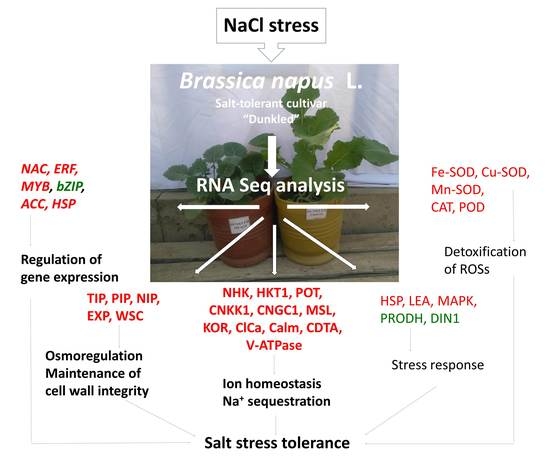
RNAseq Analysis Reveals Altered Expression of Key Ion Transporters Causing Differential Uptake of Selective Ions in Canola (Brassica napus L.) Grown under NaCl Stress
Abstract
:1. Introduction
2. Results
2.1. Evaluation of Different Cultivars of Canola (B. napus L.) for Salt Stress Tolerance
2.1.1. Growth Attributes
2.1.2. Relative Water Contents (RWC %)
2.1.3. Osmotic Potential (−MPa)
2.1.4. Leaf Mineral Nutrients (Leaf Na+, K+, Ca2+, Cl−, K+/Na+ Ratio, Ca2+/Na+ Ratio)
2.1.5. Shoot K+/Na+ and Ca2+/Na+ Ratio
2.1.6. Nutrient Utilization Efficiency (mg2/µg)
2.2. RNASeq Analysis and Differential Expression of Genes in Salt-Tolerant Cultivar Dunkled
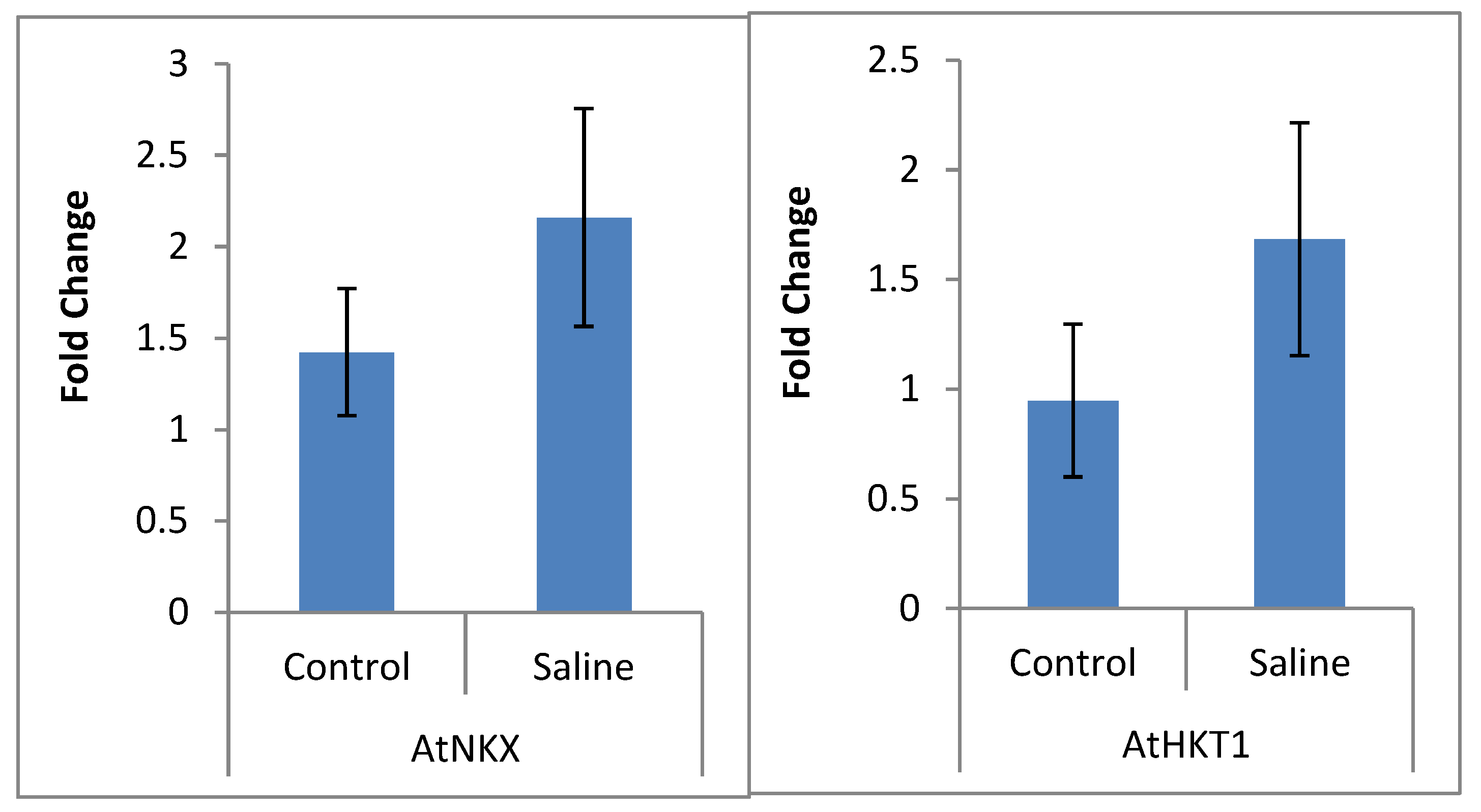
2.3. qRT-PCR
3. Discussion
4. Materials and Methods
4.1. Evaluation of Different Cultivars of Canola (B. napus L.) for Salt Stress Tolerance
4.1.1. Growth Attributes
4.1.2. Relative Water Contents (%)
4.1.3. Leaf Osmotic Potential (−MPa)
4.1.4. Mineral Contents (K+, Na+, Cl−, Ca2+)
4.1.5. Nutrient Utilization Efficiency (mg2/µg)
4.2. RNASeq Analysis and Differential Expression of Genes in Salt-Tolerant Cultivar Dunkled
4.2.1. Isolation of Total RNA
4.2.2. Next-Generation Sequencing (NGS)
4.2.3. Sequencing for Cluster Generation
4.2.4. Generation of Raw Data
4.2.5. Bioinformatics Analysis of Raw Data
4.2.6. Bioinformatics Analysis of Feature Count Matrix
4.2.7. Validation of NGS Data by qRT-PCR
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Athar, H.R.; Ashraf, M.; Wahid, A.; Jamil, A. Inducing salt tolerance in canola (Brassica napus L.) by exogenous application of glycinebetaine and proline: Response at the initial growth stages. Pak. J. Bot. 2009, 41, 1311–1319. [Google Scholar]
- Munns, R. Comparative physiology of salt and water stress. Plant Cell Environ. 2002, 25, 239–250. [Google Scholar] [CrossRef]
- Ashraf, M.; McNeilly, T. Salinity tolerance in brassica oilseeds. Crit. Rev. Plant Sci. 2004, 23, 157–174. [Google Scholar] [CrossRef]
- Ulfat, M.; Athar, H.; Ashraf, M.; Akram, N.A.; Jamil, A. Appraisal of physiological and biochemical selection criteria for evaluation of salt tolerance in canola (Brassica napus L.). Pak. J. Bot. 2007, 39, 1593–1608. [Google Scholar]
- Ashraf, M.; Athar, H.; Harris, P.; Kwon, T. Some prospective strategies for improving crop salt tolerance. Adv. Agron. 2008, 97, 45–110. [Google Scholar] [CrossRef]
- Morton, M.J.L.; Awlia, M.; Al-Tamimi, N.; Saade, S.; Pailles, Y.; Negrão, S.; Tester, M. Salt stress under the scalpel—Dissecting the genetics of salt tolerance. Plant J. 2019, 97, 148–163. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Zelm, E.; Zhang, Y.; Testerink, C. Salt tolerance mechanisms of plants. Annu. Rev. Plant Biol. 2020, 71, 403–433. [Google Scholar] [CrossRef] [Green Version]
- Munns, R.; James, R.; Gilliham, M.; Flowers, T.J.; Colmer, T.D. Tissue tolerance: An essential but elusive trait for salt-tolerant crops. Funct. Plant Biol. 2016, 43, 1103. [Google Scholar] [CrossRef] [Green Version]
- Hasegawa, P.M.; Bressan, R.A.; Zhu, J.K.; Bohnert, H.J. Plant cellular and molecular responses to high salinity. Annu. Rev. Plant Physiol. Plant Mol. Biol. 2000, 51, 463–499. [Google Scholar] [CrossRef] [Green Version]
- Munns, R.; Tester, M. Mechanisms of salinity tolerance. Annu. Rev. Plant Biol. 2008, 59, 651–681. [Google Scholar] [CrossRef] [Green Version]
- Munns, R. Genes and salt tolerance: Bringing them together. New Phytol. 2005, 167, 645–663. [Google Scholar] [CrossRef]
- Ishitani, M.; Liu, J.P.; Halfter, U.; Kim, C.S.; Shi, W.M.; Zhu, J.K. SOS3 function in plant salt tolerance requires N-myristoylation and calcium binding. Plant Cell 2000, 12, 1667–1677. [Google Scholar] [CrossRef] [Green Version]
- Zhu, J.-K. Genetic analysis of plant salt tolerance using Arabidopsis. Plant Physiol. 2000, 124, 941–948. [Google Scholar] [CrossRef] [Green Version]
- Ismail, A.M.; Horie, T. Genomics, physiology, and molecular breeding approaches for improving salt tolerance. Annu. Rev. Plant Biol. 2017, 68, 405–434. [Google Scholar] [CrossRef] [Green Version]
- Guo, J.; Shi, G.; Guo, X.; Zhang, L.; Xu, W.; Wang, Y.; Su, Z.; Hua, J. Transcriptome analysis reveals that distinct metabolic pathways operate in salt-tolerant and salt-sensitive upland cotton varieties subjected to salinity stress. Plant Sci. 2015, 238, 33–45. [Google Scholar] [CrossRef]
- Long, W.; Zou, X.; Zhang, X. Transcriptome analysis of canola (Brassica napus) under salt stress at the germination stage. PLoS ONE 2015, 10, e0116217. [Google Scholar] [CrossRef]
- Lin, J.; Li, J.; Yuan, F.; Yang, Z.; Wang, B.; Chen, M. Transcriptome profiling of genes involved in photosynthesis in Elaeagnus angustifolia L. under salt stress. Photosynthesis 2018, 56, 998–1009. [Google Scholar] [CrossRef]
- Iqbal, M.; Athar, H.-U.-R.; Ibrahim, M.; Javed, M.; Zafar, Z.U.; Ashraf, A. Leaf proteome analysis signified that photosynthesis and antioxidants are key indicators of salinity tolerance in canola (Brassica napus L.). Pak. J. Bot. 2019, 51, 52. [Google Scholar] [CrossRef] [Green Version]
- Sicilia, A.; Testa, G.; Santoro, D.F.; Cosentino, S.L.; Piero, A.R.L. RNASeq analysis of giant cane reveals the leaf transcriptome dynamics under long-term salt stress. BMC Plant Biol. 2019, 19, 355. [Google Scholar] [CrossRef] [Green Version]
- Du, X.; Wang, G.; Ji, J.; Shi, L.; Guan, C.; Jin, C. Comparative transcriptome analysis of transcription factors in different maize varieties under salt stress conditions. Plant Growth Regul. 2016, 81, 183–195. [Google Scholar] [CrossRef]
- Ashraf, M.; Oztürk, M.A.; Athar, H.R. Salinity and Water Stress: Improving Crop Efficiency; Springer: Berlin, Germany, 2009; ISBN 978-1-4020-9065-3. [Google Scholar]
- Lauchli, A.; Grattan, S. Plant growth and development under salinity stress. In Advances in Molecular Breeding Toward Drought and Salt Tolerant Crops; Jenks, M.A., Hasegawa, P.M., Jain, S.M., Eds.; Springer: Dordrecht, The Netherlands, 2007; pp. 1–32. ISBN 978-1-4020-5578-2. [Google Scholar]
- Roy, S.; Chakraborty, U. Salt tolerance mechanisms in Salt Tolerant Grasses (STGs) and their prospects in cereal crop improvement. Bot. Stud. 2014, 55, 31. [Google Scholar] [CrossRef] [Green Version]
- Roy, S.; Negrão, S.; Tester, M. Salt resistant crop plants. Curr. Opin. Biotechnol. 2014, 26, 115–124. [Google Scholar] [CrossRef]
- Cuartero, J.; Romero-Aranda, R.; Yeo, A.; Flowers, T. Variability for some physiological characters affecting salt tolerance in tomato. Acta Hortic. 2002, 573, 435–441. [Google Scholar] [CrossRef]
- Ashraf, M. Some important physiological selection criteria for salt tolerance in plants. Flora Morphol. Distrib. Funct. Ecol. Plants 2004, 199, 361–376. [Google Scholar] [CrossRef]
- Negrão, S.; Schmöckel, S.M.; Tester, M. Evaluating physiological responses of plants to salinity stress. Ann. Bot. 2016, 119, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Taiz, L.; Zeiger, E.; Moller, I.S.; Murphy, A. Plant Physiology and Development; Sinauer Associates Inc.: Boston, MA, USA, 2015. [Google Scholar]
- Ali, Q.; Athar, H.R.; Ashraf, M. Ion transport in four canola cultivars as influenced by salt stress. Pak. J. Bot. 2006, 8, 1703–1708. [Google Scholar]
- Akram, M.S.; Ali, Q.; Athar, H.R.; Bhatti, A.S. Ion uptake and distribution in Panicum antidotale Retz. under salt stress. Pak. J. Bot. 2006, 38, 1661–1669. [Google Scholar]
- Munns, R.; Passioura, J.; Colmer, T.D.; Byrt, C. Osmotic adjustment and energy limitations to plant growth in saline soil. New Phytol. 2019, 225, 1091–1096. [Google Scholar] [CrossRef] [Green Version]
- Winicov, I.; Bastola, D.R. Transgenic overexpression of the transcription factor Alfin1 enhances expression of the endogenous MsPRP2 gene in Alfalfa and improves salinity tolerance of the plants. Plant Physiol. 1999, 120, 473–480. [Google Scholar] [CrossRef] [Green Version]
- Postnikova, O.A.; Shao, J.; Nemchinov, L.G. Analysis of the Alfalfa root transcriptome in response to salinity stress. Plant Cell Physiol. 2013, 54, 1041–1055. [Google Scholar] [CrossRef]
- Lei, Y.; Xu, Y.; Hettenhausen, C.; Lu, C.; Shen, G.; Zhang, C.; Li, J.; Song, J.; Lin, H.-H.; Wu, J. Comparative analysis of alfalfa (Medicago sativa L.) leaf transcriptomes reveals genotype-specific salt tolerance mechanisms. BMC Plant Biol. 2018, 18, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Long, R.; Li, M.; Kang, J.; Zhang, T.; Sun, Y.; Yang, Q. Small RNA deep sequencing identifies novel and salt-stress-regulated microRNAs from roots of Medicago sativa and Medicago truncatula. In The Model Legume Medicago truncatula; Bruijn, F.D., Ed.; Wiley online Library: Hoboken, NJ, USA, 2020; pp. 963–976. [Google Scholar]
- Ageeva, I.F.; Prusakova, L.D.; Chizhova, S.I. Influence of brassinosteroids on stem formation and potassium and calcium ion contents in spring barley plants. Agrokhimiya 2001, 6, 49–55. [Google Scholar]
- Deinlein, U.; Stephan, A.B.; Horie, T.; Luo, W.; Xu, G.; I Schroeder, J. Plant salt-tolerance mechanisms. Trends Plant Sci. 2014, 19, 371–379. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wolf, B. A comprehensive system of leaf analyses and its use for diagnosing crop nutrient status. Commun. Soil Sci. Plant Anal. 1982, 13, 1035–1059. [Google Scholar] [CrossRef]
- Glass, A.D.M. Regulation of Ion Transport. Annu. Rev. Plant Physiol. 1983, 34, 311–326. [Google Scholar] [CrossRef]
- Wan, C.; Wilkins, T. A modified hot borate method significantly enhances the yield of high-quality RNA from cotton (Gossypium hirsutum L.). Anal. Biochem. 1994, 223, 7–12. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. Fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef] [Green Version]
- Liao, Y.; Smyth, G.K.; Shi, W. FeatureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2013, 30, 923–930. [Google Scholar] [CrossRef] [Green Version]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. EdgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2009, 26, 139–140. [Google Scholar] [CrossRef] [Green Version]





| Source of Variation | Cultivar (df = 7) | Salinity (df = 1) | Cultivar × Salinity (df = 7) | Error (df = 32) |
|---|---|---|---|---|
| Shoot fresh weight | 257.58 *** | 2254.50 *** | 26.85 ns | 11.75 |
| Shoot dry weight | 0.79 ** | 10.71 *** | 0.11 ns | 0.18 |
| Osmotic potential | 0.16 *** | 1.35 *** | 0.07 *** | 0.01 |
| Relative water contents | 15.44 ns | 685.35 *** | 32.28 * | 10.42 |
| Shoot Na+ contents | 1.58 ns | 3579.38 *** | 36.46 *** | 6.12 |
| Shoot K+ contents | 209.51 *** | 3291.80 *** | 45.73 *** | 7.57 |
| Shoot Ca2+ contents | 21.42 *** | 643.15 *** | 2.15 ns | 3.85 |
| Shoot Cl− contents | 9.13 ns | 23919.01 *** | 28.71 ** | 6.97 |
| Shoot Na+/K+ ratio | 0.18 *** | 23.13 *** | 0.10 * | 0.03 |
| Shoot Ca2+/Na+ ratio | 0.01 ns | 8.46 *** | 0.03 ns | 0.02 |
| Nutrient utilization efficiency | 3.12e−5 * | 0.004 *** | 7.53e−5 *** | 1.28e−5 |
| S.No | KEGG Orthology | K No. | Protein ID | Term Size |
|---|---|---|---|---|
| 1. | calcium_sensing_receptor,_chloroplastic-like | K01013 | XP_013677299 | 114 |
| 2. | calcium-transporting_ATPase_10,_plasma_membrane-type-like | K01537 | XP_013668846 | 345 |
| 3. | V-type_proton_ATPase_subunit_H-like | K02144 | XP_022557635 | 163 |
| 4. | V-type_proton_ATPase_subunit_G2 | K02152 | XP_013674837 | 79 |
| 5. | calmodulin-like_protein_12 | K02183 | XP_013736721 | 447 |
| 6. | potassium_transporter_4-like | K03549 | XP_013689829 | 783 |
| 7. | mitogen-activated_protein_kinase_19 | K04371 | XP_013644611 | 603 |
| 8. | probable_cyclic_nucleotide-gated_ion_channel_14 | K05391 | XP_013660045 | 733 |
| 9. | ABC_transporter_B_family_member_13-like | K05658 | XP_013650695 | 328 |
| 10. | ABC_transporter_C_family_member_7 | K05666 | XP_013668255 | 1477 |
| 11. | AP2-like_ethylene-responsive_transcription_factor_AIL1 | K09285 | XP_013746829 | 457 |
| 12. | ethylene-responsive_transcription_factor_ERF056-like | K09286 | XP_022545196 | 151 |
| 13. | dehydration-responsive_element-binding_protein_2B-like | K09287 | XP_022555207 | 394 |
| 14. | transcription_factor_MYB35-like | K09422 | XP_013655211 | 310 |
| 15. | aquaporin_TIP3-1 | K09873 | XP_013649603 | 265 |
| 16. | aquaporin_NIP6-1_XP | K09874 | 013725889 | 305 |
| 17. | V-type_proton_ATPase_subunit_G3 | K10604 | XP_013669950 | 108 |
| 18. | mitogen-activated_protein_kinase_kinase_4-like | K13413 | XP_013655549 | 353 |
| 19. | calmodulin-like_protein_8 | K13448 | XP_013710349 | 153 |
| 20. | sodium/potassium/calcium_exchanger_1-like | K13749 | XP_013717164 | 288 |
| 21. | abscisic_acid_receptor_PYL10-like | K14496 | XP_013741522 | 222 |
| 22. | mitogen-activated_protein_kinase_6-like | K14512 | XP_013656013 | 392 |
| 23. | mitogen-activated_protein_kinase_1 | K20535 | XP_013640933 | 369 |
| 24. | mitogen-activated_protein_kinase_3 | K20536 | NP_001303218 | 370 |
| 25. | mitogen-activated_protein_kinase_7 | K20537 | NP_001303162 | 368 |
| 26. | mitogen-activated_protein_kinase_kinase_9 | K20604 | XP_013648942 | 308 |
| 27. | mitogen-activated_protein_kinase_kinase_kinase_ANP1-like_isoform_X1 | K20606 | XP_013641030 | 668 |
| 28. | mitogen-activated_protein_kinase_kinase_kinase_18-like | K20716 | XP_013651099 | 456 |
| 29. | potassium_channel_KOR1-like | K21867 | XP_022549779 | 632 |
| 30. | mechanosensitive_ion_channel_protein_9-like | K22048 | XP_013659826 | 738 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ulfat, M.; Athar, H.-u.-R.; Khan, Z.-d.; Kalaji, H.M. RNAseq Analysis Reveals Altered Expression of Key Ion Transporters Causing Differential Uptake of Selective Ions in Canola (Brassica napus L.) Grown under NaCl Stress. Plants 2020, 9, 891. https://doi.org/10.3390/plants9070891
Ulfat M, Athar H-u-R, Khan Z-d, Kalaji HM. RNAseq Analysis Reveals Altered Expression of Key Ion Transporters Causing Differential Uptake of Selective Ions in Canola (Brassica napus L.) Grown under NaCl Stress. Plants. 2020; 9(7):891. https://doi.org/10.3390/plants9070891
Chicago/Turabian StyleUlfat, Mobina, Habib-ur-Rehman Athar, Zaheerud-din Khan, and Hazem M. Kalaji. 2020. "RNAseq Analysis Reveals Altered Expression of Key Ion Transporters Causing Differential Uptake of Selective Ions in Canola (Brassica napus L.) Grown under NaCl Stress" Plants 9, no. 7: 891. https://doi.org/10.3390/plants9070891
APA StyleUlfat, M., Athar, H.-u.-R., Khan, Z.-d., & Kalaji, H. M. (2020). RNAseq Analysis Reveals Altered Expression of Key Ion Transporters Causing Differential Uptake of Selective Ions in Canola (Brassica napus L.) Grown under NaCl Stress. Plants, 9(7), 891. https://doi.org/10.3390/plants9070891