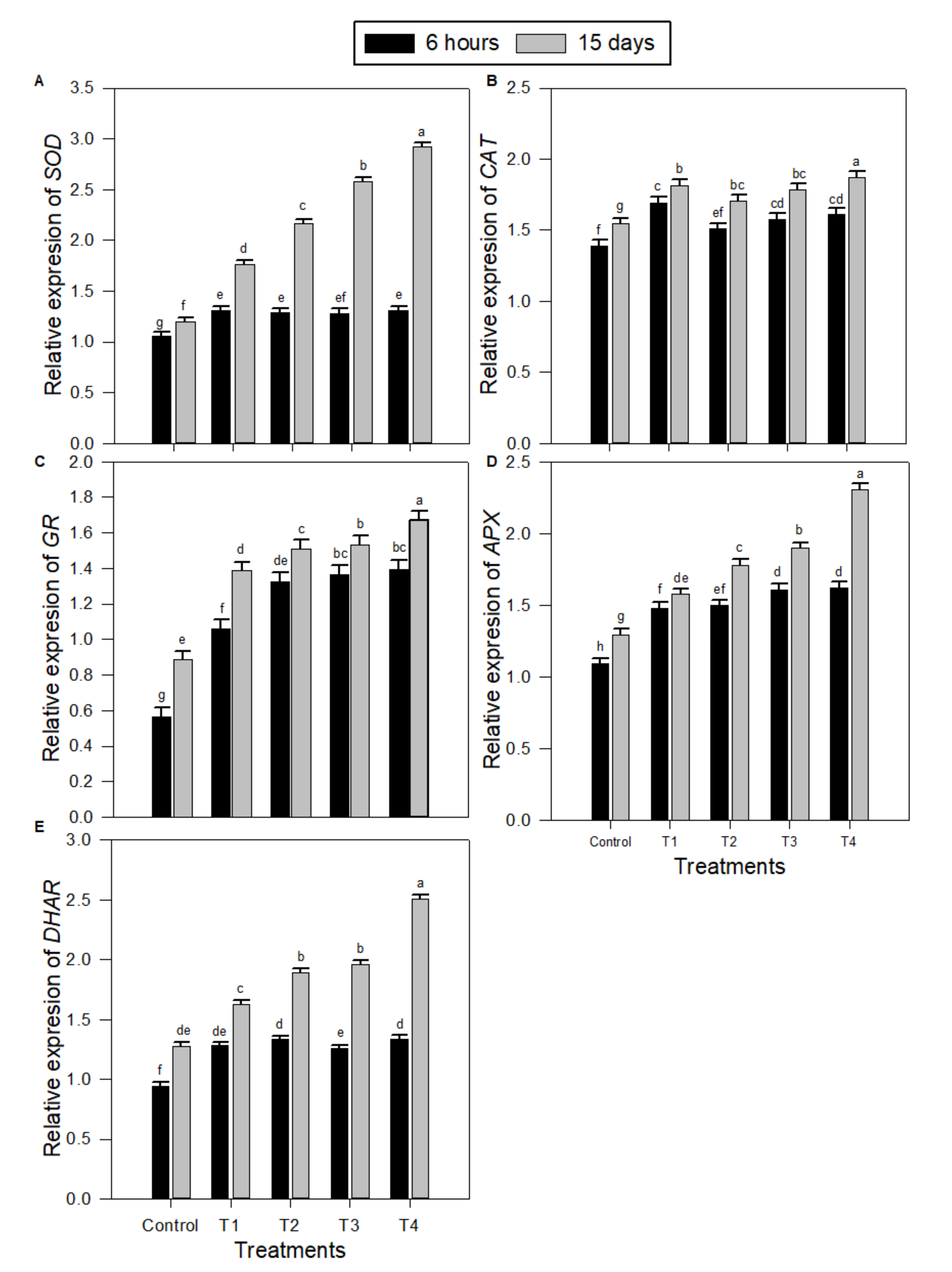
2.3. Levels of Transcription of Gene-encoding Antioxidative Enzymes in Peanut Seedlings under Salinity Stress and Melatonin (MT) Treatments
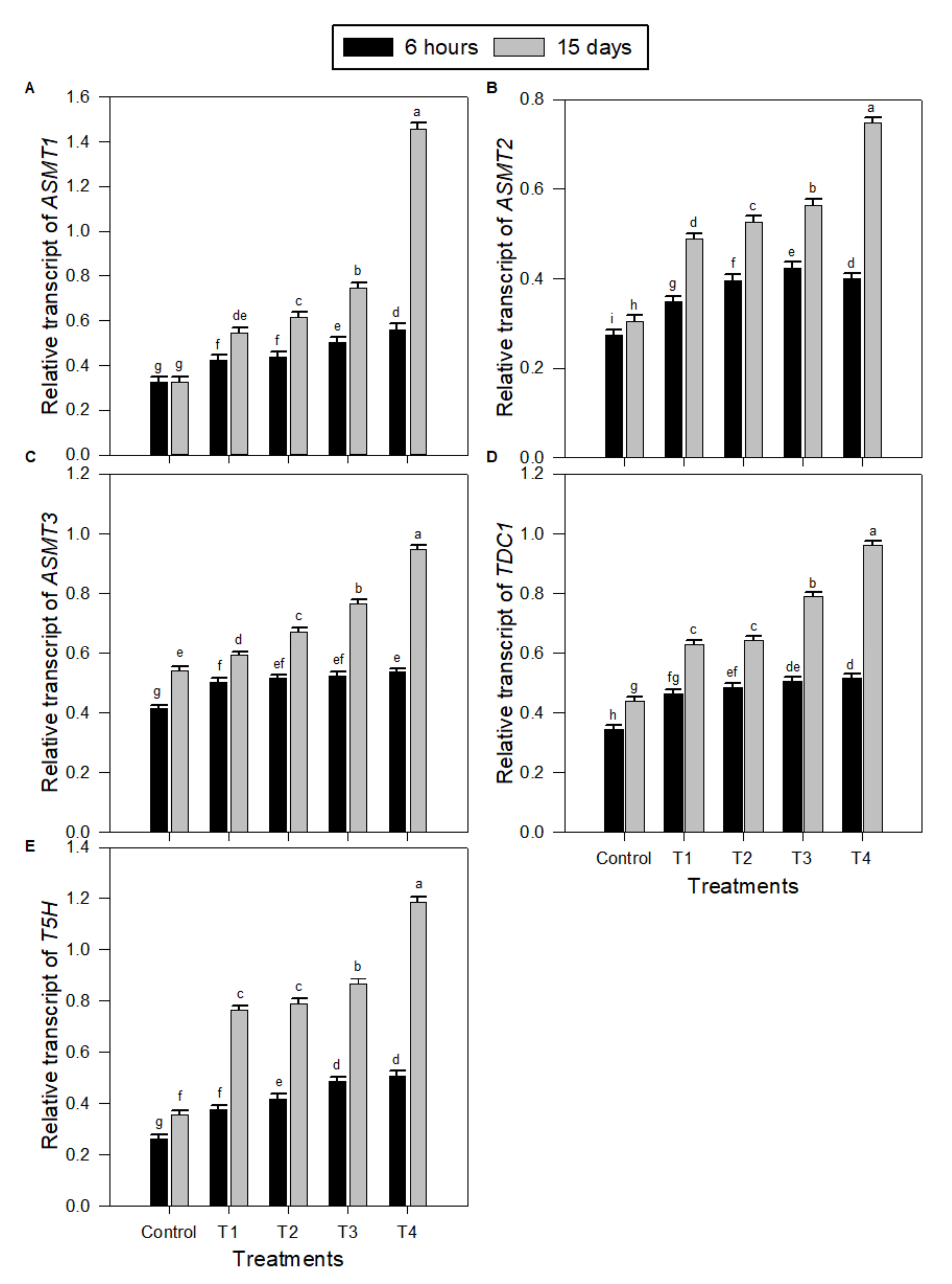
2.4. Levels of Transcription of Melatonin (MT) Biosynthesis Genes in Peanut Seedlings under Salinity Stress and MT Treatments
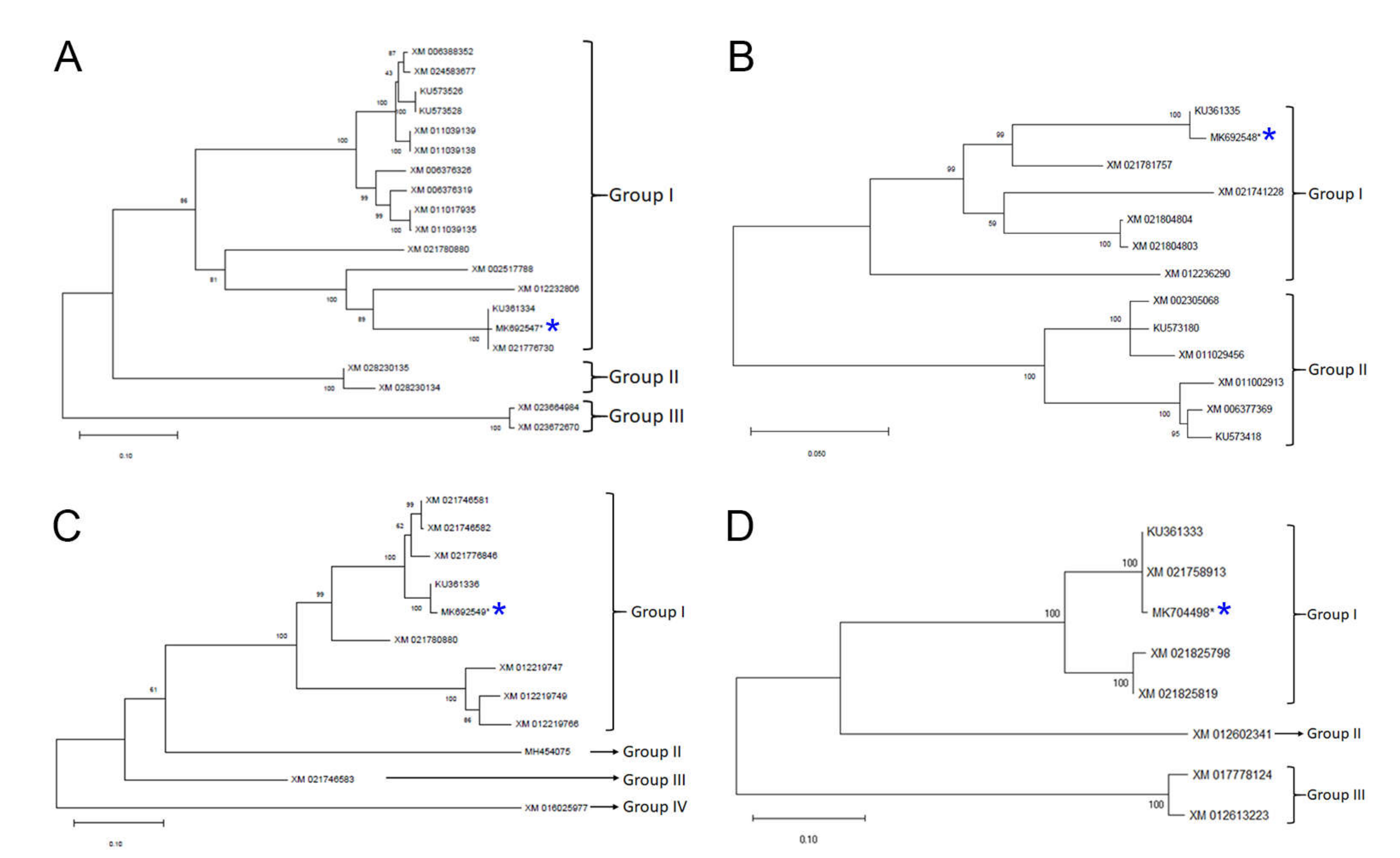
2.5. Nucleotide Identity Comparison and Phylogenetic Analyses
2.6. Maximum Likelihood Estimation of Substitution Matrix and Transition/Transversion Bias
3. Discussion
4. Materials and Methods
4.1. Plant Material and Treatments
4.2. Determination of Malondialdehyde (MDA) Contents
4.3. Determination of Hydrogen Peroxide (H2O2) Content
4.4. Determination of Proline Content
4.5. Determination of GSH, GSSG, DHA, and AsA Contents
4.6. Determination of Antioxidant Enzyme Activities
4.7. Total RNA Isolation and Quantitative Real-time PCR (qRT-PCR) Amplification
4.8. Cloning and Sequencing of Melatonin Biosynthesis Genes
4.9. Sequence Submission to GenBank
4.10. Sequences Alignment and Construction of Phylogenetic Trees
4.11. Statistical Analyses
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Zhang, H.-X.; Blumwald, E. Transgenic salt-tolerant tomato plants accumulate salt in foliage but not in fruit. Nat. Biotechnol. 2001, 19, 765–768. [Google Scholar] [CrossRef] [PubMed]
- Parida, A.K.; Das, A.B. Salt tolerance and salinity effects on plants: A review. Ecotoxicol. Environ. Saf. 2005, 60, 324–349. [Google Scholar] [CrossRef] [PubMed]
- Maurino, V.G.; Flügge, U.-I. Experimental systems to assess the effects of reactive oxygen species in plant tissues. Plant Signal. Behav. 2008, 3, 923–928. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Mittler, R. Oxidative stress, antioxidants and stress tolerance. Trends Plant Sci. 2002, 7, 405–410. [Google Scholar] [CrossRef]
- Hardeland, R.; Cardinali, D.P.; Srinivasan, V.; Spence, D.W.; Brown, G.M.; Pandi-Perumal, S.R. Melatonin—A pleiotropic, orchestrating regulator molecule. Prog. Neurobiol. 2011, 93, 350–384. [Google Scholar] [CrossRef] [PubMed]
- Kołodziejczyk, I.; Dzitko, K.; Szewczyk, R.; Posmyk, M.M. Exogenous melatonin improves corn (Zea mays L.) embryo proteome in seeds subjected to chilling stress. J. Plant Physiol. 2016, 193, 47–56. [Google Scholar] [CrossRef]
- Li, M.Q.; Hasan, M.K.; Li, C.X.; Ahammed, G.J.; Xia, X.J.; Shi, K.; Zhou, Y.H.; Reiter, R.J.; Yu, J.Q.; Xu, M.X. Melatonin mediates selenium-induced tolerance to cadmium stress in tomato plants. J. Pineal Res. 2016, 61, 291–302. [Google Scholar] [CrossRef]
- Reiter, R.J.; Tan, D.-X.; Zhou, Z.; Cruz, M.H.C.; Fuentes-Broto, L.; Galano, A. Phytomelatonin: Assisting plants to survive and thrive. Molecules 2015, 20, 7396–7437. [Google Scholar] [CrossRef]
- Turk, H.; Erdal, S. Melatonin alleviates cold-induced oxidative damage in maize seedlings by up-regulating mineral elements and enhancing antioxidant activity. J. Plant Nutr. Soil Sci. 2015, 178, 433–439. [Google Scholar] [CrossRef]
- Xu, W.; Cai, S.Y.; Zhang, Y.; Wang, Y.; Ahammed, G.J.; Xia, X.J.; Shi, K.; Zhou, Y.H.; Yu, J.Q.; Reiter, R.J. Melatonin enhances thermotolerance by promoting cellular protein protection in tomato plants. J. Pineal Res. 2016, 61, 457–469. [Google Scholar] [CrossRef]
- Kang, K.; Kong, K.; Park, S.; Natsagdorj, U.; Kim, Y.S.; Back, K. Molecular cloning of a plant N-acetylserotonin methyltransferase and its expression characteristics in rice. J. Pineal Res. 2011, 50, 304–309. [Google Scholar] [CrossRef] [PubMed]
- Kang, K.; Lee, K.; Park, S.; Byeon, Y.; Back, K. Molecular cloning of rice serotonin N-acetyltransferase, the penultimate gene in plant melatonin biosynthesis. J. Pineal Res. 2013, 55, 7–13. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Wang, P.; Wei, Z.; Liang, D.; Liu, C.; Yin, L.; Jia, D.; Fu, M.; Ma, F. The mitigation effects of exogenous melatonin on salinity-induced stress in Malus hupehensis. J. Pineal Res. 2012, 53, 298–306. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.J.; Zhang, N.; Yang, R.C.; Wang, L.; Sun, Q.Q.; Li, D.B.; Cao, Y.Y.; Weeda, S.; Zhao, B.; Ren, S. Melatonin promotes seed germination under high salinity by regulating antioxidant systems, ABA and GA4 interaction in cucumber (Cucumis sativus L.). J. Pineal Res. 2014, 57, 269–279. [Google Scholar] [CrossRef]
- Ke, Q.; Ye, J.; Wang, B.; Ren, J.; Yin, L.; Deng, X.; Wang, S. Melatonin mitigates salt stress in wheat seedlings by modulating polyamine metabolism. Front. Plant Sci. 2018, 9, 914. [Google Scholar] [CrossRef]
- Zhang, T.; Shi, Z.; Zhang, X.; Zheng, S.; Wang, J.; Mo, J. Alleviating effects of exogenous melatonin on salt stress in cucumber. Sci. Hortic. 2020, 262. [Google Scholar] [CrossRef]
- Wang, L.; Liu, J.; Wang, W.; Sun, Y. Exogenous melatonin improves growth and photosynthetic capacity of cucumber under salinity-induced stress. Photosynthetica 2016, 54, 19–27. [Google Scholar] [CrossRef]
- Arnao, M.B.; Hernández-Ruiz, J. Chemical stress by different agents affects the melatonin content of barley roots. J. Pineal Res. 2009, 46, 295–299. [Google Scholar] [CrossRef]
- Mukherjee, S.; David, A.; Yadav, S.; Baluška, F.; Bhatla, S.C. Salt stress-induced seedling growth inhibition coincides with differential distribution of serotonin and melatonin in sunflower seedling roots and cotyledons. Physiol. Plant. 2014, 152, 714–728. [Google Scholar] [CrossRef]
- Chen, L.; Liu, L.; Lu, B.; Ma, T.; Jiang, D.; Li, J.; Zhang, K.; Sun, H.; Zhang, Y.; Bai, Z. Exogenous melatonin promotes seed germination and osmotic regulation under salt stress in cotton (Gossypium hirsutum L.). PLoS ONE 2020, 15, e0228241. [Google Scholar] [CrossRef]
- Cen, H.; Wang, T.; Liu, H.; Tian, D.; Zhang, Y. Melatonin application improves salt tolerance of alfalfa (Medicago sativa L.) by enhancing antioxidant capacity. Plants 2020, 9, 220. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Chang, J.; Chen, H.; Wang, Z.; Gu, X.; Wei, C.; Zhang, Y.; Ma, J.; Yang, J.; Zhang, X. Exogenous melatonin confers salt stress tolerance to watermelon by improving photosynthesis and redox homeostasis. Front. Plant Sci. 2017, 8, 295. [Google Scholar] [CrossRef] [PubMed]
- Banavath, J.N.; Chakradhar, T.; Pandit, V.; Konduru, S.; Guduru, K.K.; Akila, C.S.; Podha, S.; Puli, C.O. Stress inducible overexpression of AtHDG11 leads to improved drought and salt stress tolerance in peanut (Arachis hypogaea L.). Front. Chem. 2018, 6, 34. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.E.; Mao, J.J.; Sun, L.Q.; Huang, B.; Ding, C.B.; Gu, Y.; Liao, J.Q.; Hu, C.; Zhang, Z.W.; Yuan, S. Exogenous melatonin enhances salt stress tolerance in maize seedlings by improving antioxidant and photosynthetic capacity. Physiol. Plant. 2018, 164, 349–363. [Google Scholar] [CrossRef] [PubMed]
- Park, S.; Byeon, Y.; Back, K. Functional analyses of three ASMT gene family members in rice plants. J. Pineal Res. 2013, 55, 409–415. [Google Scholar] [CrossRef] [PubMed]
- Shi, H.; Jiang, C.; Ye, T.; Tan, D.-X.; Reiter, R.J.; Zhang, H.; Liu, R.; Chan, Z. Comparative physiological, metabolomic, and transcriptomic analyses reveal mechanisms of improved abiotic stress resistance in bermudagrass [Cynodon dactylon (L). Pers.] by exogenous melatonin. J. Exp. Bot. 2015, 66, 681–694. [Google Scholar] [CrossRef]
- Tan, D.-X.; Hardeland, R.; Manchester, L.C.; Korkmaz, A.; Ma, S.; Rosales-Corral, S.; Reiter, R.J. Functional roles of melatonin in plants, and perspectives in nutritional and agricultural science. J. Exp. Bot. 2012, 63, 577–597. [Google Scholar] [CrossRef]
- Wei, W.; Li, Q.-T.; Chu, Y.-N.; Reiter, R.J.; Yu, X.-M.; Zhu, D.-H.; Zhang, W.-K.; Ma, B.; Lin, Q.; Zhang, J.-S. Melatonin enhances plant growth and abiotic stress tolerance in soybean plants. J. Exp. Bot. 2015, 66, 695–707. [Google Scholar] [CrossRef]
- Yin, L.; Wang, P.; Li, M.; Ke, X.; Li, C.; Liang, D.; Wu, S.; Ma, X.; Li, C.; Zou, Y. Exogenous melatonin improves Malus resistance to Marssonina apple blotch. J. Pineal Res. 2013, 54, 426–434. [Google Scholar] [CrossRef]
- Cano, A.; Hernández-Ruiz, J.; Arnao, M.B. Changes in hydrophilic antioxidant activity in Avena sativa and Triticum aestivum leaves of different age during de-etiolation and high-light treatment. J. Plant Res. 2006, 119, 321–327. [Google Scholar] [CrossRef]
- Shi, H.; Wang, X.; Tan, D.X.; Reiter, R.J.; Chan, Z. Comparative physiological and proteomic analyses reveal the actions of melatonin in the reduction of oxidative stress in Bermuda grass (Cynodon dactylon (L). Pers.). J. Pineal Res. 2015, 59, 120–131. [Google Scholar] [CrossRef] [PubMed]
- Zhang, N.; Sun, Q.; Zhang, H.; Cao, Y.; Weeda, S.; Ren, S.; Guo, Y.-D. Roles of melatonin in abiotic stress resistance in plants. J. Exp. Bot. 2015, 66, 647–656. [Google Scholar] [CrossRef] [PubMed]
- Tan, D.X.; Hardeland, R.; Manchester, L.C.; Paredes, S.D.; Korkmaz, A.; Sainz, R.M.; Mayo, J.C.; Fuentes-Broto, L.; Reiter, R.J. The changing biological roles of melatonin during evolution: From an antioxidant to signals of darkness, sexual selection and fitness. Biol. Rev. 2010, 85, 607–623. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Sun, X.; Xie, Y.; Li, M.; Chen, W.; Zhang, S.; Liang, D.; Ma, F. Melatonin regulates proteomic changes during leaf senescence in Malus hupehensis. J. Pineal Res. 2014, 57, 291–307. [Google Scholar] [CrossRef] [PubMed]
- Bonnefont-Rousselot, D.; Collin, F.; Jore, D.; Gardès-Albert, M. Reaction mechanism of melatonin oxidation by reactive oxygen species in vitro. J. Pineal Res. 2011, 50, 328–335. [Google Scholar] [CrossRef]
- Fowler, G.; Daroszewska, M.; Ingold, K.U. Melatonin does not “directly scavenge hydrogen peroxide”: Demise of another myth. Free Radic. Biol. Med. 2003, 34, 77–83. [Google Scholar] [CrossRef]
- Noctor, G.; Foyer, C.H. Ascorbate and glutathione: Keeping active oxygen under control. Annu. Rev. Plant Biol. 1998, 49, 249–279. [Google Scholar] [CrossRef]
- Reiter, R.J.; Tan, D.X.; Burkhardt, S.; Manchester, L.C. Melatonin in plants. Nutr. Rev. 2001, 59, 286–290. [Google Scholar] [CrossRef]
- Stepien, P.; Klobus, G. Antioxidant defense in the leaves of C3 and C4 plants under salinity stress. Physiol. Plant. 2005, 125, 31–40. [Google Scholar] [CrossRef]
- Hossain, M.S.; ElSayed, A.I.; Moore, M.; Dietz, K.-J. Redox and reactive oxygen species network in acclimation for salinity tolerance in sugar beet. J. Exp. Bot. 2017, 68, 1283–1298. [Google Scholar] [CrossRef]
- Byeon, Y.; Lee, H.Y.; Hwang, O.J.; Lee, H.J.; Lee, K.; Back, K. Coordinated regulation of melatonin synthesis and degradation genes in rice leaves in response to cadmium treatment. J. Pineal Res. 2015, 58, 470–478. [Google Scholar] [CrossRef] [PubMed]
- Park, S.; Lee, D.E.; Jang, H.; Byeon, Y.; Kim, Y.S.; Back, K. Melatonin-rich transgenic rice plants exhibit resistance to herbicide-induced oxidative stress. J. Pineal Res. 2013, 54, 258–263. [Google Scholar] [CrossRef] [PubMed]
- Kang, S.; Kang, K.; Lee, K.; Back, K. Characterization of rice tryptophan decarboxylases and their direct involvement in serotonin biosynthesis in transgenic rice. Planta 2007, 227, 263–272. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Tan, D.X.; Lei, Q.; Chen, H.; Wang, L.; Li, Q.t.; Gao, Y.; Kong, J. Melatonin and its potential biological functions in the fruits of sweet cherry. J. Pineal Res. 2013, 55, 79–88. [Google Scholar] [CrossRef] [PubMed]
- Stevens, L.H.; Blom, T.J.; Verpoorte, R. Subcellular localization of tryptophan decarboxylase, strictosidine synthase and strictosidine glucosidase in suspension cultured cells of Catharanthus roseus and Tabernaemontana divaricata. Plant Cell Rep. 1993, 12, 573–576. [Google Scholar] [CrossRef]
- Fujiwara, T.; Maisonneuve, S.; Isshiki, M.; Mizutani, M.; Chen, L.; Wong, H.L.; Kawasaki, T.; Shimamoto, K. Sekiguchi lesion gene encodes a cytochrome P450 monooxygenase that catalyzes conversion of tryptamine to serotonin in rice. J. Biol. Chem. 2010, 285, 11308–11313. [Google Scholar] [CrossRef]
- Byeon, Y.; Lee, H.Y.; Lee, K.; Park, S.; Back, K. Cellular localization and kinetics of the rice melatonin biosynthetic enzymes SNAT and ASMT. J. Pineal Res. 2014, 56, 107–114. [Google Scholar] [CrossRef]
- Kanjanaphachoat, P.; Wei, B.-Y.; Lo, S.-F.; Wang, I.-W.; Wang, C.-S.; Yu, S.-M.; Yen, M.-L.; Chiu, S.-H.; Lai, C.-C.; Chen, L.-J. Serotonin accumulation in transgenic rice by over-expressing tryptophan decarboxlyase results in a dark brown phenotype and stunted growth. Plant Mol. Biol. 2012, 78, 525–543. [Google Scholar] [CrossRef]
- Boulila, M.; ElSayed, A.I.; Rafudeen, M.S.; Omar, A.A. Investigating molecular evolutionary forces and phylogenetic relationships among melatonin precursor-encoding genes of different plant species. Mol. Biol. Rep. 2020, 1–12. [Google Scholar] [CrossRef]
- Hoagland, D.R.; Arnon, D.I. The water-culture method for growing plants without soil. In Circular; California Agricultural Experiment Station: Sacramento, CA, USA, 1950; Volume 347, Available online: https://www.cabdirect.org/cabdirect/abstract/19500302257 (accessed on 6 July 2020).
- Chen, Y.-E.; Cui, J.-M.; Su, Y.-Q.; Yuan, S.; Yuan, M.; Zhang, H.-Y. Influence of stripe rust infection on the photosynthetic characteristics and antioxidant system of susceptible and resistant wheat cultivars at the adult plant stage. Front. Plant Sci. 2015, 6, 779. [Google Scholar] [CrossRef] [PubMed]
- Willekens, H.; Chamnongpol, S.; Davey, M.; Schraudner, M.; Langebartels, C.; Van Montagu, M.; Inze, D.; Van Camp, W. Catalase is a sink for H2O2 and is indispensable for stress defence in C3 plants. EMBO J. 1997, 16, 4806–4816. [Google Scholar] [CrossRef] [PubMed]
- Vicente, O.; Boscaiu, M.; Naranjo, M.Á.; Estrelles, E.; Bellés, J.M.A.; Soriano, P. Responses to salt stress in the halophyte Plantago crassifolia (Plantaginaceae). J. Arid Environ. 2004, 58, 463–481. [Google Scholar] [CrossRef]
- Rao, M.V.; Hale, B.A.; Ormrod, D.P. Amelioration of ozone-induced oxidative damage in wheat plants grown under high carbon dioxide (role of antioxidant enzymes). Plant Physiol. 1995, 109, 421–432. [Google Scholar] [CrossRef] [PubMed]
- Law, M.; Charles, S.A.; Halliwell, B. Glutathione and ascorbic acid in spinach (Spinacia oleracea) chloroplasts. The effect of hydrogen peroxide and of paraquat. Biochem. J. 1983, 210, 899–903. [Google Scholar] [CrossRef] [PubMed]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Aebi, H. Catalase in vitro. In Methods in Enzymology; Elsevier: Amsterdam, The Netherlands, 1984; Volume 105, pp. 121–126. [Google Scholar]
- Dzung, N.A.; Khanh, V.T.P.; Dzung, T.T. Research on impact of chitosan oligomers on biophysical characteristics, growth, development and drought resistance of coffee. Carbohydr. Polym. 2011, 84, 751–755. [Google Scholar] [CrossRef]
- Mandhania, S.; Madan, S.; Sawhney, V. Antioxidant defense mechanism under salt stress in wheat seedlings. Biol. Plant. 2006, 50, 227–231. [Google Scholar] [CrossRef]
- Mishra, P.; Bhoomika, K.; Dubey, R. Differential responses of antioxidative defense system to prolonged salinity stress in salt-tolerant and salt-sensitive Indica rice (Oryza sativa L.) seedlings. Protoplasma 2013, 250, 3–19. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Pearson, W.R.; Lipman, D.J. Improved tools for biological sequence comparison. Proc. Natl. Acad. Sci. USA 1988, 85, 2444–2448. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Boguski, M.S.; Gish, W.; Wootton, J.C. Issues in searching molecular sequence databases. Nat. Genet. 1994, 6, 119–129. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evolut. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K. Estimation of the number of nucleotide substitutions when there are strong transition-transversion and G+ C-content biases. Mol. Biol. Evolut. 1992, 9, 678–687. [Google Scholar] [CrossRef]
- Jukes, T.H.; Cantor, C.R. Evolution of protein molecules. In Mammalian Protein Metabolism; Munro, H.M., Ed.; Academic Press: New York, NY, USA, 1969; p. 21. [Google Scholar]
- Stell, R.; Torrie, J.; Dickey, D. Principles and Procedures of Statistics: A Biometrical Approach, 3rd ed.; MacGraw-Hill: New York, NY, USA, 1997. [Google Scholar]
- M-STATC. A Microcomputer Program for the Design, Management and Analysis of Agronomic Research Experiments; Michigan State University: East Lansing, MI, USA, 1990. [Google Scholar]


| Gene | Nucleotide Substitution Matrix | ||||
|---|---|---|---|---|---|
| A | T | C | G | ||
| ASMT1 | A | - | 5.31 | 4.17 | 13.63 |
| T | 5.55 | - | 13.47 | 4.58 | |
| C | 5.55 | 17.17 | - | 4.58 | |
| G | 16.51 | 5.31 | 4.17 | - | |
| ASMT2 | A | - | 5.88 | 4.09 | 12.04 |
| T | 6.50 | - | 11.96 | 4.86 | |
| C | 6.50 | 17.22 | - | 4.86 | |
| G | 16.12 | 5.88 | 4.09 | - | |
| ASMT3 | A | - | 6.40 | 4.06 | 12.48 |
| T | 6.36 | - | 10.91 | 5.08 | |
| C | 6.36 | 17.80 | - | 5.08 | |
| G | 15.63 | 6.40 | 4.06 | - | |
| T5H | A | - | 6.39 | 4.09 | 12.32 |
| T | 6.40 | - | 10.98 | 5.02 | |
| C | 6.40 | 17.18 | - | 5.02 | |
| G | 15.72 | 6.39 | 4.09 | - | |
| TDC | A | - | 8.33 | 8.33 | 8.33 |
| T | 8.33 | - | 8.33 | 8.33 | |
| C | 8.33 | 8.33 | - | 8.33 | |
| G | 8.33 | 8.33 | 8.33 | - | |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
ElSayed, A.I.; Boulila, M.; Rafudeen, M.S.; Mohamed, A.H.; Sengupta, S.; Rady, M.; Omar, A.A. Melatonin Regulatory Mechanisms and Phylogenetic Analyses of Melatonin Biosynthesis Related Genes Extracted from Peanut under Salinity Stress. Plants 2020, 9, 854. https://doi.org/10.3390/plants9070854
ElSayed AI, Boulila M, Rafudeen MS, Mohamed AH, Sengupta S, Rady M, Omar AA. Melatonin Regulatory Mechanisms and Phylogenetic Analyses of Melatonin Biosynthesis Related Genes Extracted from Peanut under Salinity Stress. Plants. 2020; 9(7):854. https://doi.org/10.3390/plants9070854
Chicago/Turabian StyleElSayed, Abdelaleim I., Moncef Boulila, Mohammed S. Rafudeen, Azza H. Mohamed, Sonali Sengupta, Mostafa Rady, and Ahmad A. Omar. 2020. "Melatonin Regulatory Mechanisms and Phylogenetic Analyses of Melatonin Biosynthesis Related Genes Extracted from Peanut under Salinity Stress" Plants 9, no. 7: 854. https://doi.org/10.3390/plants9070854
APA StyleElSayed, A. I., Boulila, M., Rafudeen, M. S., Mohamed, A. H., Sengupta, S., Rady, M., & Omar, A. A. (2020). Melatonin Regulatory Mechanisms and Phylogenetic Analyses of Melatonin Biosynthesis Related Genes Extracted from Peanut under Salinity Stress. Plants, 9(7), 854. https://doi.org/10.3390/plants9070854








