SNP Markers and Evaluation of Duplicate Holdings of Brassica oleracea in Two European Genebanks
Abstract
:1. Introduction
2. Results
2.1. Morphological Diversity
2.2. Marker Efficiency and Accession Diversity
2.3. Accession Comparisons
2.4. Genotypic and Morphological Comparisons
2.5. Limiting the Number of SNP Markers
3. Discussion
3.1. Discrimination Power and the Number of Markers
3.2. Duplication Assessment and Genetic Similarity
3.3. Cultivation History and Naming Practices
3.4. The Effect of Genebank Conservation
3.5. Implications for Genebank Conservation
4. Materials and Methods
4.1. Plant Material, Cultivation, and Morphological Characterization
4.2. DNA Extraction
4.3. SNP Analysis and Statistics
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- FAO. The Second Report on the State of the World’s Plant Genetic Resources for Food and Agriculture; Food and Agriculture Organization of the United Nations: Rome, Italy, 2010. [Google Scholar]
- Fu, Y.B. The Vulnerability of Plant Genetic Resources Conserved Ex Situ. Crop Sci. 2017, 57, 2314–2328. [Google Scholar] [CrossRef] [Green Version]
- ECPGR. A European Genebank Integrated System AEGIS. Available online: http://www.ecpgr.cgiar.org/aegis/about-aegis/ (accessed on 11 June 2020).
- Engels, J.M.M.; Maggioni, L. Benefits of Establishing and Operating a European Collection of Unique and Important Germplasm; ECPGR Secretariat, Bioversity International: Maccarese, Rome, Italy, 2015. [Google Scholar]
- Germeier, C.U.; Frese, L.; Bücken, S. Concepts and data models for treatment of duplicate groups and sharing of responsibilities in genetic resources information systems. Genet. Resour. Crop Evol. 2003, 50, 693–705. [Google Scholar] [CrossRef]
- Vilmorin-Andrieux, M.M. The Vegetable Garden; Ten Speed Press: Berkeley, CA, USA, 1885. [Google Scholar]
- Börjesson, A. Sorter Av Köksväxter: Svenske Priskuranter Fra 1800-Talet till Ca 1930; NordGen Publication Series 2015:01; Nordic Genetic Resource Centre: Alnarp, Sweden, 2015. [Google Scholar]
- Lamm, R.; Tometorp, G.; Åvall, H. Klassificerande försök med köksväxter. Medd. Statens Trädgårdsförsök 1945, 26, 165–202. (In Swedish) [Google Scholar]
- Mackay, I.; Horwell, A.; Garner, J.; White, J.; McKee, J.; Philpott, H. Reanalyses of the historical series of UK variety trials to quantify the contributions of genetic and environmental factors to trends and variability in yield over time. Theor. Appl. Genet. 2011, 122, 225–238. [Google Scholar] [CrossRef]
- UPOV. The International Union for the Protection of New Varieties of Plants. Available online: http://www.upov.int (accessed on 1 April 2020).
- Solberg, S.Ø.; Breian, L. Commercial cultivars and farmers’ access to crop diversity: A case study from the Nordic region. Agric. Food Sci. 2015, 24, 150–163. [Google Scholar] [CrossRef]
- Alercia, A.; López, F.M.; Sackville Hamilton, N.R.; Marsella, M. Digital Object Identifiers for Food Crops-Descriptors and Guidelines of the Global Information System; Food and Agriculture Organization of the United Nations: Rome, Italy, 2018. [Google Scholar]
- Zamir, D. Where Have All the Crop Phenotypes Gone? PLoS Biol. 2013, 11, e1001595. [Google Scholar] [CrossRef] [Green Version]
- McCouch, S.R.; McNally, K.L.; Wang, W.; Sackville Hamilton, R. Genomics of gene banks: A case study in rice. Am. J. Bot. 2012, 99, 407–423. [Google Scholar] [CrossRef] [Green Version]
- Poland, J.A.; Rife, T.W. Genotyping-by-Sequencing for Plant Breeding and Genetics. Plant Genome 2012, 5, 92–102. [Google Scholar] [CrossRef] [Green Version]
- Huang, Y.F.; Poland, J.A.; Wight, C.P.; Jackson, E.W.; Tinker, N.A. Using genotyping-by-sequencing (GBS) for genomic discovery in cultivated oat. PLoS ONE 2014, 9, e102448. [Google Scholar] [CrossRef] [Green Version]
- Anglin, N.L.; Amri, A.; Kehel, Z.; Ellis, D. A case of need: Linking traits to Genebank accessions. Biopreservation Biobanking 2018, 16, 337–349. [Google Scholar] [CrossRef] [Green Version]
- Singh, N.; Wu, S.; Raupp, W.J.; Sehgal, S.; Arora, S.; Tiwari, V.; Vikram, P.; Singh, S.; Chhuneja, P.; Gill, B.S.; et al. Efficient curation of genebanks using next generation sequencing reveals substantial duplication of germplasm accessions. Sci. Rep. 2019, 9, 650. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McCouch, S.; Baute, G.J.; Bradeen, J.; Bramel, P.; Bretting, P.K.; Buckler, E.; Burke, J.M.; Charest, D.; Cloutier, S.; Cole, G.; et al. Agriculture: Feeding the future. Nature 2013, 499, 23–24. [Google Scholar] [CrossRef] [PubMed]
- Van Treuren, R.; van Hintum, T.J.L. Next-generation genebanking: Plant genetic resources management and utilization in the sequencing era. Plant Genet. Resour. 2014, 12, 298–307. [Google Scholar] [CrossRef]
- Bothmer, R.V.; Gustafsson, M.; Snogerup, S. Brassica sect. Brassica (Brassicaceae) II. Inter- and intraspecific crosses with cultivars of B. oleracea. Genet. Resour. Crop Evol. 1995, 42, 165–178. [Google Scholar] [CrossRef]
- Nagaharu, U. Genome analysis in Brassica with special reference to the experimental formation of B. napus and peculiar mode of fertilization. Japan J. Bot. 1935, 7, 389–452. [Google Scholar]
- Hintum, T.J.L.; van Sackville Hamilton, N.R.S.; Engels, J.M.M.; Treuren, R.V. Accession Management Strategies: Splitting and Lumping. In Managing Plant Genetic Diversity; Engels, J.M.M., Ramanatha, R.V., Brown, A.H.D., Jackson, T., Eds.; CABI Publishing: Wallingford, UK, 2002; pp. 113–120. [Google Scholar]
- Van Hintum, T.J.; van De Wiel, C.C.M.; Visser, D.L.; Van Treuren, R.; Vosman, B. The distribution of genetic diversity in a Brassica oleracea genebank collection related to the effects on diversity of regeneration, as measured with AFLPs. Theor. Appl. Genet. 2007, 114, 777–786. [Google Scholar] [CrossRef] [Green Version]
- Christensen, S.; von Bothmer, R.; Poulsen, G.; Maggioni, L.; Phillip, M.; Andersen, B.A.; Jørgensen, R.B. AFLP analysis of genetic diversity in leafy kale (Brassica oleracea L. convar. acephala (DC.) Alef.) landraces, cultivars and wild populations in Europe. Genet. Resour. Crop Evol. 2011, 58, 657–666. [Google Scholar]
- Faltusova, Z.; Kucera, L.; Ovesna, J. Genetic diversity of Brassica oleracea var. capitata gene bank accessions assessed by AFLP. Elect. J. Biotech. 2011, 14. [Google Scholar] [CrossRef]
- Izzah, N.K.; Lee, J.; Perumal, S.; Park, J.Y.; Ahn, K.; Fu, D.; Kim, G.B.; Nam, Y.W.; Yang, T.J. Microsatellite-based analysis of genetic diversity in 91 commercial Brassica oleracea L. cultivars belonging to six varietal groups. Genet. Resour. Crop Evol. 2013, 60, 1967–1986. [Google Scholar] [CrossRef]
- Clarke, W.E.; Higgins, E.E.; Plieske, J.; Wieseke, R.; Sidebottom, C.; Khedikar, Y.; Batley, J.; Edwards, D.; Meng, J.; Li, R.; et al. A high-density SNP genotyping array for Brassica napus and its ancestral diploid species based on optimised selection of single-locus markers in the allotetraploid genome. Theor. Appl. Genet. 2016, 129, 1887–1899. [Google Scholar] [CrossRef] [Green Version]
- Willing, E.M.; Dreyer, C.; Van Oosterhout, C. Estimates of genetic differentiation measured by FST do not necessarily require large sample sizes when using many SNP markers. PLoS ONE 2012, 7, e42649. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nelson, M.F.; Anderson, N.O. How many marker loci are necessary? Analysis of dominant marker data sets using two popular population genetic algorithms. Ecol. Evol. 2013, 3, 3455–3470. [Google Scholar] [CrossRef] [PubMed]
- Menting, F.; Bas, N. The ECPGR Brassica Database. Available online: http://ecpgr.cgn.wur.nl/Brasedb/ (accessed on 15 November 2017).
- Nordic Genetic Resource Center. Seed Information Database. Available online: https://nordgen.org/ (accessed on 15 November 2019).
- Soleri, D.; Smith, S.E. Morphological and phenological comparisons of two hopi maize varieties conserved in situ and ex situ. Econ. Bot. 1995, 49, 56–77. [Google Scholar] [CrossRef]
- Gomez, O.J.; Blair, M.W.; Frankow-Lindberg, B.E.; Gullberg, U. Comparative study of common bean (Phaseolus vulgaris L.) landraces conserved ex situ in genebanks and in situ by farmers. Genet. Res. Crop Evol. 2005, 52, 371–380. [Google Scholar] [CrossRef]
- Negri, V.; Tiranti, B. Effectiveness of in situ and ex situ conservation of crop diversity. What a Phaseolus vulgaris L. landrace case study can tell us. Genetica 2010, 138, 985–998. [Google Scholar] [CrossRef]
- Hagenblad, J.; Zie, J.; Leino, M.W. Exploring the population genetics of genebank and historical landrace varieties. Genet. Resour. Crop Evol. 2012, 59, 1185–1199. [Google Scholar] [CrossRef]
- Solberg, S.Ø.; Yndgaard, F.; Palmé, A. Morphological and phenological consequences of ex situ conservation of natural populations of red clover (Trifolium pratense L.). Plant Genet Resour. 2015, 15, 97–108. [Google Scholar] [CrossRef]
- Ellstrand, N.C.; Elam, D.R. Population genetic consequences of small population size: Implications for plant conservation. Annu. Rev. Ecol. Syst. 1993, 24, 217–242. [Google Scholar] [CrossRef]
- FAO. Genebank Standards for Plant Genetic Resources for Food and Agriculture; Food and Agriculture Organization of the United Nations: Rome, Italy, 2014. [Google Scholar]
- Wright, S. Evolution in Mendelian Populations. Genetics 1931, 16, 97–159. [Google Scholar]
- Bohart, G.E.; Todd, F.E. Pollination of Seed Crops by Insects. In Seeds. The Yearbook of Agriculture; US Government Printing Office: Washington, DC, USA, 1961; pp. 240–246. [Google Scholar]
- Cruz, V.; Nason, J.; Luhman, R.; Marek, L.; Shoemaker, R.; Brummer, E.; Gardner, C. Analysis of bulked and redundant accessions of Brassica germplasm using assignment tests of microsatellite markers. Euphytica 2006, 152, 339–349. [Google Scholar] [CrossRef]
- Diederichsen, A. Duplication assessments in Nordic Avena sativa accessions at the Canadian national genebank. Genet. Resour. Crop Evol. 2009, 56, 587–597. [Google Scholar] [CrossRef]
- Lund, B.; Ortiz, R.; Skovgaard, I.M.; Waugh, R.; Andersen, S.B. Analysis of potential duplicates in barley gene bank collections using re-sampling of microsatellite data. Theor. Appl. Genet. 2003, 106, 1129–1138. [Google Scholar] [CrossRef] [PubMed]
- Branca, F.; Bas, N.; Artemyeva, A.; De Haro, A.; Maggioni, L. Activities of the Brassica Working Group of the European Cooperative Programme for Plant Genetic Resources (ECPGR). Acta Hort. 2013, 1005, 149–155. [Google Scholar] [CrossRef] [Green Version]
- Solberg, S.Ø.; Artemyeva, A.; Yndgaard, F.; Dorre, M.; Niss, J.; Burleigh, S. Duplication assessments in Brassica vegetable accessions. Plant Genet. Resour. 2017, 16, 201–208. [Google Scholar] [CrossRef]
- UPOV. UPOV Guidelines for the Conduct of Tests for Distinctness, Uniformity and Stability TG/48/7; International Union for the Protection of New Varieties of Plants: Geneva, Switzerland, 2004. [Google Scholar]
- Crawley, M.J. The R Book; John Wiley & Sons Ltd.: Chichester, UK, 2009. [Google Scholar]
- Doyle, J.J.; Doyle, J.L. Isolation of plant DNA from fresh tissue. Focus 1990, 12, 13–15. [Google Scholar]
- Wright, S. The genetical structure of populations. Ann. Eugen. 1951, 15, 323–354. [Google Scholar] [CrossRef]
- Nei, M. Analysis of gene diversity in subdivided populations. Proc. Natl. Acad. Sci. USA 1973, 70, 3321–3323. [Google Scholar] [CrossRef] [Green Version]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria; Available online: https://www.R-project.org/ (accessed on 11 June 2020).
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar]
- Falush, D.; Stephens, M.; Pritchard, J.K. Inference of population structure using multilocus genotype data: Linked loci and correlated allele frequencies. Genetics 2003, 164, 1567–1587. [Google Scholar]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the number of clusters of individuals using the software STRUCTURE: A simulation study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef] [Green Version]
- Jakobsson, M.; Rosenberg, N.A. CLUMPP: A cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics 2007, 23, 1801–1806. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rosenberg, N.A. DISTRUCT: A program for the graphical display of population structure. Mol. Ecol. Notes 2004, 4, 137–138. [Google Scholar] [CrossRef]


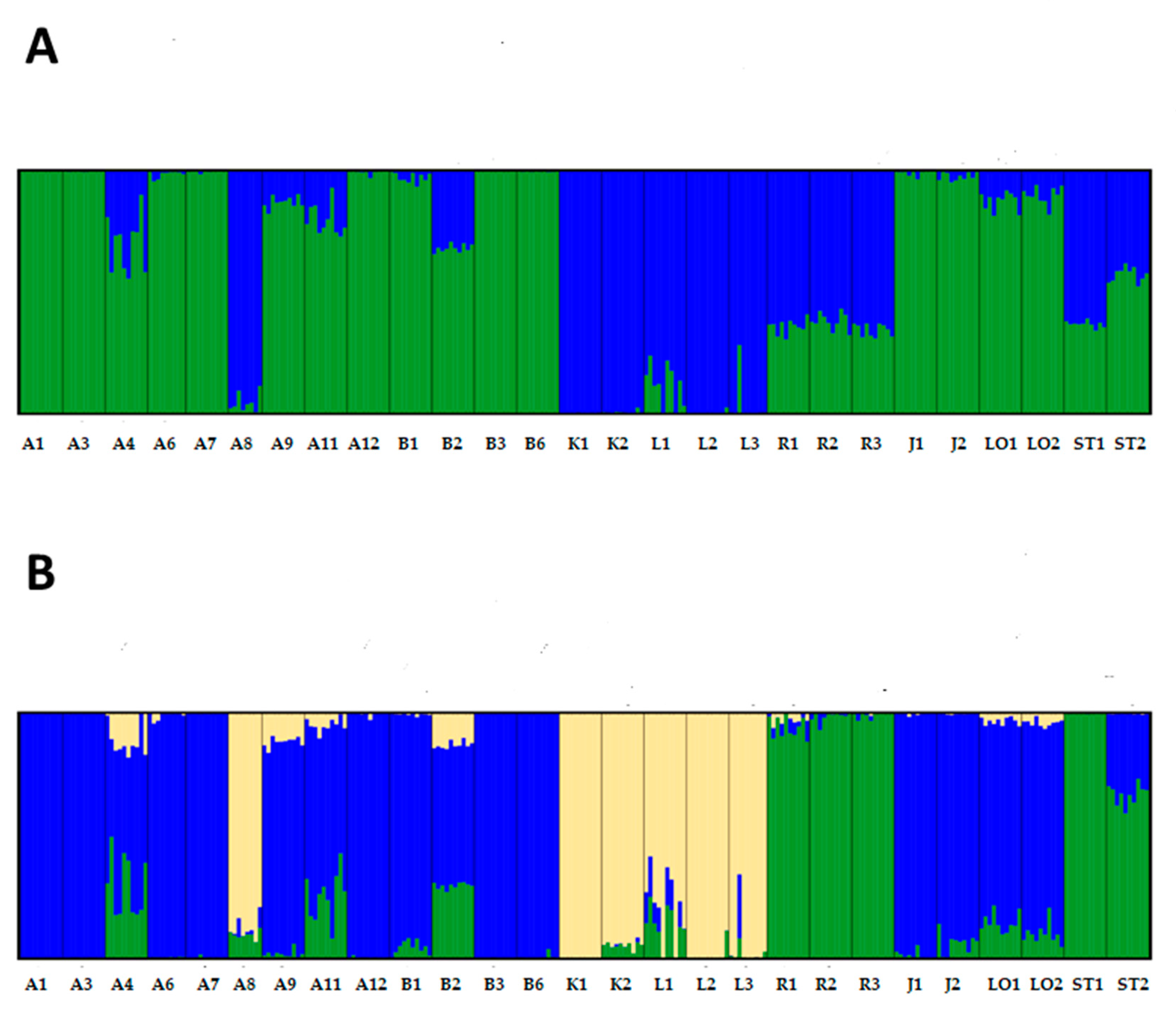
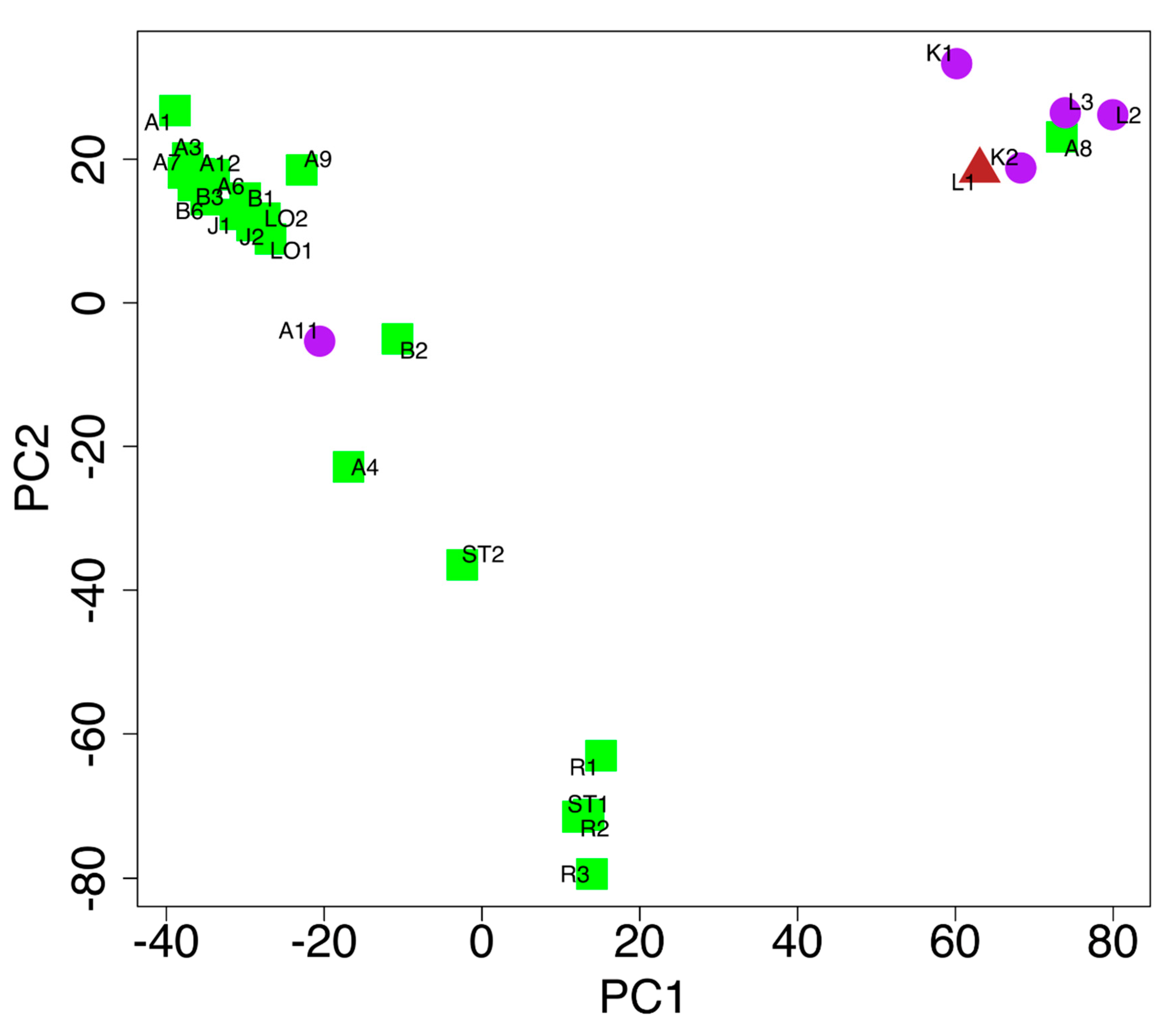
| Code | Accession Name | Accession Number | Gene Bank | Acquisition Year, Donor Institute |
|---|---|---|---|---|
| Amager Tall group | ||||
| A1 | Amager Hög | NGB11705 | NGB | 1995, Olson & Sons AB, Sweden |
| A3 | Amager Høj Grøn Grami | NGB1875 | NGB | 1980, Dæhnfeldt A/S, Denmark |
| A4 | Grami | K2537 | VIR | 1988, Unknown, Denmark |
| A6 | Amager Høj, Grøn, Toftø 67 | NGB1873 | NGB | 1980, FDB Frø, Denmark |
| A7 | Amager Tall Resistent | K2475 | VIR | 1980, Unknown, Denmark |
| Amager Short pair | ||||
| A8 | Amager Kurzstunkiger Orginal | K1485 | VIR | 1935, Ohlsens Enke, Denmark |
| A9 | Amager L NF Orginal | K2248 | VIR | 1967, Unknown, Norway |
| Amager Winter pair | ||||
| A8 | Amager Kurzstunkiger Orginal | K1485 | VIR | 1935, Ohlsens Enke, Denmark |
| A9 | Amager L NF Orginal | K2248 | VIR | 1967, Unknown, Norway |
| Blåtopp group | ||||
| B1 | Amager Faales Blatopp | K1181 | VIR | 1930, Norsk Frø, Norway |
| B2 | Blatopp | K2250 | VIR | 1967, Unknown, Norway |
| B3 | Blatopp Kvithamar | K2243 | VIR | 1967, Unknown, Norway |
| B6 | Blåtopp Kvithamar | NGB4555 | NGB | 1984, Unknown, Norway |
| Kissendrup pair | ||||
| K1 | Kissendrup | K111 | VIR | 1935, Unknown, Denmark |
| K2 | Kissendrup Tagenshus | NGB1996 | NGB | 1980, Hansens Amagerfrø, Denamrk |
| Langendijker group | ||||
| L1 | Langendijk Summer | K181 | VIR | 1959, Unknown, Denmark |
| L2 | Langendijker Sommer Debut | NGB1997 | NGB | 1980, Ohlsens Enke, Denmark |
| L3 | Langendijker Sommer Lanso | NGB1998 | NGB | 1980, Hansens Amagerfrø, Denmark |
| Ruhm v Enkhizen group | ||||
| R1 | Ruhm von Enkhizen | NGB11718 | NGB | 1996, Hansens Amagerfrø, Denamrk |
| R2 | Ruhm von Enkhizen Haba | NGB1888 | NGB | 1980, Hansens Amagerfrø, Denmark |
| R3 | Ruhm von Enkhizen, B Hunderup | NGB2431 | NGB | 1982, Unknown, Denmark |
| Jåtunsalgets pair | ||||
| J1 | Jatunsalgets Vinterkål Berbes | K2139 | VIR | 1959, Unknown, Norway |
| J2 | Jåtunsalgets Vinterkål | NGB5007 | NGB | 1983, Unknown, Norway |
| Loke pair | ||||
| LO1 | Loke | K2489 | VIR | 1982, Unknown, Sweden |
| LO2 | Loke | NGB12050 | NGB | 1997, Unknown, Denmark |
| Stavanger Torv pair | ||||
| ST1 | Stavanger Torv | K2175 | VIR | 1961, Unknown, Norway |
| ST2 | Stavanger Torg | NGB8515 | NGB | 1990, NLH, Norway |
| Code | Average No Alleles | Nei′s h | Observed Heterozygosity |
|---|---|---|---|
| A1 | 1.1 | 0.06 | 0.06 |
| A3 | 1.4 | 0.14 | 0.15 |
| A4 | 1.8 | 0.27 | 0.30 |
| A6 | 1.5 | 0.16 | 0.17 |
| A7 | 1.3 | 0.14 | 0.12 |
| A8 | 1.5 | 0.21 | 0.22 |
| A9 | 1.3 | 0.13 | 0.14 |
| A11 | 1.6 | 0.17 | 0.15 |
| A12 | 1.4 | 0.14 | 0.15 |
| B1 | 1.4 | 0.13 | 0.15 |
| B2 | 1.4 | 0.14 | 0.18 |
| B3 | 1.3 | 0.12 | 0.13 |
| B6 | 1.4 | 0.13 | 0.14 |
| K1 | 1.2 | 0.06 | 0.06 |
| K2 | 1.4 | 0.14 | 0.15 |
| L1 | 1.4 | 0.16 | 0.15 |
| L2 | 1.4 | 0.13 | 0.10 |
| L3 | 1.5 | 0.18 | 0.17 |
| R1 | 1.5 | 0.17 | 0.19 |
| R2 | 1.5 | 0.17 | 0.16 |
| R3 | 1.4 | 0.14 | 0.13 |
| J1 | 1.3 | 0.10 | 0.10 |
| J2 | 1.4 | 0.14 | 0.15 |
| LO1 | 1.6 | 0.19 | 0.22 |
| LO2 | 1.6 | 0.19 | 0.18 |
| ST1 | 1.3 | 0.11 | 0.14 |
| ST2 | 1.3 | 0.12 | 0.13 |
| Accession | Accession | Full Dataset | Subsample Average | ||
|---|---|---|---|---|---|
| 1 | 2 | FST | FST | FST − 1 SE | FST + 1 SE |
| A1 | A3 | 0.216 | 0.218 | 0.215 | 0.222 |
| A1 | A4 | 0.184 | 0.184 | 0.182 | 0.186 |
| A1 | A6 | 0.252 | 0.243 | 0.240 | 0.247 |
| A1 | A7 | 0.304 | 0.309 | 0.305 | 0.314 |
| A3 | A4 | 0.103 | 0.104 | 0.103 | 0.105 |
| A3 | A6 | 0.149 | 0.149 | 0.147 | 0.151 |
| A3 | A7 | 0.175 | 0.173 | 0.171 | 0.176 |
| A4 | A6 | 0.110 | 0.111 | 0.109 | 0.112 |
| A4 | A7 | 0.132 | 0.132 | 0.131 | 0.134 |
| A6 | A7 | 0.166 | 0.169 | 0.166 | 0.171 |
| A8 | A9 | 0.326 | 0.327 | 0.324 | 0.331 |
| A11 | A12 | 0.144 | 0.143 | 0.141 | 0.145 |
| B1 | B2 | 0.291 | 0.294 | 0.290 | 0.298 |
| B1 | B3 | 0.228 | 0.230 | 0.226 | 0.234 |
| B1 | B6 | 0.218 | 0.216 | 0.213 | 0.220 |
| B2 | B3 | 0.321 | 0.324 | 0.320 | 0.328 |
| B2 | B6 | 0.304 | 0.309 | 0.305 | 0.313 |
| B3 | B6 | 0.087 | 0.085 | 0.084 | 0.087 |
| K1 | K2 | 0.253 | 0.249 | 0.245 | 0.253 |
| L1 | L2 | 0.188 | 0.188 | 0.185 | 0.190 |
| L1 | L3 | 0.160 | 0.162 | 0.160 | 0.164 |
| L3 | L2 | 0.117 | 0.117 | 0.115 | 0.119 |
| R1 | R2 | 0.111 | 0.110 | 0.108 | 0.111 |
| R1 | R3 | 0.144 | 0.149 | 0.147 | 0.151 |
| R2 | R3 | 0.129 | 0.129 | 0.127 | 0.131 |
| J1 | J2 | 0.202 | 0.201 | 0.198 | 0.205 |
| LO1 | LO2 | 0.095 | 0.095 | 0.094 | 0.097 |
| ST1 | ST2 | 0.300 | 0.294 | 0.290 | 0.298 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Palmé, A.E.; Hagenblad, J.; Solberg, S.Ø.; Aloisi, K.; Artemyeva, A. SNP Markers and Evaluation of Duplicate Holdings of Brassica oleracea in Two European Genebanks. Plants 2020, 9, 925. https://doi.org/10.3390/plants9080925
Palmé AE, Hagenblad J, Solberg SØ, Aloisi K, Artemyeva A. SNP Markers and Evaluation of Duplicate Holdings of Brassica oleracea in Two European Genebanks. Plants. 2020; 9(8):925. https://doi.org/10.3390/plants9080925
Chicago/Turabian StylePalmé, Anna E., Jenny Hagenblad, Svein Øivind Solberg, Karolina Aloisi, and Anna Artemyeva. 2020. "SNP Markers and Evaluation of Duplicate Holdings of Brassica oleracea in Two European Genebanks" Plants 9, no. 8: 925. https://doi.org/10.3390/plants9080925
APA StylePalmé, A. E., Hagenblad, J., Solberg, S. Ø., Aloisi, K., & Artemyeva, A. (2020). SNP Markers and Evaluation of Duplicate Holdings of Brassica oleracea in Two European Genebanks. Plants, 9(8), 925. https://doi.org/10.3390/plants9080925







